bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0930_orf1
Length=185
Score E
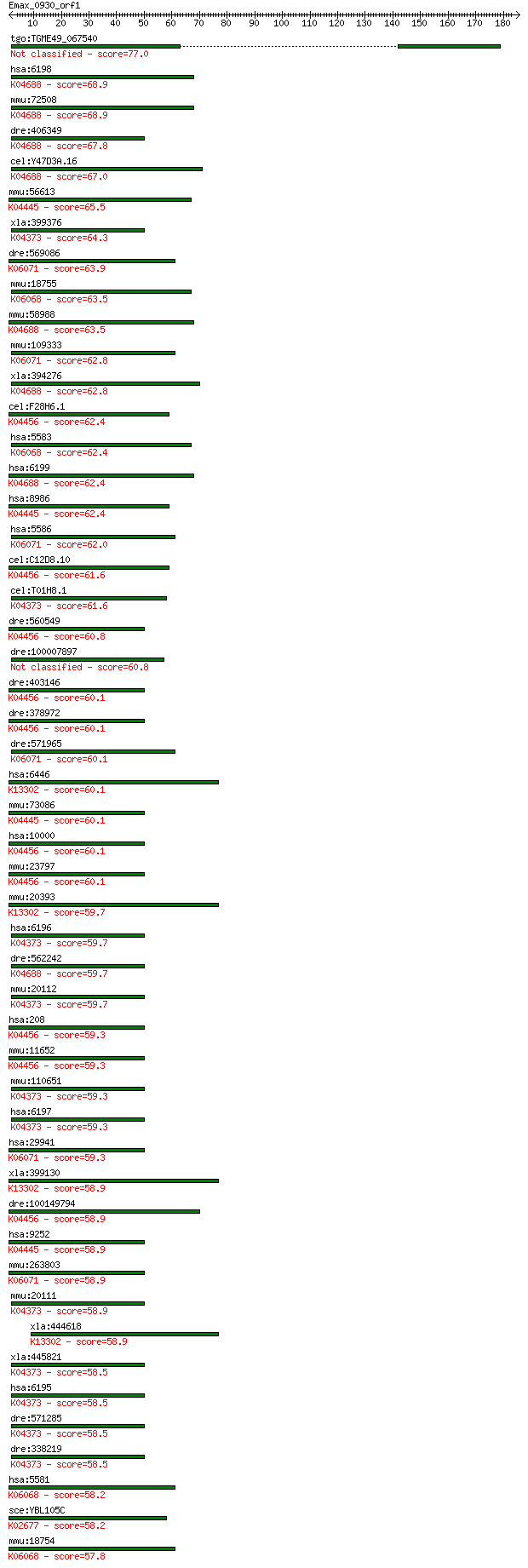
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_067540 protein kinase, putative (EC:4.1.1.70 2.7.11... 77.0 4e-14
hsa:6198 RPS6KB1, PS6K, S6K, S6K1, STK14A, p70(S6K)-alpha, p70... 68.9 1e-11
mmu:72508 Rps6kb1, 2610318I15Rik, 4732464A07Rik, 70kDa, AA9597... 68.9 1e-11
dre:406349 rps6kb1b, fc51h01, rps6kb1, wu:fc51h01, zgc:55713; ... 67.8 2e-11
cel:Y47D3A.16 rsks-1; RSK-pSeventy (RSK-p70 kinase) homolog fa... 67.0 4e-11
mmu:56613 Rps6ka4, 1110069D02Rik, 90kDa, AI848992, Msk2, mMSK2... 65.5 1e-10
xla:399376 rps6ka3, MGC82193, S6KII, p90, rsk, rsk2; ribosomal... 64.3 2e-10
dre:569086 pkn2; protein kinase N2; K06071 protein kinase N [E... 63.9 3e-10
mmu:18755 Prkch, Pkch; protein kinase C, eta (EC:2.7.11.13); K... 63.5 3e-10
mmu:58988 Rps6kb2, 70kDa, S6K2; ribosomal protein S6 kinase, p... 63.5 4e-10
mmu:109333 Pkn2, 6030436C20Rik, AI507382, PRK2, Prkcl2, Stk7; ... 62.8 6e-10
xla:394276 rps6kb1, p70-alpha, p70-s6k, p70s6k, ps6k, rps6kb1-... 62.8 6e-10
cel:F28H6.1 akt-2; AKT kinase family member (akt-2); K04456 RA... 62.4 8e-10
hsa:5583 PRKCH, MGC26269, MGC5363, PKC-L, PKCL, PRKCL, nPKC-et... 62.4 8e-10
hsa:6199 RPS6KB2, KLS, P70-beta, P70-beta-1, P70-beta-2, S6K-b... 62.4 9e-10
hsa:8986 RPS6KA4, MSK2, RSK-B; ribosomal protein S6 kinase, 90... 62.4 9e-10
hsa:5586 PKN2, MGC150606, MGC71074, PAK2, PRK2, PRKCL2, PRO204... 62.0 1e-09
cel:C12D8.10 akt-1; AKT kinase family member (akt-1); K04456 R... 61.6 1e-09
cel:T01H8.1 rskn-1; RSK-pNinety (RSK-p90 kinase) homolog famil... 61.6 1e-09
dre:560549 akt3a, akt3, si:ch211-221n23.2; v-akt murine thymom... 60.8 3e-09
dre:100007897 MGC162290; zgc:162290 60.8 3e-09
dre:403146 akt2l; v-akt murine thymoma viral oncogene homolog ... 60.1 4e-09
dre:378972 akt2, cb945; v-akt murine thymoma viral oncogene ho... 60.1 4e-09
dre:571965 protein kinase N2-like; K06071 protein kinase N [EC... 60.1 4e-09
hsa:6446 SGK1, SGK; serum/glucocorticoid regulated kinase 1 (E... 60.1 4e-09
mmu:73086 Rps6ka5, 3110005L17Rik, 6330404E13Rik, AI854034, MGC... 60.1 4e-09
hsa:10000 AKT3, DKFZp434N0250, PKB-GAMMA, PKBG, PRKBG, RAC-PK-... 60.1 5e-09
mmu:23797 Akt3, AI851531, D930002M15Rik, Nmf350; thymoma viral... 60.1 5e-09
mmu:20393 Sgk1, Sgk; serum/glucocorticoid regulated kinase 1 (... 59.7 5e-09
hsa:6196 RPS6KA2, HU-2, MAPKAPK1C, RSK, RSK3, S6K-alpha, S6K-a... 59.7 6e-09
dre:562242 ribosomal protein S6 kinase b, polypeptide 1-like; ... 59.7 6e-09
mmu:20112 Rps6ka2, 90kDa, D17Wsu134e, Rps6ka-rs1, Rsk3, p90rsk... 59.7 6e-09
hsa:208 AKT2, PKBB, PKBBETA, PRKBB, RAC-BETA; v-akt murine thy... 59.3 7e-09
mmu:11652 Akt2, 2410016A19Rik, AW554154, MGC14031, PKB, PKBbet... 59.3 7e-09
mmu:110651 Rps6ka3, MPK-9, Rsk2, S6K-alpha3, pp90RSK2; ribosom... 59.3 7e-09
hsa:6197 RPS6KA3, CLS, HU-3, ISPK-1, MAPKAPK1B, MRX19, RSK, RS... 59.3 8e-09
