bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0975_orf1
Length=120
Score E
Sequences producing significant alignments: (Bits) Value
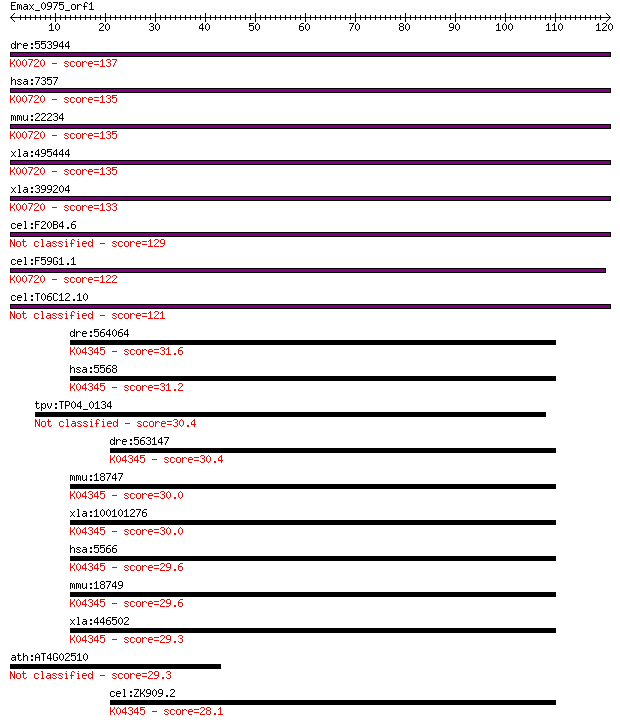
dre:553944 ugcg, cb539, sb:cb539, zgc:112506; UDP-glucose cera... 137 1e-32
hsa:7357 UGCG, GCS, GLCT1; UDP-glucose ceramide glucosyltransf... 135 2e-32
mmu:22234 Ugcg, AU043821, C80537, Epcs21, GlcT-1, Ugcgl; UDP-g... 135 2e-32
xla:495444 ugcg-b, gcs, glct1, ugcg, ugcgb, xlcgt; UDP-glucose... 135 3e-32
xla:399204 ugcg-a, MGC85211, XLCGT, gcs, glct1, ugcga; UDP-glu... 133 1e-31
cel:F20B4.6 cgt-2; Ceramide Glucosyl Transferase family member... 129 3e-30
cel:F59G1.1 cgt-3; Ceramide Glucosyl Transferase family member... 122 2e-28
cel:T06C12.10 cgt-1; Ceramide Glucosyl Transferase family memb... 121 7e-28
dre:564064 prkacaa, zgc:158799; protein kinase, cAMP-dependent... 31.6 0.64
hsa:5568 PRKACG, KAPG, PKACg; protein kinase, cAMP-dependent, ... 31.2 0.81
tpv:TP04_0134 hypothetical protein 30.4 1.3
dre:563147 prkacbb, MGC110804, prkacb, wu:fz54b03, zgc:110804;... 30.4 1.6
mmu:18747 Prkaca, Cs, PKA, PKCD, Pkaca; protein kinase, cAMP d... 30.0 1.9
xla:100101276 prkaca; protein kinase, cAMP-dependent, catalyti... 30.0 1.9
hsa:5566 PRKACA, MGC102831, MGC48865, PKACA; protein kinase, c... 29.6 2.5
mmu:18749 Prkacb, Pkacb; protein kinase, cAMP dependent, catal... 29.6 2.7
xla:446502 prkacbb, MGC80071; protein kinase, cAMP-dependent, ... 29.3 2.9
ath:AT4G02510 TOC159; TOC159 (TRANSLOCON AT THE OUTER ENVELOPE... 29.3 3.2
cel:ZK909.2 kin-1; protein KINase family member (kin-1); K0434... 28.1 6.9
> dre:553944 ugcg, cb539, sb:cb539, zgc:112506; UDP-glucose ceramide
glucosyltransferase (EC:2.4.1.80); K00720 ceramide glucosyltransferase
[EC:2.4.1.80]
Length=393
Score = 137 bits (344), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 60/120 (50%), Positives = 87/120 (72%), Gaps = 0/120 (0%)
Query 1 VSDAGILTEENTLLVLVDDMKDGVALVHQMPFVCNREGFTAILEKVYFGTAHAWIYLAAD 60
+ D+GI + +TL + + M + V LVH +P+V +R+GF A LE+VYFGT+H Y++A+
Sbjct 142 ICDSGIRVKPDTLTDMANQMTEKVGLVHGLPYVADRQGFAATLEQVYFGTSHPRSYISAN 201
Query 61 FLRINCLTGMSALMRKDVLDEVGGISAFGQYLAEDFFFAKSFTDRGWKIKLSSIPALQNS 120
I C+TGMS LMRKD+LD+ GG+ AF QY+AED+F AK+ DRGWK +++ A+QNS
Sbjct 202 VTGIKCVTGMSCLMRKDILDQAGGLIAFAQYIAEDYFMAKAIADRGWKFSMATQVAMQNS 261
> hsa:7357 UGCG, GCS, GLCT1; UDP-glucose ceramide glucosyltransferase
(EC:2.4.1.80); K00720 ceramide glucosyltransferase [EC:2.4.1.80]
Length=394
Score = 135 bits (341), Expect = 2e-32, Method: Compositional matrix adjust.
