bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0927_orf2
Length=155
Score E
Sequences producing significant alignments: (Bits) Value
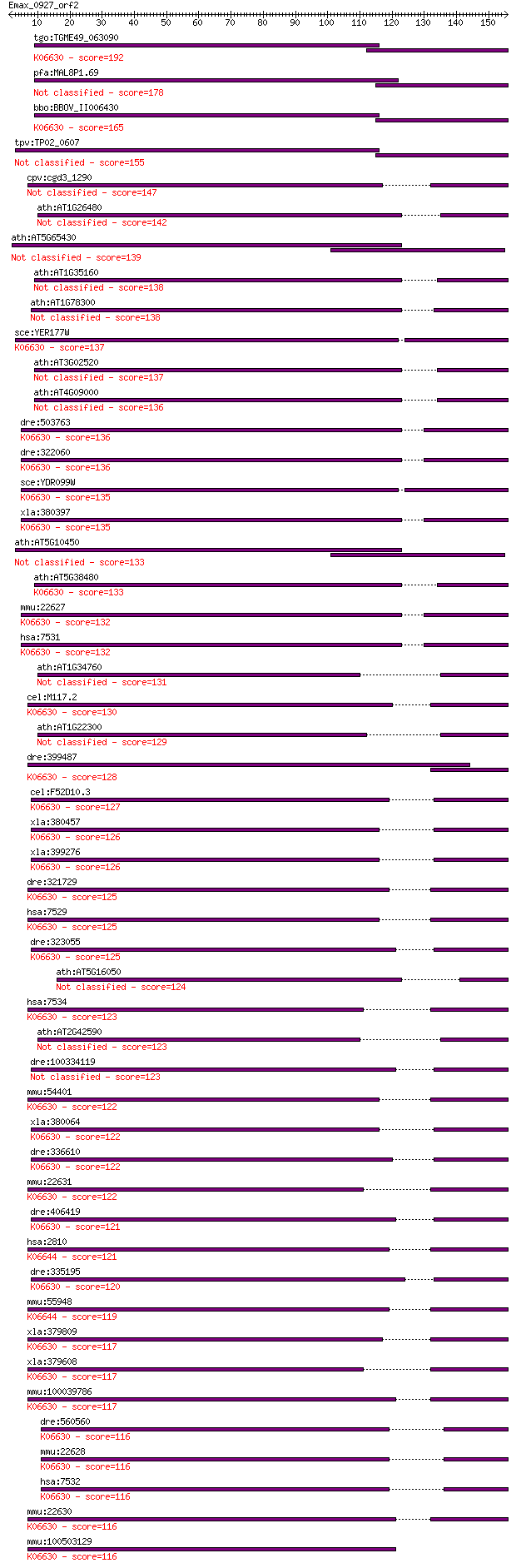
tgo:TGME49_063090 14-3-3 protein, putative ; K06630 tyrosine 3... 192 5e-49
pfa:MAL8P1.69 14-3-3 protein, putative 178 5e-45
bbo:BBOV_II006430 18.m06529; 14-3-3 protein; K06630 tyrosine 3... 165 4e-41
tpv:TP02_0607 hypothetical protein 155 4e-38
cpv:cgd3_1290 14-3-3 domain containing protein 147 2e-35
ath:AT1G26480 GRF12; GRF12 (GENERAL REGULATORY FACTOR 12); pro... 142 6e-34
ath:AT5G65430 GRF8; GRF8 (GENERAL REGULATORY FACTOR 8); protei... 139 5e-33
ath:AT1G35160 GF14 PHI (GF14 PROTEIN PHI CHAIN); protein bindi... 138 7e-33
ath:AT1G78300 GRF2; GRF2 (GENERAL REGULATORY FACTOR 2); protei... 138 9e-33
sce:YER177W BMH1, APR6; 14-3-3 protein, major isoform; control... 137 1e-32
ath:AT3G02520 GRF7; GRF7 (GENERAL REGULATORY FACTOR 7); protei... 137 2e-32
ath:AT4G09000 GRF1; 14-3-3-like protein GF14 chi / general reg... 136 2e-32
dre:503763 ywhae2, im:7138781, zgc:113329; tyrosine 3-monooxyg... 136 3e-32
dre:322060 ywhae1, fe38c12, wu:fb48f06, wu:fe38c12, wu:fi22e10... 136 3e-32
sce:YDR099W BMH2, SCD3; 14-3-3 protein, minor isoform; control... 135 5e-32
xla:380397 ywhae, 14-3-3e, MGC53170; tyrosine 3-monooxygenase/... 135 7e-32
ath:AT5G10450 GRF6; GRF6 (G-box regulating factor 6); protein ... 133 2e-31
ath:AT5G38480 GRF3; GRF3 (GENERAL REGULATORY FACTOR 3); ATP bi... 133 3e-31
mmu:22627 Ywhae, AU019196; tyrosine 3-monooxygenase/tryptophan... 132 5e-31
hsa:7531 YWHAE, 14-3-3E, FLJ45465, FLJ53559, KCIP-1, MDCR, MDS... 132 5e-31
ath:AT1G34760 GRF11; GRF11 (GENERAL REGULATORY FACTOR 11); ATP... 131 1e-30
cel:M117.2 par-5; abnormal embryonic PARtitioning of cytoplasm... 130 2e-30
ath:AT1G22300 GRF10; GRF10 (GENERAL REGULATORY FACTOR 10); ATP... 129 5e-30
dre:399487 ywhaqa, ywhaq, ywhaq2; tyrosine 3-monooxygenase/try... 128 8e-30
cel:F52D10.3 ftt-2; Fourteen-Three-Three family member (ftt-2)... 127 1e-29
xla:380457 ywhab-b, 14-3-3beta, MGC53093, ywhab; tyrosine 3-mo... 126 3e-29
xla:399276 ywhab-a, 14-3-3beta, MGC78918; tyrosine 3-monooxyge... 126 3e-29
dre:321729 ywhabl, hm:zehn0976, wu:fb33b01, zgc:73065, zgc:773... 125 4e-29
hsa:7529 YWHAB, GW128, HS1, KCIP-1, YWHAA; tyrosine 3-monooxyg... 125 7e-29
dre:323055 ywhaba, fb80c08, fk41f08, wu:fb80c08, wu:fd11d01, w... 125 7e-29
ath:AT5G16050 GRF5; GRF5 (GENERAL REGULATORY FACTOR 5); ATP bi... 124 1e-28
hsa:7534 YWHAZ, 14-3-3-zeta, KCIP-1, MGC111427, MGC126532, MGC... 123 3e-28
ath:AT2G42590 GRF9; GRF9 (GENERAL REGULATORY FACTOR 9); calciu... 123 3e-28
dre:100334119 14-3-3 protein beta/alpha-like 123 3e-28
mmu:54401 Ywhab, 1300003C17Rik; tyrosine 3-monooxygenase/trypt... 122 4e-28
xla:380064 ywhaq, 14-3-3t, MGC53015; tyrosine 3-monooxygenase/... 122 4e-28
dre:336610 ywhai, fb14h09, wu:fb05g08, wu:fb14h09, zgc:55807; ... 122 4e-28
mmu:22631 Ywhaz, 1110013I11Rik, 14-3-3zeta, AI596267, AL022924... 122 4e-28
dre:406419 ywhabb, wu:fb22g04, wu:fr86a12, ywhab, ywhab2, zgc:... 121 7e-28
hsa:2810 SFN, YWHAS; stratifin; K06644 stratifin 121 1e-27
dre:335195 ywhaqb, wu:fa91b01, wu:fb02e12, wu:fb36f06, wu:fk86... 120 2e-27
mmu:55948 Sfn, Er, Mme1, Ywhas; stratifin; K06644 stratifin 119 3e-27
xla:379809 ywhaz-a, 14-3-3z, 14-3-3zeta, MGC52760, ywhaz, ywha... 117 1e-26
xla:379608 ywhaz-b, 14-3-3z, 14-3-3zeta, MGC64423, ywhaz, ywha... 117 2e-26
mmu:100039786 Gm2423; predicted gene 2423; K06630 tyrosine 3-m... 117 2e-26
dre:560560 ywhag2, ywhagl; 3-monooxygenase/tryptophan 5-monoox... 116 3e-26
mmu:22628 Ywhag, 14-3-3gamma, D7Bwg1348e; tyrosine 3-monooxyge... 116 3e-26
hsa:7532 YWHAG, 14-3-3GAMMA; tyrosine 3-monooxygenase/tryptoph... 116 3e-26
mmu:22630 Ywhaq, 2700028P07Rik, AA409740, AU021156, MGC118161,... 116 3e-26
mmu:100503129 14-3-3 protein theta-like; K06630 tyrosine 3-mon... 116 3e-26
> tgo:TGME49_063090 14-3-3 protein, putative ; K06630 tyrosine
3-monooxygenase/tryptophan 5-monooxygenase activation protein
Length=323
Score = 192 bits (487), Expect = 5e-49, Method: Compositional matrix adjust.