hsa:29941 PKN3, RP11-545E17.1; protein kinase N3 (EC:2.7.11.13... 59.3 8e-09
xla:399130 sgk1-a, Sgk, sgk-A, sgk1; serum/glucocorticoid regu... 58.9 9e-09
dre:100149794 akt3; v-akt murine thymoma viral oncogene homolo... 58.9 9e-09
hsa:9252 RPS6KA5, MGC1911, MSK1, MSPK1, RLPK; ribosomal protei... 58.9 9e-09
mmu:263803 Pkn3, AW209115, BC034126, MGC31699; protein kinase ... 58.9 1e-08
mmu:20111 Rps6ka1, Rsk1, p90rsk, rsk; ribosomal protein S6 kin... 58.9 1e-08
xla:444618 sgk1-b, MGC84110, Sgk, sgk-B, sgk2; serum/glucocort... 58.9 1e-08
xla:445821 rps6ka1, MGC81220, hu-1, mapkapk1a, rsk, rsk1; ribo... 58.5 1e-08
hsa:6195 RPS6KA1, HU-1, MAPKAPK1A, RSK, RSK1; ribosomal protei... 58.5 1e-08
dre:571285 ribosomal protein S6 kinase, polypeptide 2-like; K0... 58.5 1e-08
dre:338219 rps6ka3a, id:ibd1103, rps6ka3, si:ch211-114c12.12, ... 58.5 1e-08
hsa:5581 PRKCE, MGC125656, MGC125657, PKCE, nPKC-epsilon; prot... 58.2 1e-08
sce:YBL105C PKC1, CLY15, HPO2, STT1; Pkc1p (EC:2.7.11.13); K02... 58.2 2e-08
mmu:18754 Prkce, 5830406C15Rik, PKC[e], PKCepsilon, Pkce, R751... 57.8 2e-08
> tgo:TGME49_067540 protein kinase, putative (EC:4.1.1.70 2.7.11.13)
Length=951
Score = 77.0 bits (188), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 33/61 (54%), Positives = 44/61 (72%), Gaps = 0/61 (0%)
Query 2 RLGAGPLDAEEIKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYFPADVKREPVLSD 61
RLG GP DAEEIKRHPFF +DW+ L AK++ PPF+P L+S D +YF + + PV++
Sbjct 699 RLGGGPGDAEEIKRHPFFGRIDWEALQAKRMRPPFRPRLQSPTDVQYFDNEFVKLPVINS 758
Query 62 E 62
E
Sbjct 759 E 759
Score = 30.0 bits (66), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 14/37 (37%), Positives = 23/37 (62%), Gaps = 0/37 (0%)
Query 142 HSSHNSQHTEDEEGEALFKGFTYDERLQGTWGKAATS 178
H + + +DE +++F+GFTYDER G+AA +
Sbjct 912 HPNMLAPPPDDESDDSMFEGFTYDERDTSALGEAAAA 948
> hsa:6198 RPS6KB1, PS6K, S6K, S6K1, STK14A, p70(S6K)-alpha, p70-S6K,
p70-alpha; ribosomal protein S6 kinase, 70kDa, polypeptide
1 (EC:2.7.11.1); K04688 p70 ribosomal S6 kinase [EC:2.7.11.1]
Length=525
Score = 68.9 bits (167), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 35/67 (52%), Positives = 45/67 (67%), Gaps = 6/67 (8%)
Query 2 RLGAGPLDAEEIKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYFPADVKRE-PVLS 60
RLGAGP DA E++ HPFF ++W+ LLA+K+EPPFKP L+S ED F + R+ PV
Sbjct 335 RLGAGPGDAGEVQAHPFFRHINWEELLARKVEPPFKPLLQSEEDVSQFDSKFTRQTPV-- 392
Query 61 DEGDGPD 67
D PD
Sbjct 393 ---DSPD 396
> mmu:72508 Rps6kb1, 2610318I15Rik, 4732464A07Rik, 70kDa, AA959758,
AI256796, AI314060, S6K1, p70/85s6k, p70s6k; ribosomal
protein S6 kinase, polypeptide 1 (EC:2.7.11.1); K04688 p70
ribosomal S6 kinase [EC:2.7.11.1]
Length=525
Score = 68.9 bits (167), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 35/67 (52%), Positives = 45/67 (67%), Gaps = 6/67 (8%)
Query 2 RLGAGPLDAEEIKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYFPADVKRE-PVLS 60
RLGAGP DA E++ HPFF ++W+ LLA+K+EPPFKP L+S ED F + R+ PV
