bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0955_orf2
Length=216
Score E
Sequences producing significant alignments: (Bits) Value
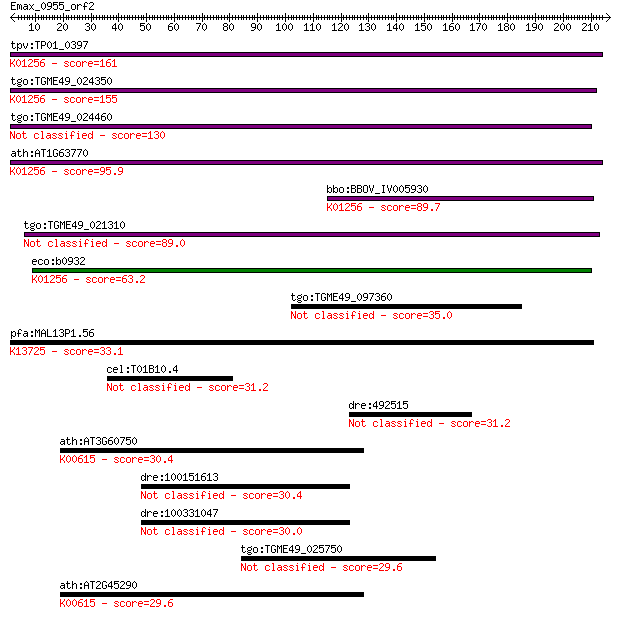
tpv:TP01_0397 alpha-aminoacylpeptide hydrolase (EC:3.4.11.2); ... 161 2e-39
tgo:TGME49_024350 aminopeptidase N, putative (EC:3.4.11.2); K0... 155 8e-38
tgo:TGME49_024460 aminopeptidase N, putative (EC:3.4.11.2) 130 4e-30
ath:AT1G63770 peptidase M1 family protein; K01256 aminopeptida... 95.9 1e-19
bbo:BBOV_IV005930 23.m06519; aminopeptidase (EC:3.4.11.2); K01... 89.7 7e-18
tgo:TGME49_021310 aminopeptidase N, putative (EC:3.4.11.2) 89.0 1e-17
eco:b0932 pepN, ECK0923, JW0915; aminopeptidase N (EC:3.4.11.2... 63.2 7e-10
tgo:TGME49_097360 hypothetical protein 35.0 0.22
pfa:MAL13P1.56 M1-family aminopeptidase (EC:3.4.11.-); K13725 ... 33.1 0.67
cel:T01B10.4 nhr-14; Nuclear Hormone Receptor family member (n... 31.2 2.7
dre:492515 ftr72, zgc:103602; finTRIM family, member 72 31.2
ath:AT3G60750 transketolase, putative (EC:2.2.1.1); K00615 tra... 30.4 4.7
dre:100151613 hypothetical LOC100151613 30.4 4.9
dre:100331047 Gag-protease-integrase-RT-RNaseH polyprotein-like 30.0 7.2
tgo:TGME49_025750 hypothetical protein 29.6 8.1
ath:AT2G45290 transketolase, putative (EC:2.2.1.1); K00615 tra... 29.6 9.1
> tpv:TP01_0397 alpha-aminoacylpeptide hydrolase (EC:3.4.11.2);
K01256 aminopeptidase N [EC:3.4.11.2]
Length=1020
Score = 161 bits (407), Expect = 2e-39, Method: Compositional matrix adjust.
Identities = 83/213 (38%), Positives = 130/213 (61%), Gaps = 9/213 (4%)
Query 1 LAYDTDPVTKWFASRALATPIVISRSKRISAKGNAQKLEQLPDDYVEGLRTILTDQATDN 60
+ YDTD + +W A+++LAT +V+ +K +A ++P Y+E + +L N
Sbjct 707 MLYDTDGLNRWNAAQSLATKVVLDLTKDPTA--------EVPKTYLEAYKKLLNSDMDHN 758
Query 61 ALKALILQLPDWNGLSTEMRPVNPEALHKAIRTVKADLATALKPEMSELYKQLTLPTDQE 120
K L + +PD + L+++M+P +P L ++R +K +L +P +E+YK LTL Q+
Sbjct 759 E-KGLCMSMPDVDILASKMKPYDPGLLFASLRKLKQELGRTFRPTFTEMYKSLTLREGQK 817
Query 121 DEVTEEATGRRKLRNTLLHFLSAMGDEEAVERAYKHFTEAKCMTDKYAGLIALSNIPVMQ 180
DE+T+E RR LRNT+ FL +M D E+VE A KH+ +AK M DKY + L ++
Sbjct 818 DELTKEDMARRFLRNTVFSFLVSMRDMESVELALKHYRDAKVMNDKYTAFVQLMHMEFQD 877
Query 181 REKAFSRFYDDAAGDSLVVDKWFKAQALSDLPS 213
R+K FY+ A GD+ +VDKWFKAQA+S+L S
Sbjct 878 RQKVVDDFYNFANGDAALVDKWFKAQAVSELES 910
> tgo:TGME49_024350 aminopeptidase N, putative (EC:3.4.11.2);
K01256 aminopeptidase N [EC:3.4.11.2]
Length=1419
Score = 155 bits (393), Expect = 8e-38, Method: Compositional matrix adjust.