Identities = 89/107 (83%), Positives = 98/107 (91%), Gaps = 0/107 (0%)
Query 9 DELTVEERNLLSVAYKNAVGARRASWRIISSVEQKEANRNHMTNKGLAASYRQKVENELN 68
DEL+VEERNLLSVAYKNAVGARRASWRIISSVEQKE ++ HM NK LAA YRQKVE ELN
Sbjct 104 DELSVEERNLLSVAYKNAVGARRASWRIISSVEQKELSKQHMQNKALAAEYRQKVEEELN 163
Query 69 KICQEILTLLTDKLLPRTTDSESRVFYFKMKGDYYRYISEFSNEEGK 115
KIC +IL LLTDKL+P+T+DSES+VFY+KMKGDYYRYISEFS EEGK
Sbjct 164 KICHDILQLLTDKLIPKTSDSESKVFYYKMKGDYYRYISEFSGEEGK 210
Score = 59.7 bits (143), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 31/44 (70%), Positives = 33/44 (75%), Gaps = 5/44 (11%)
Query 112 EEGKNLVENCLDQNSSTPGGKGDELTVEERNLLSVAYKNAVGAR 155
E KNLVENCLD+ DEL+VEERNLLSVAYKNAVGAR
Sbjct 87 EAMKNLVENCLDEQQP-----KDELSVEERNLLSVAYKNAVGAR 125
> pfa:MAL8P1.69 14-3-3 protein, putative
Length=262
Score = 178 bits (452), Expect = 5e-45, Method: Compositional matrix adjust.
Identities = 85/113 (75%), Positives = 99/113 (87%), Gaps = 0/113 (0%)
Query 9 DELTVEERNLLSVAYKNAVGARRASWRIISSVEQKEANRNHMTNKGLAASYRQKVENELN 68
DELTVEERNLLSVAYKNAVGARRASWRIISSVEQKE ++ ++ NK +AA+YR+KVE ELN
Sbjct 47 DELTVEERNLLSVAYKNAVGARRASWRIISSVEQKEMSKANVHNKNVAATYRKKVEEELN 106
Query 69 KICQEILTLLTDKLLPRTTDSESRVFYFKMKGDYYRYISEFSNEEGKNLVENC 121
ICQ+IL LLT KL+P T++SES+VFY+KMKGDYYRYISEFS +EGK NC
Sbjct 107 NICQDILNLLTKKLIPNTSESESKVFYYKMKGDYYRYISEFSCDEGKKEASNC 159
Score = 49.7 bits (117), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 26/41 (63%), Positives = 30/41 (73%), Gaps = 7/41 (17%)
Query 115 KNLVENCLDQNSSTPGGKGDELTVEERNLLSVAYKNAVGAR 155
+ LVE C++ + DELTVEERNLLSVAYKNAVGAR
Sbjct 35 RTLVEQCVNNDK-------DELTVEERNLLSVAYKNAVGAR 68
> bbo:BBOV_II006430 18.m06529; 14-3-3 protein; K06630 tyrosine
3-monooxygenase/tryptophan 5-monooxygenase activation protein
Length=256
Score = 165 bits (418), Expect = 4e-41, Method: Compositional matrix adjust.
Identities = 77/107 (71%), Positives = 93/107 (86%), Gaps = 0/107 (0%)
Query 9 DELTVEERNLLSVAYKNAVGARRASWRIISSVEQKEANRNHMTNKGLAASYRQKVENELN 68
+ELTVEERNLLSVAYKNAVG+RRASWRI+S VEQKEA++N++ + LAA+YR K+E EL
Sbjct 43 EELTVEERNLLSVAYKNAVGSRRASWRIVSGVEQKEASKNNVVQQNLAATYRLKIEKELT 102
Query 69 KICQEILTLLTDKLLPRTTDSESRVFYFKMKGDYYRYISEFSNEEGK 115
+IC ILTLL D+L+P TTDSES+VFY KMKGDYYRYISEFS ++ K
Sbjct 103 QICSNILTLLNDRLIPATTDSESQVFYHKMKGDYYRYISEFSCDQEK 149
Score = 47.4 bits (111), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 27/41 (65%), Positives = 30/41 (73%), Gaps = 8/41 (19%)
Query 115 KNLVENCLDQNSSTPGGKGDELTVEERNLLSVAYKNAVGAR 155
K LVE C+ GK +ELTVEERNLLSVAYKNAVG+R
Sbjct 32 KALVETCI-------TGK-EELTVEERNLLSVAYKNAVGSR 64
> tpv:TP02_0607 hypothetical protein
Length=256
Score = 155 bits (393), Expect = 4e-38, Method: Compositional matrix adjust.
Identities = 74/113 (65%), Positives = 93/113 (82%), Gaps = 0/113 (0%)
Query 3 TPGGKGDELTVEERNLLSVAYKNAVGARRASWRIISSVEQKEANRNHMTNKGLAASYRQK 62
T +ELTVEERNLLSVAYKN+VG+RRASWRI+SSVEQKEA++N+ ++K LA YR K
Sbjct 36 TCVSDKEELTVEERNLLSVAYKNSVGSRRASWRIVSSVEQKEASKNNTSHKALARQYRVK 95
Query 63 VENELNKICQEILTLLTDKLLPRTTDSESRVFYFKMKGDYYRYISEFSNEEGK 115
+E ELN+IC IL+LL +KL+ T +SES VFY+KMKGDYYRY+SEFS ++ K
Sbjct 96 IEGELNEICNNILSLLNNKLISSTNESESLVFYYKMKGDYYRYVSEFSCDDNK 148
Score = 45.4 bits (106), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 24/41 (58%), Positives = 28/41 (68%), Gaps = 8/41 (19%)
Query 115 KNLVENCLDQNSSTPGGKGDELTVEERNLLSVAYKNAVGAR 155
K LVE C+ +ELTVEERNLLSVAYKN+VG+R
Sbjct 31 KTLVETCV--------SDKEELTVEERNLLSVAYKNSVGSR 63
> cpv:cgd3_1290 14-3-3 domain containing protein
Length=273
Score = 147 bits (370), Expect = 2e-35, Method: Compositional matrix adjust.
Identities = 70/110 (63%), Positives = 83/110 (75%), Gaps = 0/110 (0%)
Query 7 KGDELTVEERNLLSVAYKNAVGARRASWRIISSVEQKEANRNHMTNKGLAASYRQKVENE 66
+ +ELTVEERNLLSVAYKNAVG+RR+SWRIISSVEQKE +RN + YR KVE E
Sbjct 59 ESEELTVEERNLLSVAYKNAVGSRRSSWRIISSVEQKEHSRNAEDASKMCGKYRSKVEAE 118
Query 67 LNKICQEILTLLTDKLLPRTTDSESRVFYFKMKGDYYRYISEFSNEEGKN 116
L IC +ILT+L L+P T +S+VFYFKMKGDY+RYISEFS + K
Sbjct 119 LTDICNDILTMLDKHLIPTATSPDSKVFYFKMKGDYHRYISEFSTGDSKQ 168
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 20/24 (83%), Positives = 23/24 (95%), Gaps = 0/24 (0%)
Query 132 KGDELTVEERNLLSVAYKNAVGAR 155
+ +ELTVEERNLLSVAYKNAVG+R
Sbjct 59 ESEELTVEERNLLSVAYKNAVGSR 82
> ath:AT1G26480 GRF12; GRF12 (GENERAL REGULATORY FACTOR 12); protein
binding / protein phosphorylated amino acid binding
Length=268
Score = 142 bits (357), Expect = 6e-34, Method: Compositional matrix adjust.