Sbjct 335 RLGAGPGDAGEVQAHPFFRHINWEELLARKVEPPFKPLLQSEEDVSQFDSKFTRQTPV-- 392
Query 61 DEGDGPD 67
D PD
Sbjct 393 ---DSPD 396
> dre:406349 rps6kb1b, fc51h01, rps6kb1, wu:fc51h01, zgc:55713;
ribosomal protein S6 kinase b, polypeptide 1b (EC:2.7.11.1);
K04688 p70 ribosomal S6 kinase [EC:2.7.11.1]
Length=502
Score = 67.8 bits (164), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 30/48 (62%), Positives = 36/48 (75%), Gaps = 0/48 (0%)
Query 2 RLGAGPLDAEEIKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYF 49
RLGAGP DA E++ HPFF V+WD LLA+K+EPPFKP L+ ED F
Sbjct 309 RLGAGPGDATEVQTHPFFRHVNWDDLLARKVEPPFKPFLQFAEDVSQF 356
> cel:Y47D3A.16 rsks-1; RSK-pSeventy (RSK-p70 kinase) homolog
family member (rsks-1); K04688 p70 ribosomal S6 kinase [EC:2.7.11.1]
Length=580
Score = 67.0 bits (162), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 34/77 (44%), Positives = 45/77 (58%), Gaps = 8/77 (10%)
Query 2 RLGAGPLDAEEIKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYF--------PADV 53
RLGAG DAEEIK H FF++ DW+L+ A+++E PFKP++ + ED+ F P D
Sbjct 327 RLGAGLSDAEEIKSHAFFKTTDWNLVYARQLEAPFKPNIENDEDTSLFDARFTKMTPVDS 386
Query 54 KREPVLSDEGDGPDAAF 70
E S GD P F
Sbjct 387 PCETNFSLNGDNPFVGF 403
> mmu:56613 Rps6ka4, 1110069D02Rik, 90kDa, AI848992, Msk2, mMSK2;
ribosomal protein S6 kinase, polypeptide 4 (EC:2.7.11.1);
K04445 mitogen-,stress activated protein kinases [EC:2.7.11.1]
Length=773
Score = 65.5 bits (158), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 33/67 (49%), Positives = 42/67 (62%), Gaps = 1/67 (1%)
Query 1 ERLGAGPLDAEEIKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYFPADVKR-EPVL 59
+RLGAGP A+E+K HPFF+ +DW L A+KI PF+P +RS D F + R EPV
Sbjct 283 KRLGAGPQGAQEVKSHPFFQGLDWWALAARKIPAPFRPQIRSELDVGNFAEEFTRLEPVY 342
Query 60 SDEGDGP 66
S G P
Sbjct 343 SPAGSPP 349
> xla:399376 rps6ka3, MGC82193, S6KII, p90, rsk, rsk2; ribosomal
protein S6 kinase, 90kDa, polypeptide 3 (EC:2.7.11.1); K04373
p90 ribosomal S6 kinase [EC:2.7.11.1]
Length=737
Score = 64.3 bits (155), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 28/48 (58%), Positives = 35/48 (72%), Gaps = 0/48 (0%)
Query 2 RLGAGPLDAEEIKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYF 49
RLGAGP EEIKRHPFF ++DW+ L ++I+PPFKP ED+ YF
Sbjct 307 RLGAGPDGVEEIKRHPFFVTIDWNKLFRREIQPPFKPATGGPEDTFYF 354
> dre:569086 pkn2; protein kinase N2; K06071 protein kinase N
[EC:2.7.11.13]
Length=940
Score = 63.9 bits (154), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 28/61 (45%), Positives = 43/61 (70%), Gaps = 1/61 (1%)
Query 1 ERLGAGPLDAEEIKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYFPADVKR-EPVL 59
+RLGAG DA E+K+H FF+ +DW+ LLAK+++PPF P +++ D F + R +PVL
Sbjct 855 KRLGAGEQDANEVKKHRFFQGIDWEALLAKRVKPPFLPSIKTAADVSNFDEEFTRLKPVL 914
Query 60 S 60
+
Sbjct 915 T 915
> mmu:18755 Prkch, Pkch; protein kinase C, eta (EC:2.7.11.13);
K06068 novel protein kinase C [EC:2.7.11.13]
Length=683
Score = 63.5 bits (153), Expect = 3e-10, Method: Composition-based stats.
