bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0938_orf1
Length=125
Score E
Sequences producing significant alignments: (Bits) Value
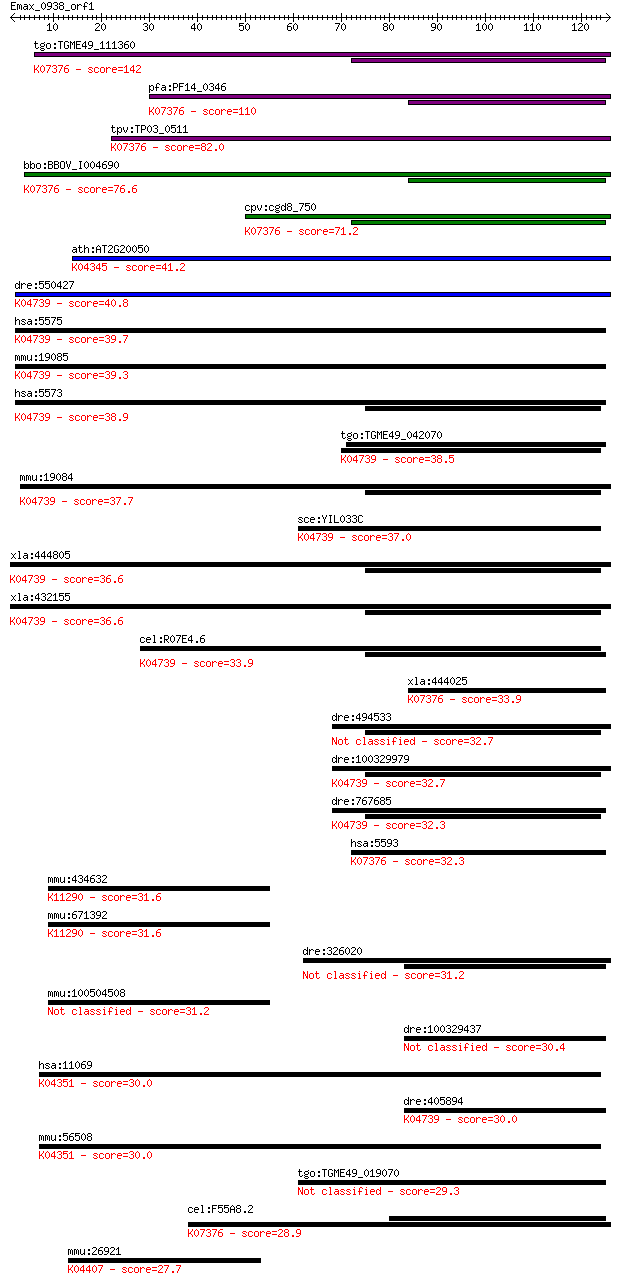
tgo:TGME49_111360 cGMP-dependent protein kinase, putative (EC:... 142 3e-34
pfa:PF14_0346 PKG; cGMP-dependent protein kinase (EC:2.7.11.12... 110 1e-24
tpv:TP03_0511 cGMP-dependent protein kinase (EC:2.7.1.37); K07... 82.0 4e-16
bbo:BBOV_I004690 19.m02237; cGMP dependent protein kinase (EC:... 76.6 2e-14
cpv:cgd8_750 cyclic nucleotide (cGMP)-dependent protein kinase... 71.2 7e-13
ath:AT2G20050 ATP binding / cAMP-dependent protein kinase regu... 41.2 0.001
dre:550427 prkar1ab, zgc:112145; protein kinase, cAMP-dependen... 40.8 0.001
hsa:5575 PRKAR1B, PRKAR1; protein kinase, cAMP-dependent, regu... 39.7 0.003
mmu:19085 Prkar1b, AI385716, RIbeta; protein kinase, cAMP depe... 39.3 0.003
hsa:5573 PRKAR1A, CAR, CNC, CNC1, DKFZp779L0468, MGC17251, PKR... 38.9 0.004
tgo:TGME49_042070 cAMP-dependent protein kinase regulatory sub... 38.5 0.005
mmu:19084 Prkar1a, 1300018C22Rik, RIalpha, Tse-1, Tse1; protei... 37.7 0.009
sce:YIL033C BCY1, SRA1; Bcy1p; K04739 cAMP-dependent protein k... 37.0 0.018
xla:444805 prkar1b, MGC82149, prkar1; protein kinase, cAMP-dep... 36.6 0.023
xla:432155 prkar1a, pkr1; protein kinase, cAMP-dependent, regu... 36.6 0.023
cel:R07E4.6 kin-2; protein KINase family member (kin-2); K0473... 33.9 0.12
xla:444025 prkg2, MGC82580; protein kinase, cGMP-dependent, ty... 33.9 0.12
dre:494533 prkar1aa, im:7047729, prkar1a, zgc:92515; protein k... 32.7 0.27
dre:100329979 protein kinase, cAMP-dependent, regulatory, type... 32.7 0.27
dre:767685 prkar1b, MGC153624, zgc:153624; protein kinase, cAM... 32.3 0.40
hsa:5593 PRKG2, PRKGR2, cGKII; protein kinase, cGMP-dependent,... 32.3 0.43
mmu:434632 MGC103372; cDNA sequence BC085271; K11290 template-... 31.6 0.59
mmu:671392 Gm9531; predicted gene 9531; K11290 template-activa... 31.6 0.63
dre:326020 fd58b04, wu:fd58b04; si:dkey-121j17.5 31.2 0.77
mmu:100504508 protein SET-like 31.2 0.88
dre:100329437 protein kinase, cAMP-dependent, regulatory, type... 30.4 1.6
hsa:11069 RAPGEF4, CAMP-GEFII, CGEF2, EPAC2, Nbla00496; Rap gu... 30.0 1.7
dre:405894 prkar2ab, MGC85886, prkar2a, zgc:85886; protein kin... 30.0 1.8
mmu:56508 Rapgef4, 1300003D15Rik, 5730402K07Rik, 6330581N18Rik... 30.0 1.8
tgo:TGME49_019070 cAMP-dependent protein kinase regulatory sub... 29.3 3.3
cel:F55A8.2 egl-4; EGg Laying defective family member (egl-4);... 28.9 4.1
mmu:26921 Map4k4, 9430080K19Rik, AU043147, AU045934, AW046177,... 27.7 9.0
> tgo:TGME49_111360 cGMP-dependent protein kinase, putative (EC:2.7.11.12);
K07376 protein kinase, cGMP-dependent [EC:2.7.11.12]
Length=994
Score = 142 bits (357), Expect = 3e-34, Method: Compositional matrix adjust.