Identities = 68/114 (59%), Positives = 84/114 (73%), Gaps = 1/114 (0%)
Query 10 ELTVEERNLLSVAYKNAVGARRASWRIISSVEQKEANRNHMTNKGLAASYRQKVENELNK 69
ELTVEERNLLSV YKN +GARRASWRI+SS+EQKE ++ + +N YRQKVE+EL
Sbjct 42 ELTVEERNLLSVGYKNVIGARRASWRIMSSIEQKEESKGNESNVKQIKGYRQKVEDELAN 101
Query 70 ICQEILTLLTDKLLPRTTDSESRVFYFKMKGDYYRYISEFSNE-EGKNLVENCL 122
ICQ+ILT++ L+P T E+ VFY+KMKGDYYRY++EF E E K E L
Sbjct 102 ICQDILTIIDQHLIPHATSGEATVFYYKMKGDYYRYLAEFKTEQERKEAAEQSL 155
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 18/21 (85%), Positives = 19/21 (90%), Gaps = 0/21 (0%)
Query 135 ELTVEERNLLSVAYKNAVGAR 155
ELTVEERNLLSV YKN +GAR
Sbjct 42 ELTVEERNLLSVGYKNVIGAR 62
> ath:AT5G65430 GRF8; GRF8 (GENERAL REGULATORY FACTOR 8); protein
binding / protein phosphorylated amino acid binding
Length=246
Score = 139 bits (349), Expect = 5e-33, Method: Compositional matrix adjust.
Identities = 69/122 (56%), Positives = 87/122 (71%), Gaps = 4/122 (3%)
Query 2 STPGGKGDELTVEERNLLSVAYKNAVGARRASWRIISSVEQKEANRNHMTNKGLAASYRQ 61
+TP G ELTVEERNLLSVAYKN +G+ RA+WRI+SS+EQKE +R + + L YR
Sbjct 37 ATPAG---ELTVEERNLLSVAYKNVIGSLRAAWRIVSSIEQKEESRKNEEHVSLVKDYRS 93
Query 62 KVENELNKICQEILTLLTDKLLPRTTDSESRVFYFKMKGDYYRYISEF-SNEEGKNLVEN 120
KVE EL+ IC IL LL L+P T SES+VFY KMKGDY+RY++EF S +E K E+
Sbjct 94 KVETELSSICSGILRLLDSHLIPSATASESKVFYLKMKGDYHRYLAEFKSGDERKTAAED 153
Query 121 CL 122
+
Sbjct 154 TM 155
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 26/59 (44%), Positives = 36/59 (61%), Gaps = 10/59 (16%)
Query 101 DYYRYISEFSN-----EEGKNLVENCLDQNSSTPGGKGDELTVEERNLLSVAYKNAVGA 154
D Y Y+++ + EE +E + + +TP G ELTVEERNLLSVAYKN +G+
Sbjct 8 DQYVYMAKLAEQAERYEEMVQFMEQLV--SGATPAG---ELTVEERNLLSVAYKNVIGS 61
> ath:AT1G35160 GF14 PHI (GF14 PROTEIN PHI CHAIN); protein binding
/ protein phosphorylated amino acid binding
Length=267
Score = 138 bits (347), Expect = 7e-33, Method: Compositional matrix adjust.
Identities = 67/115 (58%), Positives = 88/115 (76%), Gaps = 1/115 (0%)
Query 9 DELTVEERNLLSVAYKNAVGARRASWRIISSVEQKEANRNHMTNKGLAASYRQKVENELN 68
DELTVEERNLLSVAYKN +GARRASWRIISS+EQKE +R + + YR K+E+EL+
Sbjct 44 DELTVEERNLLSVAYKNVIGARRASWRIISSIEQKEESRGNDDHVTTIRDYRSKIESELS 103
Query 69 KICQEILTLLTDKLLPRTTDSESRVFYFKMKGDYYRYISEF-SNEEGKNLVENCL 122
KIC IL LL +L+P + + +S+VFY KMKGDY+RY++EF + +E K+ E+ L
Sbjct 104 KICDGILKLLDTRLVPASANGDSKVFYLKMKGDYHRYLAEFKTGQERKDAAEHTL 158
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 20/22 (90%), Positives = 21/22 (95%), Gaps = 0/22 (0%)
Query 134 DELTVEERNLLSVAYKNAVGAR 155
DELTVEERNLLSVAYKN +GAR
Sbjct 44 DELTVEERNLLSVAYKNVIGAR 65
> ath:AT1G78300 GRF2; GRF2 (GENERAL REGULATORY FACTOR 2); protein
binding / protein phosphorylated amino acid binding
Length=259
Score = 138 bits (347), Expect = 9e-33, Method: Compositional matrix adjust.
Identities = 67/116 (57%), Positives = 85/116 (73%), Gaps = 1/116 (0%)
Query 8 GDELTVEERNLLSVAYKNAVGARRASWRIISSVEQKEANRNHMTNKGLAASYRQKVENEL 67
GDELTVEERNLLSVAYKN +GARRASWRIISS+EQKE +R + + YR K+E EL
Sbjct 37 GDELTVEERNLLSVAYKNVIGARRASWRIISSIEQKEESRGNDDHVTAIREYRSKIETEL 96
Query 68 NKICQEILTLLTDKLLPRTTDSESRVFYFKMKGDYYRYISEF-SNEEGKNLVENCL 122
+ IC IL LL +L+P +S+VFY KMKGDY+RY++EF + +E K+ E+ L
Sbjct 97 SGICDGILKLLDSRLIPAAASGDSKVFYLKMKGDYHRYLAEFKTGQERKDAAEHTL 152
Score = 47.0 bits (110), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 21/23 (91%), Positives = 22/23 (95%), Gaps = 0/23 (0%)
Query 133 GDELTVEERNLLSVAYKNAVGAR 155
GDELTVEERNLLSVAYKN +GAR
Sbjct 37 GDELTVEERNLLSVAYKNVIGAR 59
> sce:YER177W BMH1, APR6; 14-3-3 protein, major isoform; controls
proteome at post-transcriptional level, binds proteins and
DNA, involved in regulation of many processes including exocytosis,
vesicle transport, Ras/MAPK signaling, and rapamycin-sensitive
signaling; K06630 tyrosine 3-monooxygenase/tryptophan
5-monooxygenase activation protein
Length=267
Score = 137 bits (345), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 65/120 (54%), Positives = 86/120 (71%), Gaps = 1/120 (0%)
Query 3 TPGGKGDELTVEERNLLSVAYKNAVGARRASWRIISSVEQKEANRNHMTNK-GLAASYRQ 61
T G EL+VEERNLLSVAYKN +GARRASWRI+SS+EQKE ++ ++ L SYR
Sbjct 30 TVASSGQELSVEERNLLSVAYKNVIGARRASWRIVSSIEQKEESKEKSEHQVELICSYRS 89
Query 62 KVENELNKICQEILTLLTDKLLPRTTDSESRVFYFKMKGDYYRYISEFSNEEGKNLVENC 121
K+E EL KI +IL++L L+P T ES+VFY+KMKGDY+RY++EFS+ + + N
Sbjct 90 KIETELTKISDDILSVLDSHLIPSATTGESKVFYYKMKGDYHRYLAEFSSGDAREKATNA 149
Score = 46.2 bits (108), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 21/32 (65%), Positives = 24/32 (75%), Gaps = 0/32 (0%)
Query 124 QNSSTPGGKGDELTVEERNLLSVAYKNAVGAR 155
+N T G EL+VEERNLLSVAYKN +GAR
Sbjct 26 ENMKTVASSGQELSVEERNLLSVAYKNVIGAR 57
> ath:AT3G02520 GRF7; GRF7 (GENERAL REGULATORY FACTOR 7); protein
binding / protein phosphorylated amino acid binding
Length=265
Score = 137 bits (345), Expect = 2e-32, Method: Compositional matrix adjust.
Identities = 68/115 (59%), Positives = 85/115 (73%), Gaps = 1/115 (0%)
Query 9 DELTVEERNLLSVAYKNAVGARRASWRIISSVEQKEANRNHMTNKGLAASYRQKVENELN 68
DELTVEERNLLSVAYKN +GARRASWRIISS+EQKE +R + + + YR K+E EL+
Sbjct 38 DELTVEERNLLSVAYKNVIGARRASWRIISSIEQKEESRGNDDHVSIIKDYRGKIETELS 97
Query 69 KICQEILTLLTDKLLPRTTDSESRVFYFKMKGDYYRYISEF-SNEEGKNLVENCL 122
KIC IL LL L+P + +ES+VFY KMKGDY+RY++EF + E K E+ L
Sbjct 98 KICDGILNLLDSHLVPTASLAESKVFYLKMKGDYHRYLAEFKTGAERKEAAESTL 152
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 20/22 (90%), Positives = 21/22 (95%), Gaps = 0/22 (0%)
Query 134 DELTVEERNLLSVAYKNAVGAR 155
DELTVEERNLLSVAYKN +GAR
Sbjct 38 DELTVEERNLLSVAYKNVIGAR 59
> ath:AT4G09000 GRF1; 14-3-3-like protein GF14 chi / general regulatory
factor 1 (GRF1)
Length=318
Score = 136 bits (343), Expect = 2e-32, Method: Compositional matrix adjust.