Identities = 33/69 (47%), Positives = 44/69 (63%), Gaps = 4/69 (5%)
Query 2 RLGAGPLDAE-EIKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYFPAD-VKREPVL 59
RLG+ E EI RHPFF+ +DW L +++EPPF+P ++S ED F D +K EPVL
Sbjct 596 RLGSLTQGGEHEILRHPFFKEIDWAQLNHRQLEPPFRPRIKSREDVSNFDPDFIKEEPVL 655
Query 60 S--DEGDGP 66
+ DEG P
Sbjct 656 TPIDEGHLP 664
> mmu:58988 Rps6kb2, 70kDa, S6K2; ribosomal protein S6 kinase,
polypeptide 2 (EC:2.7.11.1); K04688 p70 ribosomal S6 kinase
[EC:2.7.11.1]
Length=485
Score = 63.5 bits (153), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 31/68 (45%), Positives = 45/68 (66%), Gaps = 6/68 (8%)
Query 1 ERLGAGPLDAEEIKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYFPADVKRE-PVL 59
+R+G G DA +++RHPFF ++WD LLA++++PPF+P L+S ED F A R+ PV
Sbjct 310 QRIGGGLGDAADVQRHPFFRHINWDDLLARRVDPPFRPSLQSEEDVSQFDARFTRQTPV- 368
Query 60 SDEGDGPD 67
D PD
Sbjct 369 ----DSPD 372
> mmu:109333 Pkn2, 6030436C20Rik, AI507382, PRK2, Prkcl2, Stk7;
protein kinase N2 (EC:2.7.11.13); K06071 protein kinase N
[EC:2.7.11.13]
Length=983
Score = 62.8 bits (151), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 29/60 (48%), Positives = 39/60 (65%), Gaps = 1/60 (1%)
Query 2 RLGAGPLDAEEIKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYFPADVKRE-PVLS 60
RLGAG DAE++K+HPFF DW L+ KK++PPF P +R ED F + E P+L+
Sbjct 898 RLGAGEKDAEDVKKHPFFRLTDWSALMDKKVKPPFVPTIRGREDVSNFDDEFTSEAPILT 957
> xla:394276 rps6kb1, p70-alpha, p70-s6k, p70s6k, ps6k, rps6kb1-A,
s6K1, s6k, stk14a; ribosomal protein S6 kinase, 70kDa,
polypeptide 1 (EC:2.7.11.1); K04688 p70 ribosomal S6 kinase
[EC:2.7.11.1]
Length=501
Score = 62.8 bits (151), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 34/69 (49%), Positives = 44/69 (63%), Gaps = 6/69 (8%)
Query 2 RLGAGPLDAEEIKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYFPADVKRE-PVLS 60
RLGAG DA +++ H FF ++WD LLA+K+EPPFKP L+S ED F + R+ PV
Sbjct 311 RLGAGVGDAGDVQGHSFFRHINWDDLLARKVEPPFKPLLQSEEDVSQFDSKFTRQTPV-- 368
Query 61 DEGDGPDAA 69
D PD A
Sbjct 369 ---DSPDDA 374
> cel:F28H6.1 akt-2; AKT kinase family member (akt-2); K04456
RAC serine/threonine-protein kinase [EC:2.7.11.1]
Length=528
Score = 62.4 bits (150), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 28/58 (48%), Positives = 39/58 (67%), Gaps = 0/58 (0%)
Query 1 ERLGAGPLDAEEIKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYFPADVKREPV 58
+RLGAGP DA E+ R FF+ VDW+ L K++EPPFKP++ S D+ +F + PV
Sbjct 419 KRLGAGPDDAREVSRAEFFKDVDWEATLRKEVEPPFKPNVMSETDTSFFDREFTSMPV 476
> hsa:5583 PRKCH, MGC26269, MGC5363, PKC-L, PKCL, PRKCL, nPKC-eta;
protein kinase C, eta (EC:2.7.11.13); K06068 novel protein
kinase C [EC:2.7.11.13]
Length=683
Score = 62.4 bits (150), Expect = 8e-10, Method: Composition-based stats.
Identities = 33/69 (47%), Positives = 43/69 (62%), Gaps = 4/69 (5%)
Query 2 RLGAGPLDAEE-IKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYFPAD-VKREPVL 59
RLG+ E I RHPFF+ +DW L ++IEPPF+P ++S ED F D +K EPVL
Sbjct 596 RLGSLTQGGEHAILRHPFFKEIDWAQLNHRQIEPPFRPRIKSREDVSNFDPDFIKEEPVL 655
Query 60 S--DEGDGP 66
+ DEG P
Sbjct 656 TPIDEGHLP 664
> hsa:6199 RPS6KB2, KLS, P70-beta, P70-beta-1, P70-beta-2, S6K-beta2,
S6K2, SRK, STK14B, p70(S6K)-beta, p70S6Kb; ribosomal
protein S6 kinase, 70kDa, polypeptide 2 (EC:2.7.11.1); K04688
p70 ribosomal S6 kinase [EC:2.7.11.1]
Length=482
Score = 62.4 bits (150), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 31/68 (45%), Positives = 44/68 (64%), Gaps = 6/68 (8%)
Query 1 ERLGAGPLDAEEIKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYFPADVKRE-PVL 59
+R+G GP DA +++RHPFF ++WD LLA +++PPF+P L+S ED F R+ PV
Sbjct 310 QRIGGGPGDAADVQRHPFFRHMNWDDLLAWRVDPPFRPCLQSEEDVSQFDTRFTRQTPV- 368
Query 60 SDEGDGPD 67
D PD
Sbjct 369 ----DSPD 372
> hsa:8986 RPS6KA4, MSK2, RSK-B; ribosomal protein S6 kinase,
90kDa, polypeptide 4 (EC:2.7.11.1); K04445 mitogen-,stress activated
protein kinases [EC:2.7.11.1]
Length=766
Score = 62.4 bits (150), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 29/59 (49%), Positives = 39/59 (66%), Gaps = 1/59 (1%)
Query 1 ERLGAGPLDAEEIKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYFPADVKR-EPV 58
+RLGAGP A+E++ HPFF+ +DW L A+KI PF+P +RS D F + R EPV
Sbjct 283 KRLGAGPQGAQEVRNHPFFQGLDWVALAARKIPAPFRPQIRSELDVGNFAEEFTRLEPV 341
> hsa:5586 PKN2, MGC150606, MGC71074, PAK2, PRK2, PRKCL2, PRO2042,
Pak-2; protein kinase N2 (EC:2.7.11.13); K06071 protein
