bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0987_orf1
Length=186
Score E
Sequences producing significant alignments: (Bits) Value
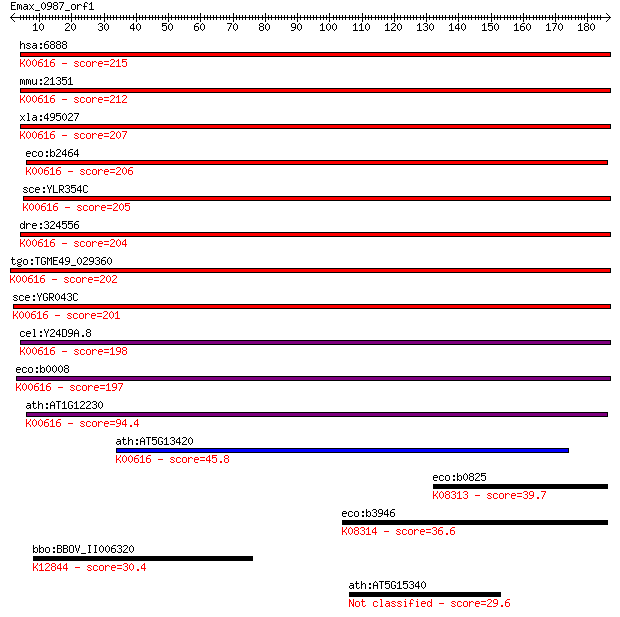
hsa:6888 TALDO1, TAL, TAL-H, TALDOR, TALH; transaldolase 1 (EC... 215 8e-56
mmu:21351 Taldo1; transaldolase 1 (EC:2.2.1.2); K00616 transal... 212 5e-55
xla:495027 taldo1; transaldolase 1 (EC:2.2.1.2); K00616 transa... 207 1e-53
eco:b2464 talA, ECK2459, JW2448; transaldolase A (EC:2.2.1.2);... 206 4e-53
sce:YLR354C TAL1; Tal1p (EC:2.2.1.2); K00616 transaldolase [EC... 205 8e-53
dre:324556 taldo1, MGC56232, Tal, wu:fc32g02, zgc:56232, zgc:6... 204 2e-52
tgo:TGME49_029360 transaldolase, putative (EC:2.2.1.2); K00616... 202 7e-52
sce:YGR043C NQM1; Nqm1p (EC:2.2.1.2); K00616 transaldolase [EC... 201 2e-51
cel:Y24D9A.8 hypothetical protein; K00616 transaldolase [EC:2.... 198 1e-50
eco:b0008 talB, ECK0008, JW0007, yaaK; transaldolase B (EC:2.2... 197 2e-50
ath:AT1G12230 transaldolase, putative; K00616 transaldolase [E... 94.4 2e-19
ath:AT5G13420 transaldolase, putative; K00616 transaldolase [E... 45.8 8e-05
eco:b0825 fsaA, ECK0815, JW5109, mipB, ybiZ; fructose-6-phosph... 39.7 0.006
eco:b3946 fsaB, ECK3938, JW3918, talC, yijG; fructose-6-phosph... 36.6 0.050
bbo:BBOV_II006320 18.m06519; pre-mRNA processing ribonucleopro... 30.4 3.7
ath:AT5G15340 pentatricopeptide (PPR) repeat-containing protein 29.6 6.2
> hsa:6888 TALDO1, TAL, TAL-H, TALDOR, TALH; transaldolase 1 (EC:2.2.1.2);
K00616 transaldolase [EC:2.2.1.2]
Length=337
Score = 215 bits (547), Expect = 8e-56, Method: Compositional matrix adjust.