Identities = 72/125 (57%), Positives = 91/125 (72%), Gaps = 5/125 (4%)
Query 6 QQEQQQQDRKTSQPQQNDDAAAPP----KPGGERK-AQKAIMQQDDTQAEDARLLNHLEK 60
+ E Q+ D K P Q+ + AP KPGG+RK AQKAI++QDD+ E+ +L HL
Sbjct 110 EAEAQEDDLKREAPNQDVPSEAPEGPKEKPGGDRKPAQKAILKQDDSHTEEEKLNAHLAY 169
Query 61 REKTDSDLSLIRSSLSGNLVCSSLNDSEVEALANAVQFFTFAKGDIVTKQGENGSYFFIV 120
REKT +D +LI+ SL NLVCSSLN+ E++ALA A+QFFTF KGD+VTKQGE GSYFFI+
Sbjct 170 REKTPADFALIQDSLKANLVCSSLNEGEIDALAVAMQFFTFKKGDVVTKQGEPGSYFFII 229
Query 121 HSGEF 125
HSG F
Sbjct 230 HSGTF 234
Score = 37.0 bits (84), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 15/53 (28%), Positives = 29/53 (54%), Gaps = 0/53 (0%)
Query 72 RSSLSGNLVCSSLNDSEVEALANAVQFFTFAKGDIVTKQGENGSYFFIVHSGE 124
R+ + + L+D ++ L A + + G+ + K+GE G+ FFI+ +GE
Sbjct 553 RAIIRKMYIFRYLSDHQMTMLIKAFKTVRYMSGEYIIKEGERGTRFFIIKAGE 605
> pfa:PF14_0346 PKG; cGMP-dependent protein kinase (EC:2.7.11.12);
K07376 protein kinase, cGMP-dependent [EC:2.7.11.12]
Length=853
Score = 110 bits (274), Expect = 1e-24, Method: Composition-based stats.
Identities = 51/96 (53%), Positives = 69/96 (71%), Gaps = 0/96 (0%)
Query 30 KPGGERKAQKAIMQQDDTQAEDARLLNHLEKREKTDSDLSLIRSSLSGNLVCSSLNDSEV 89
K G ER +KAI DD ED+ + +HLE REK D+ +I++SL NLVCS+LND+E+
Sbjct 8 KKGNERNKKKAIFSNDDFTGEDSLMEDHLELREKLSEDIDMIKTSLKNNLVCSTLNDNEI 67
Query 90 EALANAVQFFTFAKGDIVTKQGENGSYFFIVHSGEF 125
L+N +QFF F G++V KQGE GSYFFI++SG+F
Sbjct 68 LTLSNYMQFFVFKSGNLVIKQGEKGSYFFIINSGKF 103
Score = 32.0 bits (71), Expect = 0.44, Method: Composition-based stats.
Identities = 14/41 (34%), Positives = 24/41 (58%), Gaps = 0/41 (0%)
Query 84 LNDSEVEALANAVQFFTFAKGDIVTKQGENGSYFFIVHSGE 124
L D + L A + + +GD + ++GE GS F+I+ +GE
Sbjct 422 LTDKQCNLLIEAFRTTRYEEGDYIIQEGEVGSRFYIIKNGE 462
> tpv:TP03_0511 cGMP-dependent protein kinase (EC:2.7.1.37); K07376
protein kinase, cGMP-dependent [EC:2.7.11.12]
Length=892
Score = 82.0 bits (201), Expect = 4e-16, Method: Composition-based stats.
Identities = 39/105 (37%), Positives = 68/105 (64%), Gaps = 1/105 (0%)
Query 22 NDDAAAPPKPGGERKAQKAIMQQDDTQA-EDARLLNHLEKREKTDSDLSLIRSSLSGNLV 80
N + P ++ K I + +++++ ED ++ HL KREK++SD S I+ SL+ N++
Sbjct 21 NRSMSKDKNPTIRKQRSKFIKKANESESYEDTEIVKHLVKREKSESDKSFIKKSLANNVI 80
Query 81 CSSLNDSEVEALANAVQFFTFAKGDIVTKQGENGSYFFIVHSGEF 125
S+LND E+ A +++ ++ F+ G VT+QG NGSYFF+++ G F
Sbjct 81 FSALNDLEMSAFVDSMSYYVFSVGSKVTEQGTNGSYFFVINEGIF 125
> bbo:BBOV_I004690 19.m02237; cGMP dependent protein kinase (EC:2.7.11.1);
K07376 protein kinase, cGMP-dependent [EC:2.7.11.12]
Length=887
Score = 76.6 bits (187), Expect = 2e-14, Method: Composition-based stats.