kinase N [EC:2.7.11.13]
Length=984
Score = 62.0 bits (149), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 28/60 (46%), Positives = 39/60 (65%), Gaps = 1/60 (1%)
Query 2 RLGAGPLDAEEIKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYFPADVKRE-PVLS 60
RLGA DAE++K+HPFF +DW L+ KK++PPF P +R ED F + E P+L+
Sbjct 899 RLGASEKDAEDVKKHPFFRLIDWSALMDKKVKPPFIPTIRGREDVSNFDDEFTSEAPILT 958
> cel:C12D8.10 akt-1; AKT kinase family member (akt-1); K04456
RAC serine/threonine-protein kinase [EC:2.7.11.1]
Length=541
Score = 61.6 bits (148), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 28/58 (48%), Positives = 39/58 (67%), Gaps = 0/58 (0%)
Query 1 ERLGAGPLDAEEIKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYFPADVKREPV 58
+RLG GP DA EI R FF +VDW+ K+IEPP+KP+++S D+ YF + +PV
Sbjct 432 QRLGGGPEDALEICRADFFRTVDWEATYRKEIEPPYKPNVQSETDTSYFDNEFTSQPV 489
> cel:T01H8.1 rskn-1; RSK-pNinety (RSK-p90 kinase) homolog family
member (rskn-1); K04373 p90 ribosomal S6 kinase [EC:2.7.11.1]
Length=804
Score = 61.6 bits (148), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 31/57 (54%), Positives = 38/57 (66%), Gaps = 1/57 (1%)
Query 2 RLGAGPLDAEEIKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYF-PADVKREP 57
RLGAGP EEIKRH FF +D+ LL K+I+PPFKP L + + + YF P KR P
Sbjct 340 RLGAGPDGVEEIKRHAFFAKIDFVKLLNKEIDPPFKPALSTVDSTSYFDPEFTKRTP 396
> dre:560549 akt3a, akt3, si:ch211-221n23.2; v-akt murine thymoma
viral oncogene homolog 3a (EC:2.7.11.1); K04456 RAC serine/threonine-protein
kinase [EC:2.7.11.1]
Length=479
Score = 60.8 bits (146), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 26/49 (53%), Positives = 34/49 (69%), Gaps = 0/49 (0%)
Query 1 ERLGAGPLDAEEIKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYF 49
+RLG GP DA+EI RH FF +DW + KK+ PPFKP + S D++YF
Sbjct 387 KRLGGGPDDAKEIMRHSFFTGIDWQDVYDKKLIPPFKPQVSSETDTRYF 435
> dre:100007897 MGC162290; zgc:162290
Length=909
Score = 60.8 bits (146), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 27/55 (49%), Positives = 37/55 (67%), Gaps = 0/55 (0%)
Query 2 RLGAGPLDAEEIKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYFPADVKRE 56
RLG+G DAE+IK+ PFF ++DWD LL +K+ PPF P + + ED F A+ E
Sbjct 824 RLGSGEKDAEDIKKQPFFRNMDWDALLQRKVPPPFVPAVANSEDVSNFDAEFTNE 878
> dre:403146 akt2l; v-akt murine thymoma viral oncogene homolog
2, like (EC:2.7.11.1); K04456 RAC serine/threonine-protein
kinase [EC:2.7.11.1]
Length=478
Score = 60.1 bits (144), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 25/49 (51%), Positives = 33/49 (67%), Gaps = 0/49 (0%)
Query 1 ERLGAGPLDAEEIKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYF 49
+RLG GP DA ++ H FF V WD +L KK+ PPFKP + S D++YF
Sbjct 388 QRLGGGPDDARDVMMHKFFSGVQWDDVLQKKLLPPFKPQVTSETDTRYF 436
> dre:378972 akt2, cb945; v-akt murine thymoma viral oncogene
homolog 2 (EC:2.7.11.1); K04456 RAC serine/threonine-protein
kinase [EC:2.7.11.1]
Length=479
Score = 60.1 bits (144), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 24/49 (48%), Positives = 35/49 (71%), Gaps = 0/49 (0%)
Query 1 ERLGAGPLDAEEIKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYF 49
+RLG GP DA+E+ H FF +++W +L KK+ PPFKP + S D++YF
Sbjct 389 QRLGGGPEDAKEVMTHKFFNNMNWQDVLQKKLVPPFKPQVTSETDTRYF 437
> dre:571965 protein kinase N2-like; K06071 protein kinase N [EC:2.7.11.13]
Length=970
Score = 60.1 bits (144), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 29/60 (48%), Positives = 39/60 (65%), Gaps = 1/60 (1%)
Query 2 RLGAGPLDAEEIKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYFPADVKRE-PVLS 60
RLGA DAE++K+H FF +DWD LLAKK++PPF P ++ D F + E PVL+
Sbjct 885 RLGAAERDAEDVKKHLFFRDIDWDGLLAKKVKPPFVPTIQCSSDVSNFDDEFTSEAPVLT 944
> hsa:6446 SGK1, SGK; serum/glucocorticoid regulated kinase 1
(EC:2.7.11.1); K13302 serum/glucocorticoid-regulated kinase
1 [EC:2.7.11.1]
Length=526
Score = 60.1 bits (144), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 31/76 (40%), Positives = 44/76 (57%), Gaps = 1/76 (1%)
Query 1 ERLGAGPLDAEEIKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYFPADVKREPVLS 60
+RLGA D EIK H FF ++WD L+ KKI PPF P++ D ++F + EPV +
Sbjct 433 KRLGAKD-DFMEIKSHVFFSLINWDDLINKKITPPFNPNVSGPNDLRHFDPEFTEEPVPN 491
Query 61 DEGDGPDAAFGTATRR 76
G PD+ TA+ +
Sbjct 492 SIGKSPDSVLVTASVK 507
> mmu:73086 Rps6ka5, 3110005L17Rik, 6330404E13Rik, AI854034, MGC28385,