Identities = 105/184 (57%), Positives = 143/184 (77%), Gaps = 3/184 (1%)
Query 4 TALDQLAKLSTIVADSADFEAIKEYNPVDATTNPTLVLQAVNNPKYKHLLDKAIKEAKEA 63
+ALDQL + +T+VAD+ DF AI EY P DATTNP+L+L A P Y+ L+++AI ++
Sbjct 13 SALDQLKQFTTVVADTGDFHAIDEYKPQDATTNPSLILAAAQMPAYQELVEEAIAYGRKL 72
Query 64 AKGKSQEIIIEEAIDRVLVLFGIEILQIIPGLVSTEIPADLSFNQQGSVERARRIIKLYE 123
G SQE I+ AID++ VLFG EIL+ IPG VSTE+ A LSF++ V RARR+I+LY+
Sbjct 73 --GGSQEDQIKNAIDKLFVLFGAEILKKIPGRVSTEVDARLSFDKDAMVARARRLIELYK 130
Query 124 EKGINKERILIKIAATWEGIQAAKELKKEN-INCNITLLFSLCQAIAASDAGAALVSPFV 182
E GI+K+RILIK+++TWEGIQA KEL++++ I+CN+TLLFS QA+A ++AG L+SPFV
Sbjct 131 EAGISKDRILIKLSSTWEGIQAGKELEEQHGIHCNMTLLFSFAQAVACAEAGVTLISPFV 190
Query 183 GRIL 186
GRIL
Sbjct 191 GRIL 194
> mmu:21351 Taldo1; transaldolase 1 (EC:2.2.1.2); K00616 transaldolase
[EC:2.2.1.2]
Length=337
Score = 212 bits (540), Expect = 5e-55, Method: Compositional matrix adjust.
Identities = 104/184 (56%), Positives = 141/184 (76%), Gaps = 3/184 (1%)
Query 4 TALDQLAKLSTIVADSADFEAIKEYNPVDATTNPTLVLQAVNNPKYKHLLDKAIKEAKEA 63
+ALDQL + +T+VAD+ DF AI EY P DATTNP+L+L A P Y+ L+++AI K+
Sbjct 13 SALDQLKQFTTVVADTGDFNAIDEYKPQDATTNPSLILAAAQMPAYQELVEEAIAYGKKL 72
Query 64 AKGKSQEIIIEEAIDRVLVLFGIEILQIIPGLVSTEIPADLSFNQQGSVERARRIIKLYE 123
G QE I+ AID++ VLFG EIL+ IPG VSTE+ A LSF++ V RARR+I+LY+
Sbjct 73 --GGPQEEQIKNAIDKLFVLFGAEILKKIPGRVSTEVDARLSFDKDAMVARARRLIELYK 130
Query 124 EKGINKERILIKIAATWEGIQAAKELKKEN-INCNITLLFSLCQAIAASDAGAALVSPFV 182
E G+ K+RILIK+++TWEGIQA KEL++++ I+CN+TLLFS QA+A ++AG L+SPFV
Sbjct 131 EAGVGKDRILIKLSSTWEGIQAGKELEEQHGIHCNMTLLFSFAQAVACAEAGVTLISPFV 190
Query 183 GRIL 186
GRIL
Sbjct 191 GRIL 194
> xla:495027 taldo1; transaldolase 1 (EC:2.2.1.2); K00616 transaldolase
[EC:2.2.1.2]
Length=338
Score = 207 bits (528), Expect = 1e-53, Method: Compositional matrix adjust.