Identities = 61/120 (50%), Positives = 86/120 (71%), Gaps = 0/120 (0%)
Query 1 VSDAGILTEENTLLVLVDDMKDGVALVHQMPFVCNREGFTAILEKVYFGTAHAWIYLAAD 60
+ D+GI +TL +V+ M + V LVH +P+V +R+GF A LE+VYFGT+H Y++A+
Sbjct 142 ICDSGIRVIPDTLTDMVNQMTEKVGLVHGLPYVADRQGFAATLEQVYFGTSHPRYYISAN 201
Query 61 FLRINCLTGMSALMRKDVLDEVGGISAFGQYLAEDFFFAKSFTDRGWKIKLSSIPALQNS 120
C+TGMS LMRKDVLD+ GG+ AF QY+AED+F AK+ DRGW+ +S+ A+QNS
Sbjct 202 VTGFKCVTGMSCLMRKDVLDQAGGLIAFAQYIAEDYFMAKAIADRGWRFAMSTQVAMQNS 261
> mmu:22234 Ugcg, AU043821, C80537, Epcs21, GlcT-1, Ugcgl; UDP-glucose
ceramide glucosyltransferase (EC:2.4.1.80); K00720
ceramide glucosyltransferase [EC:2.4.1.80]
Length=394
Score = 135 bits (341), Expect = 2e-32, Method: Compositional matrix adjust.
Identities = 61/120 (50%), Positives = 86/120 (71%), Gaps = 0/120 (0%)
Query 1 VSDAGILTEENTLLVLVDDMKDGVALVHQMPFVCNREGFTAILEKVYFGTAHAWIYLAAD 60
+ D+GI +TL +V+ M + V LVH +P+V +R+GF A LE+VYFGT+H Y++A+
Sbjct 142 ICDSGIRVIPDTLTDMVNQMTEKVGLVHGLPYVADRQGFAATLEQVYFGTSHPRSYISAN 201
Query 61 FLRINCLTGMSALMRKDVLDEVGGISAFGQYLAEDFFFAKSFTDRGWKIKLSSIPALQNS 120
C+TGMS LMRKDVLD+ GG+ AF QY+AED+F AK+ DRGW+ +S+ A+QNS
Sbjct 202 VTGFKCVTGMSCLMRKDVLDQAGGLIAFAQYIAEDYFMAKAIADRGWRFSMSTQVAMQNS 261
> xla:495444 ugcg-b, gcs, glct1, ugcg, ugcgb, xlcgt; UDP-glucose
ceramide glucosyltransferase (EC:2.4.1.80); K00720 ceramide
glucosyltransferase [EC:2.4.1.80]
Length=394
Score = 135 bits (340), Expect = 3e-32, Method: Compositional matrix adjust.