Identities = 82/213 (38%), Positives = 131/213 (61%), Gaps = 2/213 (0%)
Query 1 LAYDTDPVTKWFASRALATPIVISRSKRISAK-GNAQKLEQLPDDYVEGLRTILTDQATD 59
+A+D+D KW A+ LA+ ++ R+++ K G + +LP YVE + L +Q D
Sbjct 1089 MAHDSDDFAKWQAAHTLASGLLKHRAEQWREKQGEDVEFARLPKIYVEAFKQTLLEQGRD 1148
Query 60 NALKALILQLPDWNGLSTEMRPVNPEALHKAIRTVKADLATALKPEMSELYKQLTLPTDQ 119
+++A L+LPD +G++ EM P++P AL +A +V+ ++ LK ++ ++Y L+ P +
Sbjct 1149 RSIQAYTLRLPDRDGVAQEMEPIDPLALKEATESVRREVGQLLKSDLLKVYASLSAPESE 1208
Query 120 EDEVTEEA-TGRRKLRNTLLHFLSAMGDEEAVERAYKHFTEAKCMTDKYAGLIALSNIPV 178
+E +++ RR+LRN +L+FL+ D+EA A HF AK MT+KYA L L +I
Sbjct 1209 AEESRDQSEVSRRRLRNVILYFLTGERDKEAAALAMNHFKSAKGMTEKYAALSILCDIEG 1268
Query 179 MQREKAFSRFYDDAAGDSLVVDKWFKAQALSDL 211
+R A +FY DA GD LV+DKWF QALSD+
Sbjct 1269 PERTAALEQFYRDAKGDPLVLDKWFAVQALSDV 1301
> tgo:TGME49_024460 aminopeptidase N, putative (EC:3.4.11.2)
Length=970
Score = 130 bits (327), Expect = 4e-30, Method: Compositional matrix adjust.
Identities = 73/221 (33%), Positives = 126/221 (57%), Gaps = 12/221 (5%)
Query 1 LAYDTDPVTKWFASRALATPIVISRSKRISAK---------GNAQKLEQLPDDYVEGLRT 51
+ YD+D V +W A++ L+ ++++R + + +LP YV+ +R
Sbjct 632 VTYDSDNVNRWQAAQTLSRKVLLTRISEYQQRVEEIEEGRVSESDLFSKLPTQYVQTVRD 691
Query 52 ILT--DQATDNALKALILQLPDWNGLSTEMRPVNPEALHKAIRTVKADLATALKPEMSEL 109
I+ + + +K+L+L LP L + ++P+A++ A+ +V+ D+ AL EM +L
Sbjct 692 IVIAPESSMGKDIKSLLLSLPTKAQLELAVDSIDPDAINAALASVRRDIVDALGEEMLQL 751
Query 110 YKQLTLPTDQEDEVTE-EATGRRKLRNTLLHFLSAMGDEEAVERAYKHFTEAKCMTDKYA 168
Y +LTLP E+ + E GRR LRN LL FL+A D+++ + A HF A M+DK A
Sbjct 752 YTELTLPAGTEESGADIEHWGRRALRNELLRFLTASFDQKSAKLASAHFDRAMVMSDKVA 811
Query 169 GLIALSNIPVMQREKAFSRFYDDAAGDSLVVDKWFKAQALS 209
L L+ IP +R++AF RF + AG++++++KW QA+S
Sbjct 812 ALTVLTEIPGQERDEAFERFRQEVAGNAVLMEKWMSLQAMS 852
> ath:AT1G63770 peptidase M1 family protein; K01256 aminopeptidase
N [EC:3.4.11.2]
Length=987
Score = 95.9 bits (237), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 66/214 (30%), Positives = 109/214 (50%), Gaps = 8/214 (3%)
Query 1 LAYDTDPVTKWFASRALATPIVISRSKRISAKGNAQKLEQLPDDYVEGLRTILTDQATDN 60
LA+D+D +W A + LA ++++ + + K L +V+GL ++L+D + D
Sbjct 671 LAHDSDEFNRWEAGQVLARKLMLN----LVSDFQQNKPLALNPKFVQGLGSVLSDSSLDK 726
Query 61 ALKALILQLPDWNGLSTEMRPVNPEALHKAIRTVKADLATALKPEMSELYKQLTLPTDQE 120
A + LP + M +P+A+H + V+ LA+ LK EL K + E
Sbjct 727 EFIAKAITLPGEGEIMDMMAVADPDAVHAVRKFVRKQLASELK---EELLKIVENNRSTE 783
Query 121 DEVTEEAT-GRRKLRNTLLHFLSAMGDEEAVERAYKHFTEAKCMTDKYAGLIALSNIPVM 179
V + + RR L+NT L +L+++ D +E A + A +TD++A L ALS P