Identities = 103/184 (55%), Positives = 140/184 (76%), Gaps = 3/184 (1%)
Query 4 TALDQLAKLSTIVADSADFEAIKEYNPVDATTNPTLVLQAVNNPKYKHLLDKAIKEAKEA 63
+ALDQL + + +VAD+ DF AI+EY P DATTNP+L+L A P Y+ L++ AI+ K
Sbjct 14 SALDQLKQHTVVVADTGDFNAIEEYKPQDATTNPSLILAAAQMPDYQGLVNDAIQYGKNL 73
Query 64 AKGKSQEIIIEEAIDRVLVLFGIEILQIIPGLVSTEIPADLSFNQQGSVERARRIIKLYE 123
G S+E I +D++ VLFG+EIL+ IPG VSTE+ A LSF++ G VERA+R+I LY+
Sbjct 74 --GGSEEEQINNIMDKLFVLFGVEILKKIPGRVSTEVDARLSFDKDGMVERAKRLIALYK 131
Query 124 EKGINKERILIKIAATWEGIQAAKELKKE-NINCNITLLFSLCQAIAASDAGAALVSPFV 182
E GI+K+RILIK+++TWEGIQA K L++E I+CN+TLLFS QA+A ++AG L+SPFV
Sbjct 132 EAGIDKKRILIKLSSTWEGIQAGKILEEEYGIHCNMTLLFSFAQAVACAEAGVTLISPFV 191
Query 183 GRIL 186
GRIL
Sbjct 192 GRIL 195
> eco:b2464 talA, ECK2459, JW2448; transaldolase A (EC:2.2.1.2);
K00616 transaldolase [EC:2.2.1.2]
Length=316
Score = 206 bits (524), Expect = 4e-53, Method: Compositional matrix adjust.
Identities = 98/180 (54%), Positives = 139/180 (77%), Gaps = 2/180 (1%)
Query 6 LDQLAKLSTIVADSADFEAIKEYNPVDATTNPTLVLQAVNNPKYKHLLDKAIKEAKEAAK 65
LD + + +T+VADS D E+I+ Y+P DATTNP+L+L+A +Y+HL+D AI K+
Sbjct 4 LDGIKQFTTVVADSGDIESIRHYHPQDATTNPSLLLKAAGLSQYEHLIDDAIAWGKK--N 61
Query 66 GKSQEIIIEEAIDRVLVLFGIEILQIIPGLVSTEIPADLSFNQQGSVERARRIIKLYEEK 125
GK+QE + A D++ V FG EIL+I+PG VSTE+ A LSF+++ S+E+AR ++ LY+++
Sbjct 62 GKTQEQQVVAACDKLAVNFGAEILKIVPGRVSTEVDARLSFDKEKSIEKARHLVDLYQQQ 121
Query 126 GINKERILIKIAATWEGIQAAKELKKENINCNITLLFSLCQAIAASDAGAALVSPFVGRI 185
G+ K RILIK+A+TWEGI+AA+EL+KE INCN+TLLFS QA A ++AG L+SPFVGRI
Sbjct 122 GVEKSRILIKLASTWEGIRAAEELEKEGINCNLTLLFSFAQARACAEAGVFLISPFVGRI 181
> sce:YLR354C TAL1; Tal1p (EC:2.2.1.2); K00616 transaldolase [EC:2.2.1.2]
Length=335
Score = 205 bits (521), Expect = 8e-53, Method: Compositional matrix adjust.
Identities = 101/184 (54%), Positives = 142/184 (77%), Gaps = 4/184 (2%)
Query 5 ALDQL-AKLSTIVADSADFEAIKEYNPVDATTNPTLVLQAVNNPKYKHLLDKAIKEAKEA 63
+L+QL A + +VAD+ DF +I ++ P D+TTNP+L+L A P Y L+D A++ K+
Sbjct 15 SLEQLKASGTVVVADTGDFGSIAKFQPQDSTTNPSLILAAAKQPTYAKLIDVAVEYGKK- 73
Query 64 AKGKSQEIIIEEAIDRVLVLFGIEILQIIPGLVSTEIPADLSFNQQGSVERARRIIKLYE 123
GK+ E +E A+DR+LV FG EIL+I+PG VSTE+ A LSF+ Q ++E+AR IIKL+E
Sbjct 74 -HGKTTEEQVENAVDRLLVEFGKEILKIVPGRVSTEVDARLSFDTQATIEKARHIIKLFE 132
Query 124 EKGINKERILIKIAATWEGIQAAKEL-KKENINCNITLLFSLCQAIAASDAGAALVSPFV 182
++G++KER+LIKIA+TWEGIQAAKEL +K+ I+CN+TLLFS QA+A ++A L+SPFV
Sbjct 133 QEGVSKERVLIKIASTWEGIQAAKELEEKDGIHCNLTLLFSFVQAVACAEAQVTLISPFV 192
Query 183 GRIL 186
GRIL
Sbjct 193 GRIL 196
> dre:324556 taldo1, MGC56232, Tal, wu:fc32g02, zgc:56232, zgc:66054;
transaldolase 1 (EC:2.2.1.2); K00616 transaldolase [EC:2.2.1.2]
Length=337
Score = 204 bits (518), Expect = 2e-52, Method: Compositional matrix adjust.