Identities = 60/120 (50%), Positives = 87/120 (72%), Gaps = 0/120 (0%)
Query 1 VSDAGILTEENTLLVLVDDMKDGVALVHQMPFVCNREGFTAILEKVYFGTAHAWIYLAAD 60
+ D+GI + +TL + + M + V LVH +P+V +R+GF A LE+VYFGT+H Y++A+
Sbjct 142 ICDSGIKVKPDTLTDMANQMTEKVGLVHGLPYVADRQGFAATLEQVYFGTSHPRSYISAN 201
Query 61 FLRINCLTGMSALMRKDVLDEVGGISAFGQYLAEDFFFAKSFTDRGWKIKLSSIPALQNS 120
I C+TGMS LMRK+VLD+ GG+ AF QY+AED+F AK+ DRGWK +++ A+QNS
Sbjct 202 VTGIKCVTGMSCLMRKEVLDQAGGLIAFAQYIAEDYFMAKAIADRGWKFSMATQVAMQNS 261
> xla:399204 ugcg-a, MGC85211, XLCGT, gcs, glct1, ugcga; UDP-glucose
ceramide glucosyltransferase (EC:2.4.1.80); K00720 ceramide
glucosyltransferase [EC:2.4.1.80]
Length=394
Score = 133 bits (335), Expect = 1e-31, Method: Compositional matrix adjust.
Identities = 59/120 (49%), Positives = 86/120 (71%), Gaps = 0/120 (0%)
Query 1 VSDAGILTEENTLLVLVDDMKDGVALVHQMPFVCNREGFTAILEKVYFGTAHAWIYLAAD 60
+ D+GI + +TL + + M + V LVH +P+V +R+GF A LE+VYFGT+H Y++A+
Sbjct 142 ICDSGIKVKPDTLTDMANQMTEKVGLVHGLPYVADRQGFAATLEQVYFGTSHPRSYISAN 201
Query 61 FLRINCLTGMSALMRKDVLDEVGGISAFGQYLAEDFFFAKSFTDRGWKIKLSSIPALQNS 120
C+TGMS LMRK+VLD+ GG+ AF QY+AED+F AK+ DRGWK +++ A+QNS
Sbjct 202 VTGFKCVTGMSCLMRKEVLDQAGGLIAFAQYIAEDYFMAKAIADRGWKFSMATQVAMQNS 261
> cel:F20B4.6 cgt-2; Ceramide Glucosyl Transferase family member
(cgt-2)
Length=443
Score = 129 bits (323), Expect = 3e-30, Method: Compositional matrix adjust.
Identities = 59/122 (48%), Positives = 86/122 (70%), Gaps = 2/122 (1%)
Query 1 VSDAGILTEENTLLVLVDDM--KDGVALVHQMPFVCNREGFTAILEKVYFGTAHAWIYLA 58
+SD+GI + + +L + M + +ALV Q P+ +R+GF ++ E++YFGT+HA IYLA
Sbjct 189 ISDSGIFMKSDAVLDMASTMMSHETMALVTQTPYCKDRKGFASVFEQIYFGTSHARIYLA 248
Query 59 ADFLRINCLTGMSALMRKDVLDEVGGISAFGQYLAEDFFFAKSFTDRGWKIKLSSIPALQ 118
+ L+ NC TGMS++M+K+ LDE GG +AF YLAED+FF K RG+K +S+ PALQ
Sbjct 249 GNCLQFNCPTGMSSMMKKEALDECGGFAAFSGYLAEDYFFGKKLASRGYKSGISTHPALQ 308
Query 119 NS 120
NS
Sbjct 309 NS 310
> cel:F59G1.1 cgt-3; Ceramide Glucosyl Transferase family member
(cgt-3); K00720 ceramide glucosyltransferase [EC:2.4.1.80]
Length=470
Score = 122 bits (307), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 60/121 (49%), Positives = 82/121 (67%), Gaps = 2/121 (1%)
Query 1 VSDAGILTEENTLLVLVDDM--KDGVALVHQMPFVCNREGFTAILEKVYFGTAHAWIYLA 58
VSD+GI + +L + M + +ALV Q P+ +REGF A E++YFGT+H IYLA
Sbjct 217 VSDSGIFMRSDGVLDMATTMMSHEKMALVTQTPYCKDREGFDAAFEQMYFGTSHGRIYLA 276
Query 59 ADFLRINCLTGMSALMRKDVLDEVGGISAFGQYLAEDFFFAKSFTDRGWKIKLSSIPALQ 118
+ + C TGMS++M+K+ LDE GGIS FG YLAED+FF + +RG+K +SS PALQ
Sbjct 277 GNCMDFVCSTGMSSMMKKEALDECGGISNFGGYLAEDYFFGRELANRGYKSAISSHPALQ 336
Query 119 N 119
N
Sbjct 337 N 337
> cel:T06C12.10 cgt-1; Ceramide Glucosyl Transferase family member
(cgt-1)
Length=466
Score = 121 bits (303), Expect = 7e-28, Method: Compositional matrix adjust.