Sbjct 784 AYVFDHSNMARRALKNTALAYLASLEDPAYMELALNEYKMATNLTDQFAALAALSQNPGK 843
Query 180 QREKAFSRFYDDAAGDSLVVDKWFKAQALSDLPS 213
R+ + FY+ D LVV+KWF Q+ SD+P
Sbjct 844 TRDDILADFYNKWQDDYLVVNKWFLLQSTSDIPG 877
> bbo:BBOV_IV005930 23.m06519; aminopeptidase (EC:3.4.11.2); K01256
aminopeptidase N [EC:3.4.11.2]
Length=846
Score = 89.7 bits (221), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 42/96 (43%), Positives = 60/96 (62%), Gaps = 0/96 (0%)
Query 115 LPTDQEDEVTEEATGRRKLRNTLLHFLSAMGDEEAVERAYKHFTEAKCMTDKYAGLIALS 174
L +++D + ++ RR LRNTLL +L D AVE A H+ A+CMTD+Y + L
Sbjct 632 LDNNEKDTLEKDDMARRYLRNTLLGYLVCRSDASAVELALGHYRAARCMTDRYYAFVQLM 691
Query 175 NIPVMQREKAFSRFYDDAAGDSLVVDKWFKAQALSD 210
N+ ++ + FY+ AAGD+LV+DKWF AQA SD
Sbjct 692 NMDFAGKDDVIADFYERAAGDALVIDKWFSAQASSD 727
> tgo:TGME49_021310 aminopeptidase N, putative (EC:3.4.11.2)
Length=966
Score = 89.0 bits (219), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 75/246 (30%), Positives = 115/246 (46%), Gaps = 45/246 (18%)
Query 6 DPVTKWFASRALATPIVISR--SKRISAKGNAQKLEQLPDDYVEGLRTILTDQATDNALK 63
DP T+W A + LA I+ R +K+ G L + +GL L D A
Sbjct 615 DPFTRWNAFQFLALQILKERMATKKKETSG------VLSPAFQQGLHFALRDMPLSPAFT 668
Query 64 ALILQLPDWNGLSTEM-RPVNPEALHKAIRTVKADLATALKPEMSELYKQLTLPT----- 117
AL+L LP ++ L + RP++P+A+ A R++ D+ + + E Y T+P
Sbjct 669 ALLLTLPSYSRLEQDAPRPLDPDAIISARRSLLRDIYYFHRNALDEAYVATTIPKVDDRE 728
Query 118 -DQEDEVTE--EATGRRKLRNTLLHFLSAMGDEEAVERAYKHFTEAKCMTDKYAGLIALS 174
D++ E E E RR LR+ LL +++A DE + + A KHF +A+ MTDK A L L
Sbjct 729 RDRQLESAEDPEQWQRRALRSILLEYVTANRDERSAKLALKHFKDARVMTDKIAALHVLV 788
Query 175 NIPV-MQREKAFSRFYDDAAG---------------------------DSLVVDKWFKAQ 206
++P +RE+A FY++A G + ++ KWF Q
Sbjct 789 DLPFNKEREEALHLFYEEARGTTRGNTARTEEQEAEAENEIKSIAVLCNPQLLTKWFALQ 848
Query 207 ALSDLP 212
A S LP
Sbjct 849 ARSSLP 854
> eco:b0932 pepN, ECK0923, JW0915; aminopeptidase N (EC:3.4.11.2);
K01256 aminopeptidase N [EC:3.4.11.2]
Length=870
Score = 63.2 bits (152), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 62/202 (30%), Positives = 90/202 (44%), Gaps = 9/202 (4%)
Query 9 TKWFASRAL-ATPIVISRSKRISAKGNAQKLEQLPDDYVEGLRTILTDQATDNALKALIL 67
++W A+++L AT I ++ ++ Q L LP + R +L D+ D AL A IL
Sbjct 563 SRWDAAQSLLATYIKLN----VARHQQGQPL-SLPVHVADAFRAVLLDEKIDPALAAEIL 617
Query 68 QLPDWNGLSTEMRPVNPEALHKAIRTVKADLATALKPEMSELYKQLTLPTDQEDEVTEEA 127
LP N ++ ++P A+ + + LAT L E+ +Y E V E
Sbjct 618 TLPSVNEMAELFDIIDPIAIAEVREALTRTLATELADELLAIYNA---NYQSEYRVEHED 674
Query 128 TGRRKLRNTLLHFLSAMGDEEAVERAYKHFTEAKCMTDKYAGLIALSNIPVMQREKAFSR 187
+R LRN L FL+ A K F EA MTD A L A + R+