Identities = 98/184 (53%), Positives = 138/184 (75%), Gaps = 3/184 (1%)
Query 4 TALDQLAKLSTIVADSADFEAIKEYNPVDATTNPTLVLQAVNNPKYKHLLDKAIKEAKEA 63
+AL+Q+ K + +VAD+ DF AI+EY P DATTNP+L+L A P Y+ L+D+AIK
Sbjct 13 SALEQIKKFTVVVADTGDFNAIEEYKPQDATTNPSLILAAAKMPAYQPLVDQAIKYG--T 70
Query 64 AKGKSQEIIIEEAIDRVLVLFGIEILQIIPGLVSTEIPADLSFNQQGSVERARRIIKLYE 123
A G +++ + A+D++ V FG+EIL+ +PG VSTE+ A LSF++ V RARR+I LYE
Sbjct 71 ANGGTEDEQVTNAMDKLFVNFGLEILKKVPGRVSTEVDARLSFDKDAMVSRARRLISLYE 130
Query 124 EKGINKERILIKIAATWEGIQAAKELKK-ENINCNITLLFSLCQAIAASDAGAALVSPFV 182
+ GI+KERILIK+++TWEGIQA +EL++ I+CN+TLLFS QA+A ++A L+SPFV
Sbjct 131 DAGISKERILIKLSSTWEGIQAGRELEEIHGIHCNMTLLFSFAQAVACAEARVTLISPFV 190
Query 183 GRIL 186
GRIL
Sbjct 191 GRIL 194
> tgo:TGME49_029360 transaldolase, putative (EC:2.2.1.2); K00616
transaldolase [EC:2.2.1.2]
Length=364
Score = 202 bits (513), Expect = 7e-52, Method: Compositional matrix adjust.
Identities = 104/187 (55%), Positives = 147/187 (78%), Gaps = 1/187 (0%)
Query 1 AGTTALDQLAKLSTIVADSADFEAIKEYNPVDATTNPTLVLQAVNNPKYKHLLDKAIKEA 60
A AL+ L LS +VAD+ DF A+K+Y P DATTNP+L+L+A + P+Y HL+D+A++ A
Sbjct 32 AAQNALEALRLLSVVVADTGDFVALKKYVPHDATTNPSLLLKAASMPQYAHLMDEAVEVA 91
Query 61 KEA-AKGKSQEIIIEEAIDRVLVLFGIEILQIIPGLVSTEIPADLSFNQQGSVERARRII 119
K+ K + + ++EE ID++ V FG+EIL+I+PG+VSTE+ A LSF+ Q SVE+AR++I
Sbjct 92 KKVHGKVEPDDELVEEVIDQLFVRFGVEILKIVPGVVSTEVSAALSFDVQASVEKARKLI 151
Query 120 KLYEEKGINKERILIKIAATWEGIQAAKELKKENINCNITLLFSLCQAIAASDAGAALVS 179
+Y+E GI K+R+LIK+A TWEG +AAK L+KE I+CN+TLLFS QA+AA++A A L+S
Sbjct 152 LMYKEHGIEKDRVLIKLATTWEGCEAAKILEKEGIHCNMTLLFSFAQAVAAAEAKATLIS 211
Query 180 PFVGRIL 186
PFVGRIL
Sbjct 212 PFVGRIL 218
> sce:YGR043C NQM1; Nqm1p (EC:2.2.1.2); K00616 transaldolase [EC:2.2.1.2]
Length=333
Score = 201 bits (510), Expect = 2e-51, Method: Compositional matrix adjust.