MSK1, MSPK1, RLPK; ribosomal protein S6 kinase, polypeptide
5 (EC:2.7.11.1); K04445 mitogen-,stress activated protein
kinases [EC:2.7.11.1]
Length=863
Score = 60.1 bits (144), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 28/49 (57%), Positives = 32/49 (65%), Gaps = 0/49 (0%)
Query 1 ERLGAGPLDAEEIKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYF 49
+RLG GP DAEEIK H FFE + WD L AKK+ PFKP +R D F
Sbjct 299 KRLGCGPRDAEEIKEHLFFEKIKWDDLAAKKVPAPFKPVIRDELDVSNF 347
> hsa:10000 AKT3, DKFZp434N0250, PKB-GAMMA, PKBG, PRKBG, RAC-PK-gamma,
RAC-gamma, STK-2; v-akt murine thymoma viral oncogene
homolog 3 (protein kinase B, gamma) (EC:2.7.11.1); K04456
RAC serine/threonine-protein kinase [EC:2.7.11.1]
Length=479
Score = 60.1 bits (144), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 26/49 (53%), Positives = 34/49 (69%), Gaps = 0/49 (0%)
Query 1 ERLGAGPLDAEEIKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYF 49
+RLG GP DA+EI RH FF V+W + KK+ PPFKP + S D++YF
Sbjct 387 KRLGGGPDDAKEIMRHSFFSGVNWQDVYDKKLVPPFKPQVTSETDTRYF 435
> mmu:23797 Akt3, AI851531, D930002M15Rik, Nmf350; thymoma viral
proto-oncogene 3 (EC:2.7.11.1); K04456 RAC serine/threonine-protein
kinase [EC:2.7.11.1]
Length=479
Score = 60.1 bits (144), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 26/49 (53%), Positives = 34/49 (69%), Gaps = 0/49 (0%)
Query 1 ERLGAGPLDAEEIKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYF 49
+RLG GP DA+EI RH FF V+W + KK+ PPFKP + S D++YF
Sbjct 387 KRLGGGPDDAKEIMRHSFFSGVNWQDVYDKKLVPPFKPQVTSETDTRYF 435
> mmu:20393 Sgk1, Sgk; serum/glucocorticoid regulated kinase 1
(EC:2.7.11.1); K13302 serum/glucocorticoid-regulated kinase
1 [EC:2.7.11.1]
Length=524
Score = 59.7 bits (143), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 32/76 (42%), Positives = 44/76 (57%), Gaps = 1/76 (1%)
Query 1 ERLGAGPLDAEEIKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYFPADVKREPVLS 60
+RLGA D EIK H FF ++WD L+ KKI PPF P++ D ++F + EPV S
Sbjct 431 KRLGAKD-DFMEIKSHIFFSLINWDDLINKKITPPFNPNVSGPSDLRHFDPEFTEEPVPS 489
Query 61 DEGDGPDAAFGTATRR 76
G PD+ TA+ +
Sbjct 490 SIGRSPDSILVTASVK 505
> hsa:6196 RPS6KA2, HU-2, MAPKAPK1C, RSK, RSK3, S6K-alpha, S6K-alpha2,
p90-RSK3, pp90RSK3; ribosomal protein S6 kinase, 90kDa,
polypeptide 2 (EC:2.7.11.1); K04373 p90 ribosomal S6 kinase
[EC:2.7.11.1]
Length=741
Score = 59.7 bits (143), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 27/48 (56%), Positives = 35/48 (72%), Gaps = 0/48 (0%)
Query 2 RLGAGPLDAEEIKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYF 49
RLGAG EEIKRHPFF ++DW+ L K+I+PPFKP + ED+ +F
Sbjct 309 RLGAGIDGVEEIKRHPFFVTIDWNTLYRKEIKPPFKPAVGRPEDTFHF 356
> dre:562242 ribosomal protein S6 kinase b, polypeptide 1-like;
K04688 p70 ribosomal S6 kinase [EC:2.7.11.1]
Length=507
Score = 59.7 bits (143), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 26/48 (54%), Positives = 34/48 (70%), Gaps = 0/48 (0%)
Query 2 RLGAGPLDAEEIKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYF 49
R+GA P D +I+RH FF ++WD LLA KI+PPFKP L+S +D F
Sbjct 310 RIGASPRDVLDIQRHSFFRHINWDDLLAFKIDPPFKPFLQSADDVSQF 357
> mmu:20112 Rps6ka2, 90kDa, D17Wsu134e, Rps6ka-rs1, Rsk3, p90rsk,
pp90rsk; ribosomal protein S6 kinase, polypeptide 2 (EC:2.7.11.1);
K04373 p90 ribosomal S6 kinase [EC:2.7.11.1]
Length=733
Score = 59.7 bits (143), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 27/48 (56%), Positives = 35/48 (72%), Gaps = 0/48 (0%)
Query 2 RLGAGPLDAEEIKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYF 49
RLGAG EEIKRHPFF ++DW+ L K+I+PPFKP + ED+ +F
Sbjct 301 RLGAGVDGVEEIKRHPFFVTIDWNKLYRKEIKPPFKPAVGRPEDTFHF 348
> hsa:208 AKT2, PKBB, PKBBETA, PRKBB, RAC-BETA; v-akt murine thymoma
viral oncogene homolog 2 (EC:2.7.11.1); K04456 RAC serine/threonine-protein
kinase [EC:2.7.11.1]
Length=481
Score = 59.3 bits (142), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 24/49 (48%), Positives = 35/49 (71%), Gaps = 0/49 (0%)
Query 1 ERLGAGPLDAEEIKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYF 49
+RLG GP DA+E+ H FF S++W ++ KK+ PPFKP + S D++YF
Sbjct 391 QRLGGGPSDAKEVMEHRFFLSINWQDVVQKKLLPPFKPQVTSEVDTRYF 439
> mmu:11652 Akt2, 2410016A19Rik, AW554154, MGC14031, PKB, PKBbeta;
thymoma viral proto-oncogene 2 (EC:2.7.11.1); K04456 RAC
serine/threonine-protein kinase [EC:2.7.11.1]
Length=481