Identities = 41/124 (33%), Positives = 68/124 (54%), Gaps = 2/124 (1%)
Query 4 QEQQEQQQQDRKTSQPQQNDDAAAPPKPGGER--KAQKAIMQQDDTQAEDARLLNHLEKR 61
+EQ E + +K + A P G + ++Q + ++ E + L + R
Sbjct 3 KEQDELSHEFKKLHIVHSSTKAWKPNNYQGSKGVRSQPVNVAHNEECDEVSTLAKYATSR 62
Query 62 EKTDSDLSLIRSSLSGNLVCSSLNDSEVEALANAVQFFTFAKGDIVTKQGENGSYFFIVH 121
+KTD D++LIR +L+ NLVC SLN+ E++A ++ F G V +QG+NG+YFFI+
Sbjct 63 DKTDKDITLIRHALANNLVCESLNEYEIDAFIGSMSSFELPAGAPVVRQGDNGTYFFIIS 122
Query 122 SGEF 125
G+F
Sbjct 123 EGDF 126
Score = 27.7 bits (60), Expect = 8.4, Method: Composition-based stats.
Identities = 11/41 (26%), Positives = 23/41 (56%), Gaps = 0/41 (0%)
Query 84 LNDSEVEALANAVQFFTFAKGDIVTKQGENGSYFFIVHSGE 124
L++ +++ L A++ + D + +GE G +I+ SGE
Sbjct 454 LSEKQIDMLIKALKTLRYKLNDTIFNEGEIGDMLYIIKSGE 494
> cpv:cgd8_750 cyclic nucleotide (cGMP)-dependent protein kinase
with 3 cNMP binding domains and a Ser/Thr kinase domain ;
K07376 protein kinase, cGMP-dependent [EC:2.7.11.12]
Length=965
Score = 71.2 bits (173), Expect = 7e-13, Method: Composition-based stats.
Identities = 29/76 (38%), Positives = 54/76 (71%), Gaps = 0/76 (0%)
Query 50 EDARLLNHLEKREKTDSDLSLIRSSLSGNLVCSSLNDSEVEALANAVQFFTFAKGDIVTK 109
E +++HL+ REKT+ D+ I +L+GN+V +SLN+SE+ L +++ ++ + G++V +
Sbjct 79 EQNMVISHLKDREKTEEDIKTISKALAGNVVGASLNESEIATLVSSMHYYEYEVGEVVIE 138
Query 110 QGENGSYFFIVHSGEF 125
QG +G YFF++ +G F
Sbjct 139 QGASGFYFFVISTGSF 154
Score = 34.3 bits (77), Expect = 0.11, Method: Composition-based stats.
Identities = 14/53 (26%), Positives = 29/53 (54%), Gaps = 0/53 (0%)
Query 72 RSSLSGNLVCSSLNDSEVEALANAVQFFTFAKGDIVTKQGENGSYFFIVHSGE 124
R ++ V +++ ++ L +++ F G+ + QG+ G+ FFI+ SGE
Sbjct 491 RDAIKSCFVFQYVSEQQLSLLVKSLRLVKFTSGEKIVVQGDKGTAFFILQSGE 543
> ath:AT2G20050 ATP binding / cAMP-dependent protein kinase regulator/
catalytic/ protein kinase/ protein serine/threonine
phosphatase; K04345 protein kinase A [EC:2.7.11.11]
Length=1094
Score = 41.2 bits (95), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 33/112 (29%), Positives = 45/112 (40%), Gaps = 28/112 (25%)
Query 14 RKTSQPQQNDDAAAPPKPGGERKAQKAIMQQDDTQAEDARLLNHLEKREKTDSDLSLIRS 73
R +N A PP P RK T E+A H+E+
Sbjct 453 RAIENSLENGHAWVPPSPA-HRK----------TWEEEA----HIER------------- 484
Query 74 SLSGNLVCSSLNDSEVEALANAVQFFTFAKGDIVTKQGENGSYFFIVHSGEF 125
L + + L DS+ + L + +Q GDIV KQG G F++V SGEF
Sbjct 485 VLRDHFLFRKLTDSQCQVLLDCMQRLEANPGDIVVKQGGEGDCFYVVGSGEF 536
> dre:550427 prkar1ab, zgc:112145; protein kinase, cAMP-dependent,
regulatory, type I, alpha (tissue specific extinguisher
1) b (EC:2.7.11.1); K04739 cAMP-dependent protein kinase regulator
Length=379
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 30/127 (23%), Positives = 66/127 (51%), Gaps = 6/127 (4%)
Query 2 QQQEQQEQQQQDRKTSQPQQNDDAAAPP-KPGGERKAQKAIMQQDDTQAEDA--RLLNHL 58
+++E ++ Q + +S+ +D +PP P + + ++ + + EDA + +
Sbjct 57 EKEESKQLLNQQKASSRSDSREDEVSPPMNPVVKGRRRRGAISAEVYTEEDAASYVRKVI 116
Query 59 EKREKTDSDLSLIRSSLSGNLVCSSLNDSEVEALANAVQFFTFAKGDIVTKQGENGSYFF 118
K KT + L+ ++ N++ S L+D+E + +A+ T+ G+IV +QG+ G F+
Sbjct 117 PKDYKTMAALA---KAIEKNVLFSHLDDNERSDIFDAMFPVTYIAGEIVIQQGDEGDNFY 173
Query 119 IVHSGEF 125
++ GE
Sbjct 174 VIDQGEM 180
> hsa:5575 PRKAR1B, PRKAR1; protein kinase, cAMP-dependent, regulatory,
type I, beta (EC:2.7.11.1); K04739 cAMP-dependent
protein kinase regulator
Length=381
Score = 39.7 bits (91), Expect = 0.003, Method: Composition-based stats.