Identities = 98/187 (52%), Positives = 140/187 (74%), Gaps = 4/187 (2%)
Query 2 GTTALDQLAKLST-IVADSADFEAIKEYNPVDATTNPTLVLQAVNNPKYKHLLDKAIKEA 60
T++L+QL K T +VADS DFEAI +Y P D+TTNP+L+L A KY +D A++
Sbjct 12 ATSSLEQLKKAGTHVVADSGDFEAISKYEPQDSTTNPSLILAASKLEKYARFIDAAVEYG 71
Query 61 KEAAKGKSQEIIIEEAIDRVLVLFGIEILQIIPGLVSTEIPADLSFNQQGSVERARRIIK 120
++ GK+ IE A+D++LV FG +IL+++PG VSTE+ A LSF+++ +V++A IIK
Sbjct 72 RK--HGKTDHEKIENAMDKILVEFGTQILKVVPGRVSTEVDARLSFDKKATVKKALHIIK 129
Query 121 LYEEKGINKERILIKIAATWEGIQAAKELK-KENINCNITLLFSLCQAIAASDAGAALVS 179
LY++ G+ KER+LIKIA+TWEGIQAA+EL+ K I+CN+TLLFS QA+A ++A L+S
Sbjct 130 LYKDAGVPKERVLIKIASTWEGIQAARELEVKHGIHCNMTLLFSFTQAVACAEANVTLIS 189
Query 180 PFVGRIL 186
PFVGRI+
Sbjct 190 PFVGRIM 196
> cel:Y24D9A.8 hypothetical protein; K00616 transaldolase [EC:2.2.1.2]
Length=319
Score = 198 bits (503), Expect = 1e-50, Method: Compositional matrix adjust.
Identities = 96/184 (52%), Positives = 138/184 (75%), Gaps = 3/184 (1%)
Query 4 TALDQLAKLSTIVADSADFEAIKEYNPVDATTNPTLVLQAVNNPKYKHLLDKAIKEAKEA 63
+ L+QL S +VAD+ DF AIKE+ P DATTNP+L+L A +Y L+D+++ AKE
Sbjct 2 SVLEQLKGASVVVADTGDFNAIKEFQPTDATTNPSLILAASKMEQYAALIDQSVAYAKEH 61
Query 64 AKGKSQEIIIEEAIDRVLVLFGIEILQIIPGLVSTEIPADLSFNQQGSVERARRIIKLYE 123
A G + +++ A+DR+ V+FG EIL+ IPG VSTE+ A LSF+ Q S++RA +I YE
Sbjct 62 ASGHQE--VLQAAMDRLFVVFGKEILKTIPGRVSTEVDARLSFDTQASIDRALGLIAQYE 119
Query 124 EKGINKERILIKIAATWEGIQAAKELK-KENINCNITLLFSLCQAIAASDAGAALVSPFV 182
++GI+K+RILIK+A+TWEGI+AAK L+ K I+CN+TLLF+ QA+A +++G L+SPFV
Sbjct 120 KEGISKDRILIKLASTWEGIRAAKFLESKHGIHCNMTLLFNFEQAVACAESGVTLISPFV 179
Query 183 GRIL 186
GRI+
Sbjct 180 GRIM 183
> eco:b0008 talB, ECK0008, JW0007, yaaK; transaldolase B (EC:2.2.1.2);
K00616 transaldolase [EC:2.2.1.2]
Length=317
Score = 197 bits (500), Expect = 2e-50, Method: Compositional matrix adjust.