Score = 59.3 bits (142), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 24/49 (48%), Positives = 35/49 (71%), Gaps = 0/49 (0%)
Query 1 ERLGAGPLDAEEIKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYF 49
+RLG GP DA+E+ H FF S++W ++ KK+ PPFKP + S D++YF
Sbjct 391 QRLGGGPSDAKEVMEHRFFLSINWQDVVQKKLLPPFKPQVTSEVDTRYF 439
> mmu:110651 Rps6ka3, MPK-9, Rsk2, S6K-alpha3, pp90RSK2; ribosomal
protein S6 kinase polypeptide 3 (EC:2.7.11.1); K04373 p90
ribosomal S6 kinase [EC:2.7.11.1]
Length=740
Score = 59.3 bits (142), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 27/48 (56%), Positives = 33/48 (68%), Gaps = 0/48 (0%)
Query 2 RLGAGPLDAEEIKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYF 49
RLGAGP EEIKRH FF ++DW+ L ++I PPFKP ED+ YF
Sbjct 310 RLGAGPDGVEEIKRHSFFSTIDWNKLYRREIHPPFKPATGRPEDTFYF 357
> hsa:6197 RPS6KA3, CLS, HU-3, ISPK-1, MAPKAPK1B, MRX19, RSK,
RSK2, S6K-alpha3, p90-RSK2, pp90RSK2; ribosomal protein S6 kinase,
90kDa, polypeptide 3 (EC:2.7.11.1); K04373 p90 ribosomal
S6 kinase [EC:2.7.11.1]
Length=740
Score = 59.3 bits (142), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 27/48 (56%), Positives = 33/48 (68%), Gaps = 0/48 (0%)
Query 2 RLGAGPLDAEEIKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYF 49
RLGAGP EEIKRH FF ++DW+ L ++I PPFKP ED+ YF
Sbjct 310 RLGAGPDGVEEIKRHSFFSTIDWNKLYRREIHPPFKPATGRPEDTFYF 357
> hsa:29941 PKN3, RP11-545E17.1; protein kinase N3 (EC:2.7.11.13);
K06071 protein kinase N [EC:2.7.11.13]
Length=889
Score = 59.3 bits (142), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 27/49 (55%), Positives = 33/49 (67%), Gaps = 0/49 (0%)
Query 1 ERLGAGPLDAEEIKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYF 49
+RLGAG DAEEIK PFF + +W LLA+ I+PPF P L D +YF
Sbjct 800 KRLGAGEQDAEEIKVQPFFRTTNWQALLARTIQPPFVPTLCGPADLRYF 848
> xla:399130 sgk1-a, Sgk, sgk-A, sgk1; serum/glucocorticoid regulated
kinase 1 (EC:2.7.11.1); K13302 serum/glucocorticoid-regulated
kinase 1 [EC:2.7.11.1]
Length=434
Score = 58.9 bits (141), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 30/76 (39%), Positives = 44/76 (57%), Gaps = 1/76 (1%)
Query 1 ERLGAGPLDAEEIKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYFPADVKREPVLS 60
+R+GA D EIK H FF ++WD L+ KKI PPF P++ D ++F + EPV +
Sbjct 341 KRIGAKN-DFMEIKNHIFFSPINWDDLINKKITPPFNPNVSGPSDLQHFDPEFTEEPVPN 399
Query 61 DEGDGPDAAFGTATRR 76
G PD+ TA+ +
Sbjct 400 SIGQSPDSILITASIK 415
> dre:100149794 akt3; v-akt murine thymoma viral oncogene homolog
3 (protein kinase B, gamma); K04456 RAC serine/threonine-protein
kinase [EC:2.7.11.1]
Length=423
Score = 58.9 bits (141), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 32/75 (42%), Positives = 44/75 (58%), Gaps = 7/75 (9%)
Query 1 ERLGAGPLDAEEIKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYFPADVKREPVLS 60
+RLG GP DA+EI RH FF ++DW + KK+ PPF P + S D++YF + + +
Sbjct 331 KRLGGGPDDAKEIMRHSFFTALDWQDVYDKKLVPPFMPQVSSETDTRYFDEEFTAQTITI 390
Query 61 ------DEGDGPDAA 69
DE DG DAA
Sbjct 391 TPPEKYDE-DGMDAA 404
> hsa:9252 RPS6KA5, MGC1911, MSK1, MSPK1, RLPK; ribosomal protein
S6 kinase, 90kDa, polypeptide 5 (EC:2.7.11.1); K04445 mitogen-,stress
activated protein kinases [EC:2.7.11.1]
Length=802
Score = 58.9 bits (141), Expect = 9e-09, Method: Composition-based stats.
Identities = 26/49 (53%), Positives = 33/49 (67%), Gaps = 0/49 (0%)
Query 1 ERLGAGPLDAEEIKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYF 49
+RLG GP DA+EIK H FF+ ++WD L AKK+ PFKP +R D F
Sbjct 300 KRLGCGPRDADEIKEHLFFQKINWDDLAAKKVPAPFKPVIRDELDVSNF 348
> mmu:263803 Pkn3, AW209115, BC034126, MGC31699; protein kinase
N3 (EC:2.7.11.13); K06071 protein kinase N [EC:2.7.11.13]
Length=878
Score = 58.9 bits (141), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 26/49 (53%), Positives = 33/49 (67%), Gaps = 0/49 (0%)
Query 1 ERLGAGPLDAEEIKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYF 49
+RLGAG DAEEIK PFF + +W LLA+ ++PPF P L D +YF
Sbjct 789 KRLGAGERDAEEIKVQPFFRTTNWQALLARTVQPPFVPTLCGPADLRYF 837
> mmu:20111 Rps6ka1, Rsk1, p90rsk, rsk; ribosomal protein S6 kinase
polypeptide 1 (EC:2.7.11.1); K04373 p90 ribosomal S6 kinase
[EC:2.7.11.1]
Length=735
Score = 58.9 bits (141), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 25/48 (52%), Positives = 36/48 (75%), Gaps = 0/48 (0%)
Query 2 RLGAGPLDAEEIKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYF 49
RLG+GP AEEIKRH F+ ++DW+ L ++I+PPFKP + +D+ YF