Identities = 28/126 (22%), Positives = 65/126 (51%), Gaps = 5/126 (3%)
Query 2 QQQEQQEQQQQDRKTSQPQQNDD--AAAPPKPGGERKAQKAIMQQDDTQAEDARLLNHLE 59
+++E ++ + + SQ +D+ + PP P + + ++ + + EDA ++++
Sbjct 58 EKEENRQILARQKSNSQSDSHDEEVSPTPPNPVVKARRRRGGVSAEVYTEEDA--VSYVR 115
Query 60 KREKTD-SDLSLIRSSLSGNLVCSSLNDSEVEALANAVQFFTFAKGDIVTKQGENGSYFF 118
K D ++ + ++S N++ + L+D+E + +A+ T G+ V +QG G F+
Sbjct 116 KVIPKDYKTMTALAKAISKNVLFAHLDDNERSDIFDAMFPVTHIAGETVIQQGNEGDNFY 175
Query 119 IVHSGE 124
+V GE
Sbjct 176 VVDQGE 181
> mmu:19085 Prkar1b, AI385716, RIbeta; protein kinase, cAMP dependent
regulatory, type I beta (EC:2.7.11.1); K04739 cAMP-dependent
protein kinase regulator
Length=381
Score = 39.3 bits (90), Expect = 0.003, Method: Composition-based stats.
Identities = 28/126 (22%), Positives = 65/126 (51%), Gaps = 5/126 (3%)
Query 2 QQQEQQEQQQQDRKTSQPQQNDD--AAAPPKPGGERKAQKAIMQQDDTQAEDARLLNHLE 59
+++E ++ + + SQ +D+ + PP P + + ++ + + EDA ++++
Sbjct 58 EKEENRQILARQKSNSQCDSHDEEISPTPPNPVVKARRRRGGVSAEVYTEEDA--VSYVR 115
Query 60 KREKTD-SDLSLIRSSLSGNLVCSSLNDSEVEALANAVQFFTFAKGDIVTKQGENGSYFF 118
K D ++ + ++S N++ S L+D+E + +A+ T G+ V +QG G F+
Sbjct 116 KVIPKDYKTMTALAKAISKNVLFSHLDDNERSDIFDAMFPVTHIGGETVIQQGNEGDNFY 175
Query 119 IVHSGE 124
++ GE
Sbjct 176 VIDQGE 181
> hsa:5573 PRKAR1A, CAR, CNC, CNC1, DKFZp779L0468, MGC17251, PKR1,
PPNAD1, PRKAR1, TSE1; protein kinase, cAMP-dependent, regulatory,
type I, alpha (tissue specific extinguisher 1) (EC:2.7.11.1);
K04739 cAMP-dependent protein kinase regulator
Length=381
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 30/127 (23%), Positives = 65/127 (51%), Gaps = 7/127 (5%)
Query 2 QQQEQQEQQQQDRKTSQPQQNDDAAAPPKPGGERKAQK---AIMQQDDTQAEDARLLNH- 57
+++E ++ Q + ++ +D +PP P K ++ AI + T+ + A +
Sbjct 58 EKEEAKQIQNLQKAGTRTDSREDEISPPPPNPVVKGRRRRGAISAEVYTEEDAASYVRKV 117
Query 58 LEKREKTDSDLSLIRSSLSGNLVCSSLNDSEVEALANAVQFFTFAKGDIVTKQGENGSYF 117
+ K KT + L+ ++ N++ S L+D+E + +A+ +F G+ V +QG+ G F
Sbjct 118 IPKDYKTMAALA---KAIEKNVLFSHLDDNERSDIFDAMFSVSFIAGETVIQQGDEGDNF 174
Query 118 FIVHSGE 124
+++ GE
Sbjct 175 YVIDQGE 181
Score = 29.3 bits (64), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 25/49 (51%), Gaps = 0/49 (0%)
Query 75 LSGNLVCSSLNDSEVEALANAVQFFTFAKGDIVTKQGENGSYFFIVHSG 123
LS + SL+ E +A+A++ F G + QGE G FFI+ G
Sbjct 250 LSKVSILESLDKWERLTVADALEPVQFEDGQKIVVQGEPGDEFFIILEG 298
> tgo:TGME49_042070 cAMP-dependent protein kinase regulatory subunit,
putative (EC:2.7.10.2); K04739 cAMP-dependent protein
kinase regulator
Length=308
Score = 38.5 bits (88), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 15/54 (27%), Positives = 32/54 (59%), Gaps = 0/54 (0%)
Query 71 IRSSLSGNLVCSSLNDSEVEALANAVQFFTFAKGDIVTKQGENGSYFFIVHSGE 124
I + + + SSL+ ++E + NA Q + KG ++ +QG++G +++ +GE
Sbjct 47 ITKVIESSFLFSSLDIEDLETVINAFQEVSVKKGTVIIRQGDDGDRLYLIETGE 100
Score = 34.3 bits (77), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 15/54 (27%), Positives = 29/54 (53%), Gaps = 0/54 (0%)
Query 70 LIRSSLSGNLVCSSLNDSEVEALANAVQFFTFAKGDIVTKQGENGSYFFIVHSG 123
+ SL + ++ E L++A++ T+ GD++ K+GE G F+I+ G
Sbjct 170 IFEESLKEVRILEDMDPYERSKLSDALRTATYEDGDVIIKEGETGDTFYILLEG 223
> mmu:19084 Prkar1a, 1300018C22Rik, RIalpha, Tse-1, Tse1; protein
kinase, cAMP dependent regulatory, type I, alpha (EC:2.7.11.1);