Identities = 92/184 (50%), Positives = 139/184 (75%), Gaps = 2/184 (1%)
Query 3 TTALDQLAKLSTIVADSADFEAIKEYNPVDATTNPTLVLQAVNNPKYKHLLDKAIKEAKE 62
T L L + +T+VAD+ D A+K Y P DATTNP+L+L A P+Y+ L+D A+ AK+
Sbjct 2 TDKLTSLRQYTTVVADTGDIAAMKLYQPQDATTNPSLILNAAQIPEYRKLIDDAVAWAKQ 61
Query 63 AAKGKSQEIIIEEAIDRVLVLFGIEILQIIPGLVSTEIPADLSFNQQGSVERARRIIKLY 122
+ ++Q+I+ +A D++ V G+EIL+++PG +STE+ A LS++ + S+ +A+R+IKLY
Sbjct 62 QSNDRAQQIV--DATDKLAVNIGLEILKLVPGRISTEVDARLSYDTEASIAKAKRLIKLY 119
Query 123 EEKGINKERILIKIAATWEGIQAAKELKKENINCNITLLFSLCQAIAASDAGAALVSPFV 182
+ GI+ +RILIK+A+TW+GI+AA++L+KE INCN+TLLFS QA A ++AG L+SPFV
Sbjct 120 NDAGISNDRILIKLASTWQGIRAAEQLEKEGINCNLTLLFSFAQARACAEAGVFLISPFV 179
Query 183 GRIL 186
GRIL
Sbjct 180 GRIL 183
> ath:AT1G12230 transaldolase, putative; K00616 transaldolase
[EC:2.2.1.2]
Length=405
Score = 94.4 bits (233), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 58/182 (31%), Positives = 110/182 (60%), Gaps = 3/182 (1%)
Query 6 LDQLAKLSTIVADSADFEAIKEYNPVDATTNPTLVLQAVNNPK--YKHLLDKAIKEAKEA 63
L+ ++ S IV D+ F+ + + P AT + L+L P +++ +D A+ ++
Sbjct 71 LNAVSAFSEIVPDTVVFDDFERFPPTAATVSSALLLGICGLPDTIFRNAVDMALADS-SC 129
Query 64 AKGKSQEIIIEEAIDRVLVLFGIEILQIIPGLVSTEIPADLSFNQQGSVERARRIIKLYE 123
A ++ E + ++ +V G ++++++PG VSTE+ A L+++ G + + +++LY
Sbjct 130 AGLETTESRLSCFFNKAIVNVGGDLVKLVPGRVSTEVDARLAYDTNGIIRKVHDLLRLYN 189
Query 124 EKGINKERILIKIAATWEGIQAAKELKKENINCNITLLFSLCQAIAASDAGAALVSPFVG 183
E + +R+L KI ATW+GI+AA+ L+ E I ++T ++S QA AAS AGA+++ FVG
Sbjct 190 EIDVPHDRLLFKIPATWQGIEAARLLESEGIQTHMTFVYSFAQAAAASQAGASVIQIFVG 249
Query 184 RI 185
R+
Sbjct 250 RL 251
> ath:AT5G13420 transaldolase, putative; K00616 transaldolase
[EC:2.2.1.2]
Length=438
Score = 45.8 bits (107), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 36/141 (25%), Positives = 72/141 (51%), Gaps = 6/141 (4%)
Query 34 TTNPTLVLQAVNNPKYKHLLDKAIKEAKEAAKGKSQEIIIEEAIDRVLVLFGI-EILQII 92
T+NP + +A++ + + + E+ + + E+++++ D + I + +
Sbjct 112 TSNPAIFQKAISTSNAYNDQFRTLVESGKDIESAYWELVVKDIQDACKLFEPIYDQTEGA 171
Query 93 PGLVSTEIPADLSFNQQGSVERARRIIKLYEEKGINKERILIKIAATWEGIQAAKELKKE 152
G VS E+ L+ + QG+VE A+ Y K +N+ + IKI AT I + +++
Sbjct 172 DGYVSVEVSPRLADDTQGTVEAAK-----YLSKVVNRRNVYIKIPATAPCIPSIRDVIAA 226