Identities = 68/115 (59%), Positives = 84/115 (73%), Gaps = 1/115 (0%)
Query 9 DELTVEERNLLSVAYKNAVGARRASWRIISSVEQKEANRNHMTNKGLAASYRQKVENELN 68
DELTVEERNLLSVAYKN +GARRASWRIISS+EQKE +R + + L YR K+E EL+
Sbjct 43 DELTVEERNLLSVAYKNVIGARRASWRIISSIEQKEESRGNDDHVSLIRDYRSKIETELS 102
Query 69 KICQEILTLLTDKLLPRTTDSESRVFYFKMKGDYYRYISEF-SNEEGKNLVENCL 122
IC IL LL L+P +S+VFY KMKGDY+RY++EF S +E K+ E+ L
Sbjct 103 DICDGILKLLDTILVPAAASGDSKVFYLKMKGDYHRYLAEFKSGQERKDAAEHTL 157
Score = 44.7 bits (104), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 20/22 (90%), Positives = 21/22 (95%), Gaps = 0/22 (0%)
Query 134 DELTVEERNLLSVAYKNAVGAR 155
DELTVEERNLLSVAYKN +GAR
Sbjct 43 DELTVEERNLLSVAYKNVIGAR 64
> dre:503763 ywhae2, im:7138781, zgc:113329; tyrosine 3-monooxygenase/tryptophan
5-monooxygenase activation protein, epsilon
polypeptide 2; K06630 tyrosine 3-monooxygenase/tryptophan
5-monooxygenase activation protein
Length=255
Score = 136 bits (343), Expect = 3e-32, Method: Compositional matrix adjust.
Identities = 66/119 (55%), Positives = 84/119 (70%), Gaps = 1/119 (0%)
Query 5 GGKGDELTVEERNLLSVAYKNAVGARRASWRIISSVEQKEANRNHMTNKGLAASYRQKVE 64
GK ++L+VEERNLLSVAYKN +GARRASWRIISS+EQKE ++ + YRQ VE
Sbjct 31 AGKDEDLSVEERNLLSVAYKNVIGARRASWRIISSIEQKEESKGGADKLKMIREYRQTVE 90
Query 65 NELNKICQEILTLLTDKLLPRTTDSESRVFYFKMKGDYYRYISEFSN-EEGKNLVENCL 122
NEL IC +IL +L L+P ES+VFY+KMKGDY+RY++EF+ + K EN L
Sbjct 91 NELKSICNDILDVLDKHLIPAANTGESKVFYYKMKGDYHRYLAEFATGNDRKEAAENSL 149
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 19/26 (73%), Positives = 23/26 (88%), Gaps = 0/26 (0%)
Query 130 GGKGDELTVEERNLLSVAYKNAVGAR 155
GK ++L+VEERNLLSVAYKN +GAR
Sbjct 31 AGKDEDLSVEERNLLSVAYKNVIGAR 56
> dre:322060 ywhae1, fe38c12, wu:fb48f06, wu:fe38c12, wu:fi22e10,
ywhae, zgc:55377, zgc:77062; tyrosine 3-monooxygenase/tryptophan
5-monooxygenase activation protein, epsilon polypeptide
1; K06630 tyrosine 3-monooxygenase/tryptophan 5-monooxygenase
activation protein
Length=255
Score = 136 bits (342), Expect = 3e-32, Method: Compositional matrix adjust.
Identities = 68/119 (57%), Positives = 82/119 (68%), Gaps = 1/119 (0%)
Query 5 GGKGDELTVEERNLLSVAYKNAVGARRASWRIISSVEQKEANRNHMTNKGLAASYRQKVE 64
G ELTVEERNLLSVAYKN +GARRASWRIISS+EQKE N+ + YRQ VE
Sbjct 31 AGMDVELTVEERNLLSVAYKNVIGARRASWRIISSIEQKEENKGGEDKLKMIREYRQTVE 90
Query 65 NELNKICQEILTLLTDKLLPRTTDSESRVFYFKMKGDYYRYISEFSN-EEGKNLVENCL 122
NEL IC +IL +L L+P ES+VFY+KMKGDY+RY++EF+ + K EN L
Sbjct 91 NELKSICNDILDVLDKHLIPAANSGESKVFYYKMKGDYHRYLAEFATGNDRKEAAENSL 149
Score = 42.7 bits (99), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 20/26 (76%), Positives = 21/26 (80%), Gaps = 0/26 (0%)
Query 130 GGKGDELTVEERNLLSVAYKNAVGAR 155
G ELTVEERNLLSVAYKN +GAR
Sbjct 31 AGMDVELTVEERNLLSVAYKNVIGAR 56
> sce:YDR099W BMH2, SCD3; 14-3-3 protein, minor isoform; controls
proteome at post-transcriptional level, binds proteins and
DNA, involved in regulation of many processes including exocytosis,
vesicle transport, Ras/MAPK signaling, and rapamycin-sensitive
signaling; K06630 tyrosine 3-monooxygenase/tryptophan
5-monooxygenase activation protein
Length=273
Score = 135 bits (340), Expect = 5e-32, Method: Compositional matrix adjust.
Identities = 64/118 (54%), Positives = 85/118 (72%), Gaps = 1/118 (0%)
Query 5 GGKGDELTVEERNLLSVAYKNAVGARRASWRIISSVEQKEANRNHMTNK-GLAASYRQKV 63
G EL+VEERNLLSVAYKN +GARRASWRI+SS+EQKE ++ ++ L SYR K+
Sbjct 32 ASSGQELSVEERNLLSVAYKNVIGARRASWRIVSSIEQKEESKEKSEHQVELIRSYRSKI 91
Query 64 ENELNKICQEILTLLTDKLLPRTTDSESRVFYFKMKGDYYRYISEFSNEEGKNLVENC 121
E EL KI +IL++L L+P T ES+VFY+KMKGDY+RY++EFS+ + + N
Sbjct 92 ETELTKISDDILSVLDSHLIPSATTGESKVFYYKMKGDYHRYLAEFSSGDAREKATNS 149
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 20/32 (62%), Positives = 23/32 (71%), Gaps = 0/32 (0%)
Query 124 QNSSTPGGKGDELTVEERNLLSVAYKNAVGAR 155
+N G EL+VEERNLLSVAYKN +GAR
Sbjct 26 ENMKAVASSGQELSVEERNLLSVAYKNVIGAR 57
> xla:380397 ywhae, 14-3-3e, MGC53170; tyrosine 3-monooxygenase/tryptophan
5-monooxygenase activation protein, epsilon polypeptide;
K06630 tyrosine 3-monooxygenase/tryptophan 5-monooxygenase
activation protein
Length=255
Score = 135 bits (339), Expect = 7e-32, Method: Compositional matrix adjust.