K04739 cAMP-dependent protein kinase regulator
Length=381
Score = 37.7 bits (86), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 33/128 (25%), Positives = 65/128 (50%), Gaps = 8/128 (6%)
Query 3 QQEQQEQQQQDRKTS-QPQQNDDAAAPPKPGGERKAQK---AIMQQDDTQAEDARLLNH- 57
++E+ Q Q +KT + +D +PP P K ++ AI + T+ + A +
Sbjct 58 EKEEARQIQCLQKTGIRTDSREDEISPPPPNPVVKGRRRRGAISAEVYTEEDAASYVRKV 117
Query 58 LEKREKTDSDLSLIRSSLSGNLVCSSLNDSEVEALANAVQFFTFAKGDIVTKQGENGSYF 117
+ K KT + L+ ++ N++ S L+D+E + +A+ +F G+ V +QG+ G F
Sbjct 118 IPKDYKTMAALA---KAIEKNVLFSHLDDNERSDIFDAMFPVSFIAGETVIQQGDEGDNF 174
Query 118 FIVHSGEF 125
+++ GE
Sbjct 175 YVIDQGEM 182
Score = 29.3 bits (64), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 25/49 (51%), Gaps = 0/49 (0%)
Query 75 LSGNLVCSSLNDSEVEALANAVQFFTFAKGDIVTKQGENGSYFFIVHSG 123
LS + SL+ E +A+A++ F G + QGE G FFI+ G
Sbjct 250 LSKVSILESLDKWERLTVADALEPVQFEDGQKIVVQGEPGDEFFIILEG 298
> sce:YIL033C BCY1, SRA1; Bcy1p; K04739 cAMP-dependent protein
kinase regulator
Length=416
Score = 37.0 bits (84), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 17/63 (26%), Positives = 33/63 (52%), Gaps = 0/63 (0%)
Query 61 REKTDSDLSLIRSSLSGNLVCSSLNDSEVEALANAVQFFTFAKGDIVTKQGENGSYFFIV 120
+EK++ L + S+ N + + L+ + N ++ + KG + KQG+ G YF++V
Sbjct 165 KEKSEQQLQRLEKSIRNNFLFNKLDSDSKRLVINCLEEKSVPKGATIIKQGDQGDYFYVV 224
Query 121 HSG 123
G
Sbjct 225 EKG 227
> xla:444805 prkar1b, MGC82149, prkar1; protein kinase, cAMP-dependent,
regulatory, type I,beta; K04739 cAMP-dependent protein
kinase regulator
Length=381
Score = 36.6 bits (83), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 30/129 (23%), Positives = 64/129 (49%), Gaps = 7/129 (5%)
Query 1 QQQQEQQEQQQQDRKTSQPQQNDDAAAPPKPGGERKAQK---AIMQQDDTQAEDARLLNH 57
++++ +Q QQ + + D+ + PP K ++ AI + T+ + A +
Sbjct 57 EKEETRQTLNQQKSGSRSDSREDEISPPPHMNSVVKGRRRRGAISAEVYTEEDAASYVRK 116
Query 58 -LEKREKTDSDLSLIRSSLSGNLVCSSLNDSEVEALANAVQFFTFAKGDIVTKQGENGSY 116
+ K KT + L+ ++ N++ + L+D+E + +A+ T+ G+ V +QG+ G
Sbjct 117 VIPKDYKTMAALA---KAIEKNVLFAHLDDNERSDIFDAMFSVTYIAGETVIQQGDEGDN 173
Query 117 FFIVHSGEF 125
F++V GE
Sbjct 174 FYVVDQGEM 182
Score = 29.3 bits (64), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 25/49 (51%), Gaps = 0/49 (0%)
Query 75 LSGNLVCSSLNDSEVEALANAVQFFTFAKGDIVTKQGENGSYFFIVHSG 123
LS + SL+ E +A+A++ F G + QGE G FFI+ G
Sbjct 250 LSKVSILESLDKWERLTVADALEPVQFEDGQKIVVQGEPGDEFFIILEG 298
> xla:432155 prkar1a, pkr1; protein kinase, cAMP-dependent, regulatory,
type I, alpha (tissue specific extinguisher 1) (EC:2.7.11.1);
K04739 cAMP-dependent protein kinase regulator
Length=381
Score = 36.6 bits (83), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 30/129 (23%), Positives = 64/129 (49%), Gaps = 7/129 (5%)
Query 1 QQQQEQQEQQQQDRKTSQPQQNDDAAAPPKPGGERKAQK---AIMQQDDTQAEDARLLNH 57
++++ +Q QQ + + D+ + PP K ++ AI + T+ + A +
Sbjct 57 EKEEARQILNQQKSGSRSDSREDEISPPPHMNSVVKGRRRRGAISAEVYTEEDAASYVRK 116
Query 58 -LEKREKTDSDLSLIRSSLSGNLVCSSLNDSEVEALANAVQFFTFAKGDIVTKQGENGSY 116
+ K KT + L+ ++ N++ + L+D+E + +A+ T+ G+ V +QG+ G
Sbjct 117 VIPKDYKTMAALA---KAIEKNVLFAHLDDTERSDIFDAMFSVTYISGETVIQQGDEGDN 173
Query 117 FFIVHSGEF 125
F++V GE
Sbjct 174 FYVVDQGEM 182
Score = 29.3 bits (64), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 25/49 (51%), Gaps = 0/49 (0%)
Query 75 LSGNLVCSSLNDSEVEALANAVQFFTFAKGDIVTKQGENGSYFFIVHSG 123