Identities = 56/122 (45%), Positives = 84/122 (68%), Gaps = 2/122 (1%)
Query 1 VSDAGILTEENTLLVLVDDM--KDGVALVHQMPFVCNREGFTAILEKVYFGTAHAWIYLA 58
+SD+ I + +L + M + +A V Q+P+ +R+GF A E+++FGT+HA +YL
Sbjct 198 ISDSAIFMRPDGILDMATTMMSHEKMASVTQIPYCKDRQGFHAAFEQIFFGTSHARLYLV 257
Query 59 ADFLRINCLTGMSALMRKDVLDEVGGISAFGQYLAEDFFFAKSFTDRGWKIKLSSIPALQ 118
+FL + C +GMS++M+K LDE GG+ FG+YLAED+FFAK+ T RG K +S+ PALQ
Sbjct 258 GNFLGVVCSSGMSSMMKKSALDECGGMEKFGEYLAEDYFFAKALTSRGCKAAISTHPALQ 317
Query 119 NS 120
NS
Sbjct 318 NS 319
> dre:564064 prkacaa, zgc:158799; protein kinase, cAMP-dependent,
catalytic, alpha, genome duplicate a (EC:2.7.11.11); K04345
protein kinase A [EC:2.7.11.11]
Length=352
Score = 31.6 bits (70), Expect = 0.64, Method: Compositional matrix adjust.
Identities = 28/97 (28%), Positives = 37/97 (38%), Gaps = 1/97 (1%)
Query 13 LLVLVDDMKDGVALVHQMPFVCNREGFTAILEKVYFGTAHAWIYLAADFLRINCLTGMSA 72
L+ L KD L M +V E F+ + F HA Y A L L +
Sbjct 105 LVRLEHSFKDNTNLYMVMEYVPGGEMFSHLRRIGRFSEPHARFYAAQIVLTFEYLHSLD- 163
Query 73 LMRKDVLDEVGGISAFGQYLAEDFFFAKSFTDRGWKI 109
L+ +D+ E I G DF FAK R W +
Sbjct 164 LIYRDLKPENLLIDQQGYIQVTDFGFAKRVKGRTWTL 200
> hsa:5568 PRKACG, KAPG, PKACg; protein kinase, cAMP-dependent,
catalytic, gamma (EC:2.7.11.11); K04345 protein kinase A [EC:2.7.11.11]
Length=351
Score = 31.2 bits (69), Expect = 0.81, Method: Compositional matrix adjust.