Identities = 67/119 (56%), Positives = 82/119 (68%), Gaps = 1/119 (0%)
Query 5 GGKGDELTVEERNLLSVAYKNAVGARRASWRIISSVEQKEANRNHMTNKGLAASYRQKVE 64
G ELTVEERNLLSVAYKN +GARRASWRIISS+EQKE N+ + YRQ VE
Sbjct 31 AGMDVELTVEERNLLSVAYKNVIGARRASWRIISSIEQKEENKGGEDKLKMIREYRQMVE 90
Query 65 NELNKICQEILTLLTDKLLPRTTDSESRVFYFKMKGDYYRYISEFSN-EEGKNLVENCL 122
EL IC +IL +L L+P + ES+VFY+KMKGDY+RY++EF+ + K EN L
Sbjct 91 TELKSICNDILDVLDKHLIPAASSGESKVFYYKMKGDYHRYLAEFAQGNDRKEAAENSL 149
Score = 42.7 bits (99), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 20/26 (76%), Positives = 21/26 (80%), Gaps = 0/26 (0%)
Query 130 GGKGDELTVEERNLLSVAYKNAVGAR 155
G ELTVEERNLLSVAYKN +GAR
Sbjct 31 AGMDVELTVEERNLLSVAYKNVIGAR 56
> ath:AT5G10450 GRF6; GRF6 (G-box regulating factor 6); protein
binding / protein phosphorylated amino acid binding
Length=246
Score = 133 bits (335), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 65/121 (53%), Positives = 85/121 (70%), Gaps = 1/121 (0%)
Query 3 TPGGKGDELTVEERNLLSVAYKNAVGARRASWRIISSVEQKEANRNHMTNKGLAASYRQK 62
T +ELTVEERNLLSVAYKN +G+ RA+WRI+SS+EQKE +R + + L YR K
Sbjct 35 TGATPAEELTVEERNLLSVAYKNVIGSLRAAWRIVSSIEQKEESRKNDEHVSLVKDYRSK 94
Query 63 VENELNKICQEILTLLTDKLLPRTTDSESRVFYFKMKGDYYRYISEF-SNEEGKNLVENC 121
VE+EL+ +C IL LL L+P SES+VFY KMKGDY+RY++EF S +E K E+
Sbjct 95 VESELSSVCSGILKLLDSHLIPSAGASESKVFYLKMKGDYHRYMAEFKSGDERKTAAEDT 154
Query 122 L 122
+
Sbjct 155 M 155
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 25/59 (42%), Positives = 35/59 (59%), Gaps = 10/59 (16%)
Query 101 DYYRYISEFSN-----EEGKNLVENCLDQNSSTPGGKGDELTVEERNLLSVAYKNAVGA 154
D Y Y+++ + EE +E + +TP +ELTVEERNLLSVAYKN +G+
Sbjct 8 DQYVYMAKLAEQAERYEEMVQFMEQLV--TGATPA---EELTVEERNLLSVAYKNVIGS 61
> ath:AT5G38480 GRF3; GRF3 (GENERAL REGULATORY FACTOR 3); ATP
binding / protein binding / protein phosphorylated amino acid
binding; K06630 tyrosine 3-monooxygenase/tryptophan 5-monooxygenase
activation protein
Length=254
Score = 133 bits (334), Expect = 3e-31, Method: Compositional matrix adjust.
Identities = 64/115 (55%), Positives = 86/115 (74%), Gaps = 1/115 (0%)
Query 9 DELTVEERNLLSVAYKNAVGARRASWRIISSVEQKEANRNHMTNKGLAASYRQKVENELN 68
+EL+VEERNLLSVAYKN +GARRASWRIISS+EQKE ++ + + + YR K+E+EL+
Sbjct 37 EELSVEERNLLSVAYKNVIGARRASWRIISSIEQKEESKGNEDHVAIIKDYRGKIESELS 96
Query 69 KICQEILTLLTDKLLPRTTDSESRVFYFKMKGDYYRYISEF-SNEEGKNLVENCL 122
KIC IL +L L+P + +ES+VFY KMKGDY+RY++EF + E K E+ L
Sbjct 97 KICDGILNVLEAHLIPSASPAESKVFYLKMKGDYHRYLAEFKAGAERKEAAESTL 151
Score = 41.2 bits (95), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 18/22 (81%), Positives = 21/22 (95%), Gaps = 0/22 (0%)
Query 134 DELTVEERNLLSVAYKNAVGAR 155
+EL+VEERNLLSVAYKN +GAR
Sbjct 37 EELSVEERNLLSVAYKNVIGAR 58
> mmu:22627 Ywhae, AU019196; tyrosine 3-monooxygenase/tryptophan
5-monooxygenase activation protein, epsilon polypeptide;
K06630 tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation
protein
Length=255
Score = 132 bits (332), Expect = 5e-31, Method: Compositional matrix adjust.
Identities = 67/119 (56%), Positives = 81/119 (68%), Gaps = 1/119 (0%)
Query 5 GGKGDELTVEERNLLSVAYKNAVGARRASWRIISSVEQKEANRNHMTNKGLAASYRQKVE 64
G ELTVEERNLLSVAYKN +GARRASWRIISS+EQKE N+ + YRQ VE
Sbjct 31 AGMDVELTVEERNLLSVAYKNVIGARRASWRIISSIEQKEENKGGEDKLKMIREYRQMVE 90
Query 65 NELNKICQEILTLLTDKLLPRTTDSESRVFYFKMKGDYYRYISEFSN-EEGKNLVENCL 122
EL IC +IL +L L+P ES+VFY+KMKGDY+RY++EF+ + K EN L
Sbjct 91 TELKLICCDILDVLDKHLIPAANTGESKVFYYKMKGDYHRYLAEFATGNDRKEAAENSL 149
Score = 42.7 bits (99), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 20/26 (76%), Positives = 21/26 (80%), Gaps = 0/26 (0%)
Query 130 GGKGDELTVEERNLLSVAYKNAVGAR 155
G ELTVEERNLLSVAYKN +GAR
Sbjct 31 AGMDVELTVEERNLLSVAYKNVIGAR 56
> hsa:7531 YWHAE, 14-3-3E, FLJ45465, FLJ53559, KCIP-1, MDCR, MDS;
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation
protein, epsilon polypeptide; K06630 tyrosine 3-monooxygenase/tryptophan
5-monooxygenase activation protein
Length=255
Score = 132 bits (332), Expect = 5e-31, Method: Compositional matrix adjust.
Identities = 67/119 (56%), Positives = 81/119 (68%), Gaps = 1/119 (0%)
Query 5 GGKGDELTVEERNLLSVAYKNAVGARRASWRIISSVEQKEANRNHMTNKGLAASYRQKVE 64
G ELTVEERNLLSVAYKN +GARRASWRIISS+EQKE N+ + YRQ VE
Sbjct 31 AGMDVELTVEERNLLSVAYKNVIGARRASWRIISSIEQKEENKGGEDKLKMIREYRQMVE 90
Query 65 NELNKICQEILTLLTDKLLPRTTDSESRVFYFKMKGDYYRYISEFSN-EEGKNLVENCL 122
EL IC +IL +L L+P ES+VFY+KMKGDY+RY++EF+ + K EN L
Sbjct 91 TELKLICCDILDVLDKHLIPAANTGESKVFYYKMKGDYHRYLAEFATGNDRKEAAENSL 149
Score = 42.7 bits (99), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 20/26 (76%), Positives = 21/26 (80%), Gaps = 0/26 (0%)
Query 130 GGKGDELTVEERNLLSVAYKNAVGAR 155
G ELTVEERNLLSVAYKN +GAR
Sbjct 31 AGMDVELTVEERNLLSVAYKNVIGAR 56
> ath:AT1G34760 GRF11; GRF11 (GENERAL REGULATORY FACTOR 11); ATPase
binding / amino acid binding / protein binding / protein
phosphorylated amino acid binding
Length=252
Score = 131 bits (329), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 60/100 (60%), Positives = 76/100 (76%), Gaps = 0/100 (0%)
Query 10 ELTVEERNLLSVAYKNAVGARRASWRIISSVEQKEANRNHMTNKGLAASYRQKVENELNK 69
ELT+EERNLLSV YKN +GARRASWRI+SS+EQKE ++ + N YR KVE EL+K
Sbjct 37 ELTIEERNLLSVGYKNVIGARRASWRILSSIEQKEESKGNEQNAKRIKDYRTKVEEELSK 96
Query 70 ICQEILTLLTDKLLPRTTDSESRVFYFKMKGDYYRYISEF 109
IC +IL ++ L+P T ES VFY+KMKGDY+RY++EF
Sbjct 97 ICYDILAVIDKHLVPFATSGESTVFYYKMKGDYFRYLAEF 136
Score = 39.7 bits (91), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 17/21 (80%), Positives = 19/21 (90%), Gaps = 0/21 (0%)
Query 135 ELTVEERNLLSVAYKNAVGAR 155
ELT+EERNLLSV YKN +GAR
Sbjct 37 ELTIEERNLLSVGYKNVIGAR 57
> cel:M117.2 par-5; abnormal embryonic PARtitioning of cytoplasm
family member (par-5); K06630 tyrosine 3-monooxygenase/tryptophan
5-monooxygenase activation protein
Length=248
Score = 130 bits (326), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 64/113 (56%), Positives = 83/113 (73%), Gaps = 2/113 (1%)
Query 7 KGDELTVEERNLLSVAYKNAVGARRASWRIISSVEQKEANRNHMTNKGLAASYRQKVENE 66
+G EL+ EERNLLSVAYKN VGARR+SWR+ISS+EQK + LA YR KVE E
Sbjct 34 QGQELSNEERNLLSVAYKNVVGARRSSWRVISSIEQK--TEGSEKKQQLAKEYRVKVEQE 91
Query 67 LNKICQEILTLLTDKLLPRTTDSESRVFYFKMKGDYYRYISEFSNEEGKNLVE 119
LN ICQ++L LL + L+ + +ES+VFY KMKGDYYRY++E ++E+ +VE
Sbjct 92 LNDICQDVLKLLDEFLIVKAGAAESKVFYLKMKGDYYRYLAEVASEDRAAVVE 144
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 19/24 (79%), Positives = 21/24 (87%), Gaps = 0/24 (0%)
Query 132 KGDELTVEERNLLSVAYKNAVGAR 155
+G EL+ EERNLLSVAYKN VGAR
Sbjct 34 QGQELSNEERNLLSVAYKNVVGAR 57
> ath:AT1G22300 GRF10; GRF10 (GENERAL REGULATORY FACTOR 10); ATP
binding / protein binding / protein phosphorylated amino
acid binding
Length=254
Score = 129 bits (323), Expect = 5e-30, Method: Compositional matrix adjust.