LS + SL+ E +A+A++ F G + QGE G FFI+ G
Sbjct 250 LSKVSILESLDKWERLTVADALEPVQFEDGQKIVVQGEPGDEFFIILEG 298
> cel:R07E4.6 kin-2; protein KINase family member (kin-2); K04739
cAMP-dependent protein kinase regulator
Length=376
Score = 33.9 bits (76), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 23/98 (23%), Positives = 49/98 (50%), Gaps = 9/98 (9%)
Query 28 PPKPGGERKAQKAI--MQQDDTQAEDARLLNHLEKREKTDSDLSLIRSSLSGNLVCSSLN 85
PPK G R+ + +++DDT+ + + K D+ + S++ NL+ + L
Sbjct 85 PPKRSGGRRTGISAEPIKEDDTEYKKVVI-------PKDDATRRSLESAMRKNLLFAHLE 137
Query 86 DSEVEALANAVQFFTFAKGDIVTKQGENGSYFFIVHSG 123
+ E + + +A+ + G+ + +QGE G F+++ G
Sbjct 138 EDEQKTMYDAMFPVEKSAGETIIEQGEEGDNFYVIDKG 175
Score = 28.5 bits (62), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 17/50 (34%), Positives = 27/50 (54%), Gaps = 0/50 (0%)
Query 75 LSGNLVCSSLNDSEVEALANAVQFFTFAKGDIVTKQGENGSYFFIVHSGE 124
LS + + L+ E +A+A++ F G V +QG+ G FFI+ GE
Sbjct 245 LSKVQILADLDQWERANVADALERCDFEPGTHVVEQGQPGDEFFIILEGE 294
> xla:444025 prkg2, MGC82580; protein kinase, cGMP-dependent,
type II (EC:2.7.11.1); K07376 protein kinase, cGMP-dependent
[EC:2.7.11.12]
Length=783
Score = 33.9 bits (76), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 13/41 (31%), Positives = 25/41 (60%), Gaps = 0/41 (0%)
Query 84 LNDSEVEALANAVQFFTFAKGDIVTKQGENGSYFFIVHSGE 124
L D ++ +A+ ++ + GD + ++GE GS FFI+ G+
Sbjct 312 LPDHKLMKIADCLELEYYETGDYIIREGEEGSTFFIIAKGK 352
> dre:494533 prkar1aa, im:7047729, prkar1a, zgc:92515; protein
kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific
extinguisher 1) a
Length=379
Score = 32.7 bits (73), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 14/58 (24%), Positives = 34/58 (58%), Gaps = 0/58 (0%)
Query 68 LSLIRSSLSGNLVCSSLNDSEVEALANAVQFFTFAKGDIVTKQGENGSYFFIVHSGEF 125
++ + ++ N++ + L+D+E + +A+ T+ G+ V +QG+ G F+++ GE
Sbjct 123 MAALAKAIEKNVLFAHLDDNERSDIFDAMFSVTYIAGETVIQQGDEGDNFYVIDQGEM 180
Score = 28.1 bits (61), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 16/49 (32%), Positives = 25/49 (51%), Gaps = 0/49 (0%)
Query 75 LSGNLVCSSLNDSEVEALANAVQFFTFAKGDIVTKQGENGSYFFIVHSG 123
LS + SL+ E +A+A++ F G + QG+ G FFI+ G
Sbjct 248 LSKVSILESLDKWERLTVADALETVQFEDGQKIVVQGQPGDEFFIILEG 296
> dre:100329979 protein kinase, cAMP-dependent, regulatory, type
I, alpha (tissue specific extinguisher 1) a-like; K04739
cAMP-dependent protein kinase regulator
Length=379
Score = 32.7 bits (73), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 14/58 (24%), Positives = 34/58 (58%), Gaps = 0/58 (0%)
Query 68 LSLIRSSLSGNLVCSSLNDSEVEALANAVQFFTFAKGDIVTKQGENGSYFFIVHSGEF 125
++ + ++ N++ + L+D+E + +A+ T+ G+ V +QG+ G F+++ GE
Sbjct 123 MAALAKAIEKNVLFAHLDDNERSDIFDAMFSVTYIAGETVIQQGDEGDNFYVIDQGEM 180
Score = 28.1 bits (61), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 16/49 (32%), Positives = 25/49 (51%), Gaps = 0/49 (0%)
Query 75 LSGNLVCSSLNDSEVEALANAVQFFTFAKGDIVTKQGENGSYFFIVHSG 123
LS + SL+ E +A+A++ F G + QG+ G FFI+ G
Sbjct 248 LSKVSILESLDKWERLTVADALETVQFEDGQKIVVQGQPGDEFFIILEG 296
> dre:767685 prkar1b, MGC153624, zgc:153624; protein kinase, cAMP-dependent,
regulatory, type I, beta (EC:2.7.11.1); K04739
cAMP-dependent protein kinase regulator
Length=380
Score = 32.3 bits (72), Expect = 0.40, Method: Compositional matrix adjust.