Identities = 28/97 (28%), Positives = 38/97 (39%), Gaps = 1/97 (1%)
Query 13 LLVLVDDMKDGVALVHQMPFVCNREGFTAILEKVYFGTAHAWIYLAADFLRINCLTGMSA 72
L+ L KD L M +V E F+ + F HA Y A L + L +
Sbjct 104 LVKLQFSFKDNSYLYLVMEYVPGGEMFSRLQRVGRFSEPHACFYAAQVVLAVQYLHSLD- 162
Query 73 LMRKDVLDEVGGISAFGQYLAEDFFFAKSFTDRGWKI 109
L+ +D+ E I G DF FAK R W +
Sbjct 163 LIHRDLKPENLLIDQQGYLQVTDFGFAKRVKGRTWTL 199
> tpv:TP04_0134 hypothetical protein
Length=307
Score = 30.4 bits (67), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 27/103 (26%), Positives = 50/103 (48%), Gaps = 9/103 (8%)
Query 6 ILTEENTLLVLVDDMKDGVALVHQMPFVC-NREGFTAILEKVYFGTAHAWIYLAADFLRI 64
+LT+ N L + DG+ ++ V + E ++ + FG + +L FL
Sbjct 113 LLTKSNGFEFLNYQVVDGLVKSNKYTMVAMDFEDLESVYKLEKFGLSR---HLPTVFLSE 169
Query 65 NCLTGMSALMRKDVLDEVGGISAFGQYLAEDFFFAKSFTDRGW 107
CLT ++ D D++G +AFGQ+ ++ F F + + + GW
Sbjct 170 FCLT----YVQNDTSDQIGSWTAFGQWYSKLFEFIR-YKELGW 207
> dre:563147 prkacbb, MGC110804, prkacb, wu:fz54b03, zgc:110804;
protein kinase, cAMP-dependent, catalytic, beta b; K04345
protein kinase A [EC:2.7.11.11]
Length=351
Score = 30.4 bits (67), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 26/89 (29%), Positives = 34/89 (38%), Gaps = 1/89 (1%)
Query 21 KDGVALVHQMPFVCNREGFTAILEKVYFGTAHAWIYLAADFLRINCLTGMSALMRKDVLD 80
KD L M +V E F+ + F HA Y A L L + L+ +D+
Sbjct 112 KDNSNLYMVMEYVPGGEMFSHLRRIGRFSEPHARFYAAQIVLTFEYLHSLD-LIYRDLKP 170
Query 81 EVGGISAFGQYLAEDFFFAKSFTDRGWKI 109
E I G DF FAK R W +
Sbjct 171 ENLLIDQHGYIQVTDFGFAKRVKGRTWTL 199
> mmu:18747 Prkaca, Cs, PKA, PKCD, Pkaca; protein kinase, cAMP
dependent, catalytic, alpha (EC:2.7.11.11); K04345 protein
kinase A [EC:2.7.11.11]
Length=351
Score = 30.0 bits (66), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 28/97 (28%), Positives = 37/97 (38%), Gaps = 1/97 (1%)
Query 13 LLVLVDDMKDGVALVHQMPFVCNREGFTAILEKVYFGTAHAWIYLAADFLRINCLTGMSA 72
L+ L KD L M +V E F+ + F HA Y A L L +
Sbjct 104 LVKLEFSFKDNSNLYMVMEYVAGGEMFSHLRRIGRFSEPHARFYAAQIVLTFEYLHSLD- 162
Query 73 LMRKDVLDEVGGISAFGQYLAEDFFFAKSFTDRGWKI 109
L+ +D+ E I G DF FAK R W +
Sbjct 163 LIYRDLKPENLLIDQQGYIQVTDFGFAKRVKGRTWTL 199
> xla:100101276 prkaca; protein kinase, cAMP-dependent, catalytic,
alpha (EC:2.7.11.1); K04345 protein kinase A [EC:2.7.11.11]
Length=351
Score = 30.0 bits (66), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 27/97 (27%), Positives = 37/97 (38%), Gaps = 1/97 (1%)
Query 13 LLVLVDDMKDGVALVHQMPFVCNREGFTAILEKVYFGTAHAWIYLAADFLRINCLTGMSA 72
L+ L KD L M +V E F+ + F HA Y + L L +
Sbjct 104 LVRLEYSFKDNTNLYMVMEYVAGGEMFSHLRRIGRFSEPHARFYASQIVLTFEYLHALD- 162
Query 73 LMRKDVLDEVGGISAFGQYLAEDFFFAKSFTDRGWKI 109
L+ +D+ E I G DF FAK R W +
Sbjct 163 LIYRDLKPENLLIDQQGYIQVTDFGFAKRVKGRTWTL 199
> hsa:5566 PRKACA, MGC102831, MGC48865, PKACA; protein kinase,