Identities = 58/102 (56%), Positives = 81/102 (79%), Gaps = 0/102 (0%)
Query 10 ELTVEERNLLSVAYKNAVGARRASWRIISSVEQKEANRNHMTNKGLAASYRQKVENELNK 69
ELTVEERNL+SV YKN +GARRASWRI+SS+EQKE ++ + N +YR++VE+EL K
Sbjct 37 ELTVEERNLVSVGYKNVIGARRASWRILSSIEQKEESKGNDENVKRLKNYRKRVEDELAK 96
Query 70 ICQEILTLLTDKLLPRTTDSESRVFYFKMKGDYYRYISEFSN 111
+C +IL+++ L+P + ES VF++KMKGDYYRY++EFS+
Sbjct 97 VCNDILSVIDKHLIPSSNAVESTVFFYKMKGDYYRYLAEFSS 138
Score = 38.9 bits (89), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 17/21 (80%), Positives = 19/21 (90%), Gaps = 0/21 (0%)
Query 135 ELTVEERNLLSVAYKNAVGAR 155
ELTVEERNL+SV YKN +GAR
Sbjct 37 ELTVEERNLVSVGYKNVIGAR 57
> dre:399487 ywhaqa, ywhaq, ywhaq2; tyrosine 3-monooxygenase/tryptophan
5-monooxygenase activation protein, theta polypeptide
a; K06630 tyrosine 3-monooxygenase/tryptophan 5-monooxygenase
activation protein
Length=245
Score = 128 bits (321), Expect = 8e-30, Method: Compositional matrix adjust.
Identities = 66/138 (47%), Positives = 94/138 (68%), Gaps = 10/138 (7%)
Query 7 KGDELTVEERNLLSVAYKNAVGARRASWRIISSVEQK-EANRNHMTNKGLAASYRQKVEN 65
+G+EL+ EERNLLSVAYKN VGARR++WR+ISS+EQK E N + + YR+KVE
Sbjct 32 QGEELSNEERNLLSVAYKNVVGARRSAWRVISSIEQKTEGNDKKLQ---MVKEYREKVEG 88
Query 66 ELNKICQEILTLLTDKLLPRTTDSESRVFYFKMKGDYYRYISEFSNEEGKNLVENCLDQN 125
EL IC E+LTLL L+ +T+SES+VFY KMKGDYYRY++E + + K +D
Sbjct 89 ELRDICNEVLTLLGKYLIKNSTNSESKVFYLKMKGDYYRYLAEVAAADDK------MDTI 142
Query 126 SSTPGGKGDELTVEERNL 143
+++ G D + ++++
Sbjct 143 TNSQGAYQDAFEISKKDM 160
Score = 41.2 bits (95), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 19/24 (79%), Positives = 22/24 (91%), Gaps = 0/24 (0%)
Query 132 KGDELTVEERNLLSVAYKNAVGAR 155
+G+EL+ EERNLLSVAYKN VGAR
Sbjct 32 QGEELSNEERNLLSVAYKNVVGAR 55
> cel:F52D10.3 ftt-2; Fourteen-Three-Three family member (ftt-2);
K06630 tyrosine 3-monooxygenase/tryptophan 5-monooxygenase
activation protein
Length=248
Score = 127 bits (320), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 62/111 (55%), Positives = 81/111 (72%), Gaps = 2/111 (1%)
Query 8 GDELTVEERNLLSVAYKNAVGARRASWRIISSVEQKEANRNHMTNKGLAASYRQKVENEL 67
G EL+ EERNLLSVAYKN VGARR+SWR+ISS+EQK + +A YR+KVE EL
Sbjct 35 GAELSNEERNLLSVAYKNVVGARRSSWRVISSIEQK--TEGSEKKQQMAKEYREKVEKEL 92
Query 68 NKICQEILTLLTDKLLPRTTDSESRVFYFKMKGDYYRYISEFSNEEGKNLV 118
ICQ++L LL L+P+ +ES+VFY KMKGDYYRY++E ++ + +N V
Sbjct 93 RDICQDVLNLLDKFLIPKAGAAESKVFYLKMKGDYYRYLAEVASGDDRNSV 143
Score = 39.3 bits (90), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 19/23 (82%), Positives = 20/23 (86%), Gaps = 0/23 (0%)
Query 133 GDELTVEERNLLSVAYKNAVGAR 155
G EL+ EERNLLSVAYKN VGAR
Sbjct 35 GAELSNEERNLLSVAYKNVVGAR 57
> xla:380457 ywhab-b, 14-3-3beta, MGC53093, ywhab; tyrosine 3-monooxygenase/tryptophan
5-monooxygenase activation protein,
beta polypeptide; K06630 tyrosine 3-monooxygenase/tryptophan
5-monooxygenase activation protein
Length=244
Score = 126 bits (317), Expect = 3e-29, Method: Compositional matrix adjust.
Identities = 62/108 (57%), Positives = 79/108 (73%), Gaps = 2/108 (1%)
Query 8 GDELTVEERNLLSVAYKNAVGARRASWRIISSVEQKEANRNHMTNKGLAASYRQKVENEL 67
G EL+ EERNLLSVAYKN VGARR+SWR+ISS+EQK + + +A YR+KVE EL
Sbjct 33 GAELSNEERNLLSVAYKNVVGARRSSWRVISSIEQKTEGNDK--RQQMAREYREKVETEL 90
Query 68 NKICQEILTLLTDKLLPRTTDSESRVFYFKMKGDYYRYISEFSNEEGK 115
IC+++L LL L+P T ES+VFY KMKGDYYRY+SE ++ + K
Sbjct 91 QDICKDVLDLLDRFLVPNATPPESKVFYLKMKGDYYRYLSEVASGDSK 138
Score = 39.7 bits (91), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 19/23 (82%), Positives = 20/23 (86%), Gaps = 0/23 (0%)
Query 133 GDELTVEERNLLSVAYKNAVGAR 155
G EL+ EERNLLSVAYKN VGAR
Sbjct 33 GAELSNEERNLLSVAYKNVVGAR 55
> xla:399276 ywhab-a, 14-3-3beta, MGC78918; tyrosine 3-monooxygenase/tryptophan
5-monooxygenase activation protein, beta polypeptide;
K06630 tyrosine 3-monooxygenase/tryptophan 5-monooxygenase
activation protein
Length=244
Score = 126 bits (317), Expect = 3e-29, Method: Compositional matrix adjust.
Identities = 62/108 (57%), Positives = 79/108 (73%), Gaps = 2/108 (1%)
Query 8 GDELTVEERNLLSVAYKNAVGARRASWRIISSVEQKEANRNHMTNKGLAASYRQKVENEL 67
G EL+ EERNLLSVAYKN VGARR+SWR+ISS+EQK + + +A YR+KVE EL
Sbjct 33 GAELSNEERNLLSVAYKNVVGARRSSWRVISSIEQKTEGNDK--RQQMAREYREKVETEL 90
Query 68 NKICQEILTLLTDKLLPRTTDSESRVFYFKMKGDYYRYISEFSNEEGK 115
IC+++L LL L+P T ES+VFY KMKGDYYRY+SE ++ + K
Sbjct 91 QDICKDVLDLLDRFLVPNATPPESKVFYLKMKGDYYRYLSEVASGDSK 138
Score = 39.7 bits (91), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 19/23 (82%), Positives = 20/23 (86%), Gaps = 0/23 (0%)
Query 133 GDELTVEERNLLSVAYKNAVGAR 155
G EL+ EERNLLSVAYKN VGAR
Sbjct 33 GAELSNEERNLLSVAYKNVVGAR 55
> dre:321729 ywhabl, hm:zehn0976, wu:fb33b01, zgc:73065, zgc:77351;
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation
protein, beta polypeptide like; K06630 tyrosine 3-monooxygenase/tryptophan
5-monooxygenase activation protein
Length=245
Score = 125 bits (315), Expect = 4e-29, Method: Compositional matrix adjust.