Identities = 15/57 (26%), Positives = 34/57 (59%), Gaps = 0/57 (0%)
Query 68 LSLIRSSLSGNLVCSSLNDSEVEALANAVQFFTFAKGDIVTKQGENGSYFFIVHSGE 124
++ + ++S N++ + L+D+E + +A+ T G+ V +QG+ G F+++ GE
Sbjct 124 MTALAKAISKNVLFAHLDDNERSDIFDAMFPVTHIAGETVIQQGDEGDNFYVIDQGE 180
Score = 29.6 bits (65), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 26/49 (53%), Gaps = 0/49 (0%)
Query 75 LSGNLVCSSLNDSEVEALANAVQFFTFAKGDIVTKQGENGSYFFIVHSG 123
LS + SL+ E +A+A++ F G+ + QGE G FFI+ G
Sbjct 249 LSKVSILESLDKWERLTVADALEPVQFEDGEKIVVQGEPGDDFFIITEG 297
> hsa:5593 PRKG2, PRKGR2, cGKII; protein kinase, cGMP-dependent,
type II (EC:2.7.11.12); K07376 protein kinase, cGMP-dependent
[EC:2.7.11.12]
Length=762
Score = 32.3 bits (72), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 14/53 (26%), Positives = 30/53 (56%), Gaps = 0/53 (0%)
Query 72 RSSLSGNLVCSSLNDSEVEALANAVQFFTFAKGDIVTKQGENGSYFFIVHSGE 124
R+ L + +L + ++ + + ++ + KGD + ++GE GS FFI+ G+
Sbjct 278 RNFLRSVSLLKNLPEDKLTKIIDCLEVEYYDKGDYIIREGEEGSTFFILAKGK 330
> mmu:434632 MGC103372; cDNA sequence BC085271; K11290 template-activating
factor I
Length=289
Score = 31.6 bits (70), Expect = 0.59, Method: Compositional matrix adjust.
Identities = 17/46 (36%), Positives = 27/46 (58%), Gaps = 0/46 (0%)
Query 9 QQQQDRKTSQPQQNDDAAAPPKPGGERKAQKAIMQQDDTQAEDARL 54
Q ++ R + P+ D +A+P P GE++ Q+AI D+ Q E RL
Sbjct 12 QPKKPRPAAAPKLEDKSASPGLPKGEKEQQEAIEHIDEVQNEIDRL 57
> mmu:671392 Gm9531; predicted gene 9531; K11290 template-activating
factor I
Length=289
Score = 31.6 bits (70), Expect = 0.63, Method: Compositional matrix adjust.
Identities = 17/46 (36%), Positives = 27/46 (58%), Gaps = 0/46 (0%)
Query 9 QQQQDRKTSQPQQNDDAAAPPKPGGERKAQKAIMQQDDTQAEDARL 54
Q ++ R + P+ D +A+P P GE++ Q+AI D+ Q E RL
Sbjct 12 QPKKPRPAAAPKLEDKSASPGLPKGEKEQQEAIEHIDEVQNEIDRL 57
> dre:326020 fd58b04, wu:fd58b04; si:dkey-121j17.5
Length=663
Score = 31.2 bits (69), Expect = 0.77, Method: Compositional matrix adjust.
Identities = 16/64 (25%), Positives = 32/64 (50%), Gaps = 0/64 (0%)
Query 62 EKTDSDLSLIRSSLSGNLVCSSLNDSEVEALANAVQFFTFAKGDIVTKQGENGSYFFIVH 121
+KT S+ S I ++ N L++ ++ + + ++ GD + K+G G +IV
Sbjct 89 QKTSSETSQIVKAIGKNDFLRRLDEEQISMMVDLMKVLDCRAGDEIIKEGTEGDSMYIVA 148
Query 122 SGEF 125
+GE
Sbjct 149 AGEL 152
Score = 30.0 bits (66), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 10/42 (23%), Positives = 26/42 (61%), Gaps = 0/42 (0%)
Query 83 SLNDSEVEALANAVQFFTFAKGDIVTKQGENGSYFFIVHSGE 124
+LND ++ + ++++ F +++ ++G G+ F+I+ GE
Sbjct 228 ALNDVQLSKIIDSMEEVRFQDNEVIVREGAEGNTFYIILKGE 269
> mmu:100504508 protein SET-like
Length=289
Score = 31.2 bits (69), Expect = 0.88, Method: Compositional matrix adjust.
Identities = 17/46 (36%), Positives = 26/46 (56%), Gaps = 0/46 (0%)
Query 9 QQQQDRKTSQPQQNDDAAAPPKPGGERKAQKAIMQQDDTQAEDARL 54
Q ++ R + P+ D A+P P GE++ Q+AI D+ Q E RL
Sbjct 12 QPKKPRPAAAPKLEDKLASPGLPKGEKEQQEAIEHIDEVQNEIDRL 57
> dre:100329437 protein kinase, cAMP-dependent, regulatory, type
II, alpha-like
Length=385
Score = 30.4 bits (67), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 14/42 (33%), Positives = 25/42 (59%), Gaps = 0/42 (0%)
Query 83 SLNDSEVEALANAVQFFTFAKGDIVTKQGENGSYFFIVHSGE 124
SL SE + + + +F+ G+ + +QG++ F+IV SGE
Sbjct 286 SLQLSERMKIVDVLGMRSFSDGERIIQQGDSADCFYIVESGE 327
> hsa:11069 RAPGEF4, CAMP-GEFII, CGEF2, EPAC2, Nbla00496; Rap
guanine nucleotide exchange factor (GEF) 4; K04351 Rap guanine
nucleotide exchange factor (GEF) 4
Length=867
Score = 30.0 bits (66), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 34/125 (27%), Positives = 52/125 (41%), Gaps = 8/125 (6%)
Query 7 QEQQQQDRKTSQPQQNDDAAAPPKPGGERKAQKAIMQQDDT-----QAEDARLLNHLEKR 61