Sbjct 675 IAKRTLRNACLRFLAFGETHLADVLVSKQFHEANNMTDALAALSAAVAAQLPCRDALMQE 734
Query 188 FYDDAAGDSLVVDKWFKAQALS 209
+ D + LV+DKWF QA S
Sbjct 735 YDDKWHQNGLVMDKWFILQATS 756
> tgo:TGME49_097360 hypothetical protein
Length=910
Score = 35.0 bits (79), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 28/83 (33%), Positives = 40/83 (48%), Gaps = 4/83 (4%)
Query 102 LKPEMSELYKQLTLPTDQEDEVTEEATGRRKLRNTLLHFLSAMGDEEAVERAYKHFTEAK 161
L E L+KQL L D E ++T E +R+LR + L A+ E +++ H K
Sbjct 759 LIEEQERLFKQLILLLDHEQQLTAE---QRRLRENVKENLEAVNRAEKLQQELDHLRREK 815
Query 162 CMTDKYAGLIALSNIPVMQREKA 184
T K AG +A S +Q E A
Sbjct 816 ENTGKEAGQMA-SRTATLQTEVA 837
> pfa:MAL13P1.56 M1-family aminopeptidase (EC:3.4.11.-); K13725
M1-family aminopeptidase [EC:3.4.11.-]
Length=1085
Score = 33.1 bits (74), Expect = 0.67, Method: Composition-based stats.
Identities = 45/225 (20%), Positives = 89/225 (39%), Gaps = 23/225 (10%)
Query 1 LAYDTDPVTKWFA-SRALATPIVISRSKRISAKGNAQKLEQ-----LPDDYVEGLRTILT 54
L YD+D ++ + + I+++ ++ + AK +KLE + +++ ++ +L
Sbjct 755 LKYDSDAFVRYNSCTNIYMKQILMNYNEFLKAKN--EKLESFNLTPVNAQFIDAIKYLLE 812
Query 55 DQATDNALKALILQLPDWNGLSTEMRPVNPEALHKAIRTVKADLATALKPEMSELYKQLT 114
D D K+ I+ LP + + ++ + L + + L +++K L
Sbjct 813 DPHADAGFKSYIVSLPQDRYIINFVSNLDTDVLADTKEYIYKQIGDKLNDVYYKMFKSLE 872
Query 115 LPTDQ------EDEVTEEATGRRKLRNTLLHFLSAMGDEEAVERAYKHFTEAKCMTDKYA 168
D E V + R LRNTLL LS + +H Y
Sbjct 873 AKADDLTYFNDESHVDFDQMNMRTLRNTLLSLLSKAQYPNILNEIIEH------SKSPYP 926
Query 169 G--LIALSNIPVMQRE-KAFSRFYDDAAGDSLVVDKWFKAQALSD 210
L +LS + + + + Y + D L++ +W K + SD
Sbjct 927 SNWLTSLSVSAYFDKYFELYDKTYKLSKDDELLLQEWLKTVSRSD 971
> cel:T01B10.4 nhr-14; Nuclear Hormone Receptor family member
(nhr-14)
Length=435
Score = 31.2 bits (69), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 17/45 (37%), Positives = 26/45 (57%), Gaps = 0/45 (0%)
Query 36 QKLEQLPDDYVEGLRTILTDQATDNALKALILQLPDWNGLSTEMR 80
+ L ++ D+ + LR + TD A + LKAL+L PD G+S R
Sbjct 237 EALTRIIDELLTPLRRLRTDHAEFSCLKALLLLNPDVVGISNNTR 281
> dre:492515 ftr72, zgc:103602; finTRIM family, member 72
Length=550
Score = 31.2 bits (69), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 19/44 (43%), Positives = 24/44 (54%), Gaps = 2/44 (4%)
Query 123 VTEEATGRRKLRNTLLHFLSAMGDEEAVERAYKHFTEAKCMTDK 166
+ E G+R+LR H S+ EEAVER K FTE C +K
Sbjct 206 IKEREKGQRQLRTATKHLKSSA--EEAVERMEKVFTELICSLEK 247
> ath:AT3G60750 transketolase, putative (EC:2.2.1.1); K00615 transketolase
[EC:2.2.1.1]
Length=741
Score = 30.4 bits (67), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 30/114 (26%), Positives = 52/114 (45%), Gaps = 17/114 (14%)