Sbjct 304 RLGSGPDGAEEIKRHIFYSTIDWNKLYRREIKPPFKPAVAQPDDTFYF 351
> xla:444618 sgk1-b, MGC84110, Sgk, sgk-B, sgk2; serum/glucocorticoid
regulated kinase 1 (EC:2.7.11.1); K13302 serum/glucocorticoid-regulated
kinase 1 [EC:2.7.11.1]
Length=434
Score = 58.9 bits (141), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 28/68 (41%), Positives = 39/68 (57%), Gaps = 0/68 (0%)
Query 9 DAEEIKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYFPADVKREPVLSDEGDGPDA 68
D EIK H FF +DWD L+ KKI PPF P++ D ++F + EPV + G PD+
Sbjct 348 DFMEIKNHIFFSPIDWDDLINKKITPPFNPNVSGPSDLQHFDPEFTDEPVPNSIGQSPDS 407
Query 69 AFGTATRR 76
TA+ +
Sbjct 408 ILITASIK 415
> xla:445821 rps6ka1, MGC81220, hu-1, mapkapk1a, rsk, rsk1; ribosomal
protein S6 kinase, 90kDa, polypeptide 1 (EC:2.7.11.1);
K04373 p90 ribosomal S6 kinase [EC:2.7.11.1]
Length=733
Score = 58.5 bits (140), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 24/48 (50%), Positives = 35/48 (72%), Gaps = 0/48 (0%)
Query 2 RLGAGPLDAEEIKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYF 49
RLG+G AEE+KRHPFF ++DW+ L +++ PPFKP + +D+ YF
Sbjct 304 RLGSGVEGAEELKRHPFFSTIDWNKLYRRELSPPFKPSVTQPDDTYYF 351
> hsa:6195 RPS6KA1, HU-1, MAPKAPK1A, RSK, RSK1; ribosomal protein
S6 kinase, 90kDa, polypeptide 1 (EC:2.7.11.1); K04373 p90
ribosomal S6 kinase [EC:2.7.11.1]
Length=744
Score = 58.5 bits (140), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 25/48 (52%), Positives = 36/48 (75%), Gaps = 0/48 (0%)
Query 2 RLGAGPLDAEEIKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYF 49
RLG+GP AEEIKRH F+ ++DW+ L ++I+PPFKP + +D+ YF
Sbjct 313 RLGSGPDGAEEIKRHVFYSTIDWNKLYRREIKPPFKPAVAQPDDTFYF 360
> dre:571285 ribosomal protein S6 kinase, polypeptide 2-like;
K04373 p90 ribosomal S6 kinase [EC:2.7.11.1]
Length=733
Score = 58.5 bits (140), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 26/48 (54%), Positives = 35/48 (72%), Gaps = 0/48 (0%)
Query 2 RLGAGPLDAEEIKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYF 49
RLGAGP EEIKRH FF ++DW+ L ++I+PPFKP + ED+ +F
Sbjct 301 RLGAGPDGVEEIKRHIFFATIDWNKLYRREIKPPFKPAVGRPEDTFHF 348
> dre:338219 rps6ka3a, id:ibd1103, rps6ka3, si:ch211-114c12.12,
wu:fk86e02, zgc:56006; ribosomal protein S6 kinase, polypeptide
3a (EC:2.7.11.1); K04373 p90 ribosomal S6 kinase [EC:2.7.11.1]
Length=732
Score = 58.5 bits (140), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 25/48 (52%), Positives = 33/48 (68%), Gaps = 0/48 (0%)
Query 2 RLGAGPLDAEEIKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYF 49
RLGAGP E IKRH FF +DW+ L K++ PPFKP ++ +D+ YF
Sbjct 301 RLGAGPDGVEGIKRHSFFTRIDWNKLFRKEVPPPFKPAIKKPDDTFYF 348
> hsa:5581 PRKCE, MGC125656, MGC125657, PKCE, nPKC-epsilon; protein
kinase C, epsilon (EC:2.7.11.13); K06068 novel protein
kinase C [EC:2.7.11.13]
Length=737
Score = 58.2 bits (139), Expect = 1e-08, Method: Composition-based stats.
Identities = 30/63 (47%), Positives = 40/63 (63%), Gaps = 3/63 (4%)
Query 1 ERLG--AGPLDAEEIKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYFPADVKR-EP 57
+RLG A + IK+HPFF+ +DW LL KKI+PPFKP +++ D F D R EP
Sbjct 648 KRLGCVASQNGEDAIKQHPFFKEIDWVLLEQKKIKPPFKPRIKTKRDVNNFDQDFTREEP 707
Query 58 VLS 60
VL+
Sbjct 708 VLT 710
> sce:YBL105C PKC1, CLY15, HPO2, STT1; Pkc1p (EC:2.7.11.13); K02677
classical protein kinase C [EC:2.7.11.13]
Length=1151
Score = 58.2 bits (139), Expect = 2e-08, Method: Composition-based stats.
Identities = 23/57 (40%), Positives = 39/57 (68%), Gaps = 0/57 (0%)
Query 1 ERLGAGPLDAEEIKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYFPADVKREP 57
+RLGAGP DA+E+ PFF ++++D +L +++PP+ P ++S ED+ YF + P
Sbjct 1065 KRLGAGPRDADEVMEEPFFRNINFDDILNLRVKPPYIPEIKSPEDTSYFEQEFTSAP 1121
> mmu:18754 Prkce, 5830406C15Rik, PKC[e], PKCepsilon, Pkce, R75156;
protein kinase C, epsilon (EC:2.7.11.13); K06068 novel
protein kinase C [EC:2.7.11.13]
Length=737
Score = 57.8 bits (138), Expect = 2e-08, Method: Composition-based stats.
Identities = 29/63 (46%), Positives = 40/63 (63%), Gaps = 3/63 (4%)
Query 1 ERLG--AGPLDAEEIKRHPFFESVDWDLLLAKKIEPPFKPHLRSHEDSKYFPADVKR-EP 57
+RLG A + IK+HPFF+ +DW LL KKI+PPFKP +++ D F D R EP
Sbjct 648 KRLGCVAAQNGEDAIKQHPFFKEIDWVLLEQKKIKPPFKPRIKTKRDVNNFDQDFTREEP 707
Query 58 VLS 60
+L+
Sbjct 708 ILT 710
Lambda K H
0.311 0.127 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5106150148
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40