cAMP-dependent, catalytic, alpha (EC:2.7.11.11); K04345 protein
kinase A [EC:2.7.11.11]
Length=351
Score = 29.6 bits (65), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 28/97 (28%), Positives = 37/97 (38%), Gaps = 1/97 (1%)
Query 13 LLVLVDDMKDGVALVHQMPFVCNREGFTAILEKVYFGTAHAWIYLAADFLRINCLTGMSA 72
L+ L KD L M +V E F+ + F HA Y A L L +
Sbjct 104 LVKLEFSFKDNSNLYMVMEYVPGGEMFSHLRRIGRFSEPHARFYAAQIVLTFEYLHSLD- 162
Query 73 LMRKDVLDEVGGISAFGQYLAEDFFFAKSFTDRGWKI 109
L+ +D+ E I G DF FAK R W +
Sbjct 163 LIYRDLKPENLLIDQQGYIQVTDFGFAKRVKGRTWTL 199
> mmu:18749 Prkacb, Pkacb; protein kinase, cAMP dependent, catalytic,
beta (EC:2.7.11.11); K04345 protein kinase A [EC:2.7.11.11]
Length=338
Score = 29.6 bits (65), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 28/97 (28%), Positives = 37/97 (38%), Gaps = 1/97 (1%)
Query 13 LLVLVDDMKDGVALVHQMPFVCNREGFTAILEKVYFGTAHAWIYLAADFLRINCLTGMSA 72
L+ L KD L M +V E F+ + F HA Y A L L +
Sbjct 91 LVRLEYSFKDNSNLYMVMEYVPGGEMFSHLRRIGRFSEPHARFYAAQIVLTFEYLHSLD- 149
Query 73 LMRKDVLDEVGGISAFGQYLAEDFFFAKSFTDRGWKI 109
L+ +D+ E I G DF FAK R W +
Sbjct 150 LIYRDLKPENLLIDHQGYIQVTDFGFAKRVKGRTWTL 186
> xla:446502 prkacbb, MGC80071; protein kinase, cAMP-dependent,
catalytic, beta b; K04345 protein kinase A [EC:2.7.11.11]
Length=351
Score = 29.3 bits (64), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 27/97 (27%), Positives = 37/97 (38%), Gaps = 1/97 (1%)
Query 13 LLVLVDDMKDGVALVHQMPFVCNREGFTAILEKVYFGTAHAWIYLAADFLRINCLTGMSA 72
L+ L KD L M +V E F+ + F HA Y + L L +
Sbjct 104 LVRLEYSFKDNSNLYMVMEYVAGGEMFSHLRRIGRFSEPHARFYASQIVLTFEYLHALD- 162
Query 73 LMRKDVLDEVGGISAFGQYLAEDFFFAKSFTDRGWKI 109
L+ +D+ E I G DF FAK R W +
Sbjct 163 LIYRDLKPENLLIDQQGYIQVTDFGFAKRVKGRTWTL 199
> ath:AT4G02510 TOC159; TOC159 (TRANSLOCON AT THE OUTER ENVELOPE
MEMBRANE OF CHLOROPLASTS 159); transmembrane receptor
Length=1503
Score = 29.3 bits (64), Expect = 3.2, Method: Composition-based stats.
Identities = 15/42 (35%), Positives = 24/42 (57%), Gaps = 0/42 (0%)
Query 1 VSDAGILTEENTLLVLVDDMKDGVALVHQMPFVCNREGFTAI 42
V+D G EE+ L +VDD ++GV L ++ FV + A+
Sbjct 358 VADNGTKEEESVLGGIVDDAEEGVKLNNKGDFVVDSSAIEAV 399
> cel:ZK909.2 kin-1; protein KINase family member (kin-1); K04345
protein kinase A [EC:2.7.11.11]
Length=381
Score = 28.1 bits (61), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 24/89 (26%), Positives = 35/89 (39%), Gaps = 1/89 (1%)
Query 21 KDGVALVHQMPFVCNREGFTAILEKVYFGTAHAWIYLAADFLRINCLTGMSALMRKDVLD 80
KD L + F+ E F+ + F H+ Y A L L + L+ +D+
Sbjct 142 KDNSNLYMVLEFISGGEMFSHLRRIGRFSEPHSRFYAAQIVLAFEYLHSLD-LIYRDLKP 200
Query 81 EVGGISAFGQYLAEDFFFAKSFTDRGWKI 109
E I + G DF FAK R W +
Sbjct 201 ENLLIDSTGYLKITDFGFAKRVKGRTWTL 229
Lambda K H
0.325 0.139 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2018002440
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40