Query 153 NINCNITLLFSLCQAIAASDA 173
I+ N+TL+FS+ + A DA
Sbjct 227 GISVNVTLIFSIARYEAVIDA 247
> eco:b0825 fsaA, ECK0815, JW5109, mipB, ybiZ; fructose-6-phosphate
aldolase 1 (EC:4.1.2.-); K08313 fructose-6-phosphate aldolase
1 [EC:4.1.2.-]
Length=220
Score = 39.7 bits (91), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 21/54 (38%), Positives = 32/54 (59%), Gaps = 0/54 (0%)
Query 132 ILIKIAATWEGIQAAKELKKENINCNITLLFSLCQAIAASDAGAALVSPFVGRI 185
I++K+ T EG+ A K LK E I T ++ Q + ++ AGA V+P+V RI
Sbjct 82 IVVKVPVTAEGLAAIKMLKAEGIPTLGTAVYGAAQGLLSALAGAEYVAPYVNRI 135
> eco:b3946 fsaB, ECK3938, JW3918, talC, yijG; fructose-6-phosphate
aldolase 2 (EC:4.1.2.-); K08314 fructose-6-phosphate aldolase
2 [EC:4.1.2.-]
Length=220
Score = 36.6 bits (83), Expect = 0.050, Method: Compositional matrix adjust.
Identities = 32/82 (39%), Positives = 46/82 (56%), Gaps = 7/82 (8%)
Query 104 LSFNQQGSVERARRIIKLYEEKGINKERILIKIAATWEGIQAAKELKKENINCNITLLFS 163
+S + QG VE A+R+ I I++KI T EG+ A K LKKE I T ++S
Sbjct 61 MSRDAQGMVEEAKRL-----RDAIPG--IVVKIPVTSEGLAAIKILKKEGITTLGTAVYS 113
Query 164 LCQAIAASDAGAALVSPFVGRI 185
Q + A+ AGA V+P+V R+
Sbjct 114 AAQGLLAALAGAKYVAPYVNRV 135
> bbo:BBOV_II006320 18.m06519; pre-mRNA processing ribonucleoprotein
binding region-containing protein; K12844 U4/U6 small
nuclear ribonucleoprotein PRP31
Length=483
Score = 30.4 bits (67), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 22/69 (31%), Positives = 37/69 (53%), Gaps = 3/69 (4%)
Query 8 QLAKLSTIVADSADFEAIKEYNPVDATTNPTLVL-QAVNNPKYKHLLDKAIKEAKEAAKG 66
Q+++L D+ +A++EY + +NP V + VN+PK LL+K + A E
Sbjct 31 QISELCESDDDAPIIDAVEEY--FSSVSNPHFVFSKKVNDPKLNGLLEKVRQLALEKDWS 88
Query 67 KSQEIIIEE 75
S+ +IEE
Sbjct 89 HSELTLIEE 97
> ath:AT5G15340 pentatricopeptide (PPR) repeat-containing protein
Length=623
Score = 29.6 bits (65), Expect = 6.2, Method: Composition-based stats.
Identities = 12/47 (25%), Positives = 24/47 (51%), Gaps = 0/47 (0%)
Query 106 FNQQGSVERARRIIKLYEEKGINKERILIKIAATWEGIQAAKELKKE 152
+ + G V +RI + EEK + +++ WEG++ +E+ E
Sbjct 156 YGKCGLVSEVKRIFEELEEKSVVSWTVVLDTVVKWEGLERGREVFHE 202
Lambda K H
0.316 0.133 0.358
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5170784960
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40