Identities = 61/113 (53%), Positives = 83/113 (73%), Gaps = 3/113 (2%)
Query 7 KGD-ELTVEERNLLSVAYKNAVGARRASWRIISSVEQKEANRNHMTNKGLAASYRQKVEN 65
+GD EL+ EERNLLSVAYKN VGARR+SWR++SS+EQK + + + YR+K+E
Sbjct 31 EGDIELSNEERNLLSVAYKNVVGARRSSWRVVSSIEQKMEGSDK--KQQMVKEYREKIEK 88
Query 66 ELNKICQEILTLLTDKLLPRTTDSESRVFYFKMKGDYYRYISEFSNEEGKNLV 118
EL +IC ++L LL L+P+ T +ESRVFY KMKGDY+RY++E + E KN +
Sbjct 89 ELKEICNDVLVLLDKYLIPKATPAESRVFYLKMKGDYFRYLAEVAVGEEKNSI 141
Score = 38.1 bits (87), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 20/25 (80%), Positives = 22/25 (88%), Gaps = 1/25 (4%)
Query 132 KGD-ELTVEERNLLSVAYKNAVGAR 155
+GD EL+ EERNLLSVAYKN VGAR
Sbjct 31 EGDIELSNEERNLLSVAYKNVVGAR 55
> hsa:7529 YWHAB, GW128, HS1, KCIP-1, YWHAA; tyrosine 3-monooxygenase/tryptophan
5-monooxygenase activation protein, beta
polypeptide; K06630 tyrosine 3-monooxygenase/tryptophan 5-monooxygenase
activation protein
Length=246
Score = 125 bits (313), Expect = 7e-29, Method: Compositional matrix adjust.
Identities = 61/109 (55%), Positives = 79/109 (72%), Gaps = 2/109 (1%)
Query 7 KGDELTVEERNLLSVAYKNAVGARRASWRIISSVEQKEANRNHMTNKGLAASYRQKVENE 66
+G EL+ EERNLLSVAYKN VGARR+SWR+ISS+EQK RN + + YR+K+E E
Sbjct 34 QGHELSNEERNLLSVAYKNVVGARRSSWRVISSIEQK-TERNE-KKQQMGKEYREKIEAE 91
Query 67 LNKICQEILTLLTDKLLPRTTDSESRVFYFKMKGDYYRYISEFSNEEGK 115
L IC ++L LL L+P T ES+VFY KMKGDY+RY+SE ++ + K
Sbjct 92 LQDICNDVLELLDKYLIPNATQPESKVFYLKMKGDYFRYLSEVASGDNK 140
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 19/24 (79%), Positives = 21/24 (87%), Gaps = 0/24 (0%)
Query 132 KGDELTVEERNLLSVAYKNAVGAR 155
+G EL+ EERNLLSVAYKN VGAR
Sbjct 34 QGHELSNEERNLLSVAYKNVVGAR 57
> dre:323055 ywhaba, fb80c08, fk41f08, wu:fb80c08, wu:fd11d01,
wu:fk41f08, ywhab1; tyrosine 3-monooxygenase/tryptophan 5-monooxygenase
activation protein, beta polypeptide a; K06630
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation
protein
Length=244
Score = 125 bits (313), Expect = 7e-29, Method: Compositional matrix adjust.
Identities = 65/115 (56%), Positives = 84/115 (73%), Gaps = 5/115 (4%)
Query 8 GDELTVEERNLLSVAYKNAVGARRASWRIISSVEQK-EANRNHMTNKGLAASYRQKVENE 66
G EL+ EERNLLSVAYKN VGARR+SWR+ISS+EQK E N + +A YR+K+E E
Sbjct 33 GVELSNEERNLLSVAYKNVVGARRSSWRVISSIEQKTEGNEKK---QQMAREYREKIEAE 89
Query 67 LNKICQEILTLLTDKLLPRTTDSESRVFYFKMKGDYYRYISEFSNEEGK-NLVEN 120
L +IC ++L LL L+P + +ES+VFY KMKGDYYRY+SE ++ + K VEN
Sbjct 90 LQEICNDVLGLLEKYLIPNASQAESKVFYLKMKGDYYRYLSEVASGDSKRTTVEN 144
Score = 38.9 bits (89), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 19/23 (82%), Positives = 20/23 (86%), Gaps = 0/23 (0%)
Query 133 GDELTVEERNLLSVAYKNAVGAR 155
G EL+ EERNLLSVAYKN VGAR
Sbjct 33 GVELSNEERNLLSVAYKNVVGAR 55
> ath:AT5G16050 GRF5; GRF5 (GENERAL REGULATORY FACTOR 5); ATP
binding / protein binding / protein phosphorylated amino acid
binding
Length=268
Score = 124 bits (312), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 61/108 (56%), Positives = 78/108 (72%), Gaps = 1/108 (0%)
Query 16 RNLLSVAYKNAVGARRASWRIISSVEQKEANRNHMTNKGLAASYRQKVENELNKICQEIL 75
RNLLSVAYKN +GARRASWRIISS+EQKE +R + + + YR K+E EL+KIC IL
Sbjct 47 RNLLSVAYKNVIGARRASWRIISSIEQKEDSRGNSDHVSIIKDYRGKIETELSKICDGIL 106
Query 76 TLLTDKLLPRTTDSESRVFYFKMKGDYYRYISEF-SNEEGKNLVENCL 122
LL L+P + +ES+VFY KMKGDY+RY++EF + E K E+ L
Sbjct 107 NLLEAHLIPAASLAESKVFYLKMKGDYHRYLAEFKTGAERKEAAESTL 154
Score = 31.6 bits (70), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 13/15 (86%), Positives = 14/15 (93%), Gaps = 0/15 (0%)
Query 141 RNLLSVAYKNAVGAR 155
RNLLSVAYKN +GAR
Sbjct 47 RNLLSVAYKNVIGAR 61
> hsa:7534 YWHAZ, 14-3-3-zeta, KCIP-1, MGC111427, MGC126532, MGC138156,
YWHAD; tyrosine 3-monooxygenase/tryptophan 5-monooxygenase
activation protein, zeta polypeptide; K06630 tyrosine
3-monooxygenase/tryptophan 5-monooxygenase activation protein
Length=245
Score = 123 bits (308), Expect = 3e-28, Method: Compositional matrix adjust.
Identities = 57/104 (54%), Positives = 77/104 (74%), Gaps = 2/104 (1%)
Query 7 KGDELTVEERNLLSVAYKNAVGARRASWRIISSVEQKEANRNHMTNKGLAASYRQKVENE 66
+G EL+ EERNLLSVAYKN VGARR+SWR++SS+EQK + +A YR+K+E E
Sbjct 32 QGAELSNEERNLLSVAYKNVVGARRSSWRVVSSIEQK--TEGAEKKQQMAREYREKIETE 89
Query 67 LNKICQEILTLLTDKLLPRTTDSESRVFYFKMKGDYYRYISEFS 110
L IC ++L+LL L+P + +ES+VFY KMKGDYYRY++E +
Sbjct 90 LRDICNDVLSLLEKFLIPNASQAESKVFYLKMKGDYYRYLAEVA 133
Score = 40.0 bits (92), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 19/24 (79%), Positives = 21/24 (87%), Gaps = 0/24 (0%)
Query 132 KGDELTVEERNLLSVAYKNAVGAR 155
+G EL+ EERNLLSVAYKN VGAR
Sbjct 32 QGAELSNEERNLLSVAYKNVVGAR 55
> ath:AT2G42590 GRF9; GRF9 (GENERAL REGULATORY FACTOR 9); calcium
ion binding / protein binding / protein phosphorylated amino
acid binding
Length=262
Score = 123 bits (308), Expect = 3e-28, Method: Compositional matrix adjust.
Identities = 57/100 (57%), Positives = 76/100 (76%), Gaps = 0/100 (0%)
Query 10 ELTVEERNLLSVAYKNAVGARRASWRIISSVEQKEANRNHMTNKGLAASYRQKVENELNK 69
+LTVEERNLLSV YKN +G+RRASWRI SS+EQKEA + + N Y +KVE EL+
Sbjct 39 DLTVEERNLLSVGYKNVIGSRRASWRIFSSIEQKEAVKGNDVNVKRIKEYMEKVELELSN 98
Query 70 ICQEILTLLTDKLLPRTTDSESRVFYFKMKGDYYRYISEF 109
IC +I+++L + L+P ++ ES VF+ KMKGDYYRY++EF
Sbjct 99 ICIDIMSVLDEHLIPSASEGESTVFFNKMKGDYYRYLAEF 138
Score = 37.7 bits (86), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 16/21 (76%), Positives = 19/21 (90%), Gaps = 0/21 (0%)
Query 135 ELTVEERNLLSVAYKNAVGAR 155
+LTVEERNLLSV YKN +G+R
Sbjct 39 DLTVEERNLLSVGYKNVIGSR 59
> dre:100334119 14-3-3 protein beta/alpha-like
Length=242
Score = 123 bits (308), Expect = 3e-28, Method: Compositional matrix adjust.