Query 19 TPIVISRSKRISAKGNAQKLEQLPDDYVEGLRT---ILTDQATDNALKALILQLPDWNGL 75
TP +++ S+ QKL LP +EG+ ++D ++ N +++ G
Sbjct 590 TPSILALSR--------QKLPHLPGTSIEGVEKGGYTISDDSSGNKPDVILIG----TGS 637
Query 76 STEMRPVNPEALHKAIRTVK--ADLATALKPEMSELYKQLTLPTDQEDEVTEEA 127
E+ E L K +TV+ + + L E S+ YK+ LP+D V+ EA
Sbjct 638 ELEIAAQAAEVLRKDGKTVRVVSFVCWELFDEQSDEYKESVLPSDVSARVSIEA 691
> dre:100151613 hypothetical LOC100151613
Length=1273
Score = 30.4 bits (67), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 25/80 (31%), Positives = 40/80 (50%), Gaps = 6/80 (7%)
Query 48 GLR-TILT----DQATDNALKALILQLPDWNGLSTEMRPVNPEALHKAIRTVKADLATAL 102
GL+ TIL+ D+ T+ A ++Q D LS MR + KA++ ++ A+
Sbjct 2 GLKETILSTGDIDEKTNEECYAKLIQFLDDKSLSLVMREATDDG-RKALQILRNHYASQG 60
Query 103 KPEMSELYKQLTLPTDQEDE 122
KP + LY +LT + DE
Sbjct 61 KPRIITLYTELTSLKKESDE 80
> dre:100331047 Gag-protease-integrase-RT-RNaseH polyprotein-like
Length=1170
Score = 30.0 bits (66), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 25/80 (31%), Positives = 40/80 (50%), Gaps = 6/80 (7%)
Query 48 GLR-TILT----DQATDNALKALILQLPDWNGLSTEMRPVNPEALHKAIRTVKADLATAL 102
GL+ TIL+ D+ T+ A ++Q D LS MR + KA++ ++ A+
Sbjct 2 GLKETILSTGDIDEKTNEECYAELIQFLDDKSLSLVMREATDDG-RKALQILRNHYASQG 60
Query 103 KPEMSELYKQLTLPTDQEDE 122
KP + LY +LT + DE
Sbjct 61 KPRIIALYTELTSLKKESDE 80
> tgo:TGME49_025750 hypothetical protein
Length=7371
Score = 29.6 bits (65), Expect = 8.1, Method: Composition-based stats.
Identities = 18/70 (25%), Positives = 32/70 (45%), Gaps = 0/70 (0%)
Query 84 PEALHKAIRTVKADLATALKPEMSELYKQLTLPTDQEDEVTEEATGRRKLRNTLLHFLSA 143
P A++ A R + + S L + +P + +TE +GRRK R +++H +
Sbjct 6037 PLAMYNAGRFLTQGSQPRHRQSRSRLLCSIGVPRRTKYGITESVSGRRKWRRSVVHGIKC 6096
Query 144 MGDEEAVERA 153
A+ RA
Sbjct 6097 SNGAPAIGRA 6106
> ath:AT2G45290 transketolase, putative (EC:2.2.1.1); K00615 transketolase
[EC:2.2.1.1]
Length=741
Score = 29.6 bits (65), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 32/115 (27%), Positives = 52/115 (45%), Gaps = 19/115 (16%)
Query 19 TPIVISRSKRISAKGNAQKLEQLPDDYVEGLRT---ILTDQATDNALKALILQLPDWNGL 75
TP V++ S+ QKL QLP +E + ++D +T N +++ G
Sbjct 590 TPSVLALSR--------QKLPQLPGTSIESVEKGGYTISDNSTGNKPDVILIG----TGS 637
Query 76 STEMRPVNPEALH---KAIRTVKADLATALKPEMSELYKQLTLPTDQEDEVTEEA 127
E+ E L K++R V + + L E S+ YK+ LP+D V+ EA
Sbjct 638 ELEIAAQAAEKLREQGKSVRVV-SFVCWELFDEQSDAYKESVLPSDVSARVSIEA 691
Lambda K H
0.315 0.130 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6945028560
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40