QE QD+ +D+ P P E K + QD DA + L K
Sbjct 132 QEHHFQDKYLFYRFLDDEHEDAPLPTEEEKKECDEELQDTMLLLSQMGPDAHMRMILRKP 191
Query 62 --EKTDSDLSLIRSSLSGNLVCSSLNDSEVEALANAVQFFTFAKG-DIVTKQGENGSYFF 118
++T DL +I L S L+ + LA + F + AKG ++ QGE G+ ++
Sbjct 192 PGQRTVDDLEIIYEELLHIKALSHLSTTVKRELAGVLIFESHAKGGTVLFNQGEEGTSWY 251
Query 119 IVHSG 123
I+ G
Sbjct 252 IILKG 256
> dre:405894 prkar2ab, MGC85886, prkar2a, zgc:85886; protein kinase,
cAMP-dependent, regulatory, type II, alpha, B; K04739
cAMP-dependent protein kinase regulator
Length=422
Score = 30.0 bits (66), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 14/42 (33%), Positives = 25/42 (59%), Gaps = 0/42 (0%)
Query 83 SLNDSEVEALANAVQFFTFAKGDIVTKQGENGSYFFIVHSGE 124
SL SE + + + +F+ G+ + +QG++ F+IV SGE
Sbjct 286 SLQLSERMKIVDVLGMRSFSDGERIIQQGDSADCFYIVESGE 327
> mmu:56508 Rapgef4, 1300003D15Rik, 5730402K07Rik, 6330581N18Rik,
EPAC_2, Epac-2, Epac2, KIAA4040, mKIAA4040; Rap guanine
nucleotide exchange factor (GEF) 4; K04351 Rap guanine nucleotide
exchange factor (GEF) 4
Length=1011
Score = 30.0 bits (66), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 34/125 (27%), Positives = 53/125 (42%), Gaps = 8/125 (6%)
Query 7 QEQQQQDRKTSQPQQNDDAAAPPKPGGERKAQKAIMQQDDT-----QAEDARLLNHLEKR 61
QE+ QD+ +D+ P P E K + QD DA + L K
Sbjct 276 QERHFQDKYLFYRFLDDEREDAPLPTEEEKKECDEELQDTMLLLSQMGPDAHMRMILRKP 335
Query 62 --EKTDSDLSLIRSSLSGNLVCSSLNDSEVEALANAVQFFTFAKG-DIVTKQGENGSYFF 118
++T DL +I L S L+ + LA + F + AKG ++ QGE G+ ++
Sbjct 336 PGQRTVDDLEIIYDELLHIKALSHLSTTVKRELAGVLIFESHAKGGTVLFNQGEEGTSWY 395
Query 119 IVHSG 123
I+ G
Sbjct 396 IILKG 400
> tgo:TGME49_019070 cAMP-dependent protein kinase regulatory subunit,
putative (EC:3.6.3.14 2.7.10.2 2.7.11.12)
Length=2634
Score = 29.3 bits (64), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 15/64 (23%), Positives = 33/64 (51%), Gaps = 0/64 (0%)
Query 61 REKTDSDLSLIRSSLSGNLVCSSLNDSEVEALANAVQFFTFAKGDIVTKQGENGSYFFIV 120
RE+ +D I + L V + L++ ++ L A + + + +++ +G +FFI+
Sbjct 1633 REEPTADYGEIMNLLVQVPVINMLDEDALQQLTRAFKVECYQEREVIIHEGTPAEHFFII 1692
Query 121 HSGE 124
SG+
Sbjct 1693 FSGD 1696
> cel:F55A8.2 egl-4; EGg Laying defective family member (egl-4);
K07376 protein kinase, cGMP-dependent [EC:2.7.11.12]
Length=780
Score = 28.9 bits (63), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 11/45 (24%), Positives = 26/45 (57%), Gaps = 0/45 (0%)
Query 80 VCSSLNDSEVEALANAVQFFTFAKGDIVTKQGENGSYFFIVHSGE 124
+ +L++ + +A+ + + G + +QGE G FF+++SG+
Sbjct 319 IFQNLSEDRISKMADVMDQDYYDGGHYIIRQGEKGDAFFVINSGQ 363
Score = 27.7 bits (60), Expect = 9.8, Method: Compositional matrix adjust.
Identities = 26/99 (26%), Positives = 38/99 (38%), Gaps = 11/99 (11%)
Query 38 QKAIMQQDDTQ--------AEDARLLNH---LEKREKTDSDLSLIRSSLSGNLVCSSLND 86
QKA++ D Q AE N L+ KT +IR ++ N L
Sbjct 148 QKAVLPADGVQRAKKIAVSAEPTNFENKPATLQHYNKTVGAKQMIRDAVQKNDFLKQLAK 207
Query 87 SEVEALANAVQFFTFAKGDIVTKQGENGSYFFIVHSGEF 125
++ L N + G V ++GE G F+V GE
Sbjct 208 EQIIELVNCMYEMRARAGQWVIQEGEPGDRLFVVAEGEL 246
> mmu:26921 Map4k4, 9430080K19Rik, AU043147, AU045934, AW046177,
HGK, Nik; mitogen-activated protein kinase kinase kinase
kinase 4 (EC:2.7.11.1); K04407 mitogen-activated protein kinase
kinase kinase kinase 4 [EC:2.7.11.1]
Length=1234
Score = 27.7 bits (60), Expect = 9.0, Method: Composition-based stats.
Identities = 13/40 (32%), Positives = 22/40 (55%), Gaps = 0/40 (0%)
Query 13 DRKTSQPQQNDDAAAPPKPGGERKAQKAIMQQDDTQAEDA 52
D TS P+ A++P GE ++ K ++ DD ++E A
Sbjct 782 DDSTSGPEDTRAASSPNLSNGETESVKTMIVHDDVESEPA 821
Lambda K H
0.308 0.124 0.333
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2064871684
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40