Identities = 65/115 (56%), Positives = 82/115 (71%), Gaps = 5/115 (4%)
Query 8 GDELTVEERNLLSVAYKNAVGARRASWRIISSVEQK-EANRNHMTNKGLAASYRQKVENE 66
G EL+ EERNLLSVAYKN VGARR+SWR+ISS+EQK E N + +A YR+K+E E
Sbjct 33 GVELSNEERNLLSVAYKNVVGARRSSWRVISSIEQKTEGNEKK---QQMAREYREKIETE 89
Query 67 LNKICQEILTLLTDKLLPRTTDSESRVFYFKMKGDYYRYISEFSNEEGK-NLVEN 120
L IC ++L LL L+ + +ES+VFY KMKGDYYRY+SE ++ E K VEN
Sbjct 90 LQDICSDVLGLLEKYLIANASQAESKVFYLKMKGDYYRYLSEVASGESKATTVEN 144
Score = 38.9 bits (89), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 19/23 (82%), Positives = 20/23 (86%), Gaps = 0/23 (0%)
Query 133 GDELTVEERNLLSVAYKNAVGAR 155
G EL+ EERNLLSVAYKN VGAR
Sbjct 33 GVELSNEERNLLSVAYKNVVGAR 55
> mmu:54401 Ywhab, 1300003C17Rik; tyrosine 3-monooxygenase/tryptophan
5-monooxygenase activation protein, beta polypeptide;
K06630 tyrosine 3-monooxygenase/tryptophan 5-monooxygenase
activation protein
Length=246
Score = 122 bits (307), Expect = 4e-28, Method: Compositional matrix adjust.
Identities = 61/109 (55%), Positives = 79/109 (72%), Gaps = 2/109 (1%)
Query 7 KGDELTVEERNLLSVAYKNAVGARRASWRIISSVEQKEANRNHMTNKGLAASYRQKVENE 66
+G EL+ EERNLLSVAYKN VGARR+SWR+ISS+EQK RN + + YR+K+E E
Sbjct 34 QGHELSNEERNLLSVAYKNVVGARRSSWRVISSIEQK-TERNE-KKQQMGKEYREKIEAE 91
Query 67 LNKICQEILTLLTDKLLPRTTDSESRVFYFKMKGDYYRYISEFSNEEGK 115
L IC ++L LL L+ T +ES+VFY KMKGDY+RY+SE ++ E K
Sbjct 92 LQDICNDVLELLDKYLILNATQAESKVFYLKMKGDYFRYLSEVASGENK 140
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 19/24 (79%), Positives = 21/24 (87%), Gaps = 0/24 (0%)
Query 132 KGDELTVEERNLLSVAYKNAVGAR 155
+G EL+ EERNLLSVAYKN VGAR
Sbjct 34 QGHELSNEERNLLSVAYKNVVGAR 57
> xla:380064 ywhaq, 14-3-3t, MGC53015; tyrosine 3-monooxygenase/tryptophan
5-monooxygenase activation protein, theta polypeptide;
K06630 tyrosine 3-monooxygenase/tryptophan 5-monooxygenase
activation protein
Length=246
Score = 122 bits (307), Expect = 4e-28, Method: Compositional matrix adjust.
Identities = 59/108 (54%), Positives = 80/108 (74%), Gaps = 2/108 (1%)
Query 8 GDELTVEERNLLSVAYKNAVGARRASWRIISSVEQKEANRNHMTNKGLAASYRQKVENEL 67
G EL+ EERNLLSVAYKN VGARR++WR+ISS+EQK + + T +A Y++KVE+EL
Sbjct 33 GGELSSEERNLLSVAYKNVVGARRSAWRVISSIEQK--SDSEETKLKIAKEYKEKVESEL 90
Query 68 NKICQEILTLLTDKLLPRTTDSESRVFYFKMKGDYYRYISEFSNEEGK 115
IC+ +L LL L+ +T +ES+VFY KMKGDYYRY++E + E +
Sbjct 91 QSICETVLNLLDKHLISASTATESQVFYLKMKGDYYRYLAEVATGENR 138
Score = 40.4 bits (93), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 19/23 (82%), Positives = 20/23 (86%), Gaps = 0/23 (0%)
Query 133 GDELTVEERNLLSVAYKNAVGAR 155
G EL+ EERNLLSVAYKN VGAR
Sbjct 33 GGELSSEERNLLSVAYKNVVGAR 55
> dre:336610 ywhai, fb14h09, wu:fb05g08, wu:fb14h09, zgc:55807;
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation
protein, iota polypeptide; K06630 tyrosine 3-monooxygenase/tryptophan
5-monooxygenase activation protein
Length=229
Score = 122 bits (307), Expect = 4e-28, Method: Compositional matrix adjust.
Identities = 59/113 (52%), Positives = 83/113 (73%), Gaps = 4/113 (3%)
Query 8 GDELTVEERNLLSVAYKNAVGARRASWRIISSVEQKEANRNHMTNKGLAASYRQKVENEL 67
G+EL+ EERNLLSVAYKN VGARR++WR++SS+EQK + K +A YR+K+E EL
Sbjct 33 GEELSDEERNLLSVAYKNVVGARRSAWRVVSSIEQKTEGTD---KKQMAQEYREKIEAEL 89
Query 68 NKICQEILTLLTDKLLPRTTDSESRVFYFKMKGDYYRYISEFSN-EEGKNLVE 119
IC ++L LL L+ T+ +ES+VFY KMKGDYYRY++E + +E N+++
Sbjct 90 KAICNDVLHLLDKFLIRSTSPAESQVFYLKMKGDYYRYLAEVATGDEKTNIIQ 142
Score = 41.2 bits (95), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 19/23 (82%), Positives = 21/23 (91%), Gaps = 0/23 (0%)
Query 133 GDELTVEERNLLSVAYKNAVGAR 155
G+EL+ EERNLLSVAYKN VGAR
Sbjct 33 GEELSDEERNLLSVAYKNVVGAR 55
> mmu:22631 Ywhaz, 1110013I11Rik, 14-3-3zeta, AI596267, AL022924,
AU020854; tyrosine 3-monooxygenase/tryptophan 5-monooxygenase
activation protein, zeta polypeptide; K06630 tyrosine
3-monooxygenase/tryptophan 5-monooxygenase activation protein
Length=245
Score = 122 bits (306), Expect = 4e-28, Method: Compositional matrix adjust.
Identities = 57/104 (54%), Positives = 76/104 (73%), Gaps = 2/104 (1%)
Query 7 KGDELTVEERNLLSVAYKNAVGARRASWRIISSVEQKEANRNHMTNKGLAASYRQKVENE 66
+G EL+ EERNLLSVAYKN VGARR+SWR++SS+EQK + +A YR+K+E E
Sbjct 32 QGAELSNEERNLLSVAYKNVVGARRSSWRVVSSIEQK--TEGAEKKQQMAREYREKIETE 89
Query 67 LNKICQEILTLLTDKLLPRTTDSESRVFYFKMKGDYYRYISEFS 110
L IC ++L+LL L+P + ES+VFY KMKGDYYRY++E +
Sbjct 90 LRDICNDVLSLLEKFLIPNASQPESKVFYLKMKGDYYRYLAEVA 133
Score = 40.0 bits (92), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 19/24 (79%), Positives = 21/24 (87%), Gaps = 0/24 (0%)
Query 132 KGDELTVEERNLLSVAYKNAVGAR 155
+G EL+ EERNLLSVAYKN VGAR
Sbjct 32 QGAELSNEERNLLSVAYKNVVGAR 55
> dre:406419 ywhabb, wu:fb22g04, wu:fr86a12, ywhab, ywhab2, zgc:64092;
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase
activation protein, beta polypeptide b; K06630 tyrosine 3-monooxygenase/tryptophan
5-monooxygenase activation protein
Length=242
Score = 121 bits (304), Expect = 7e-28, Method: Compositional matrix adjust.
Identities = 64/115 (55%), Positives = 82/115 (71%), Gaps = 5/115 (4%)
Query 8 GDELTVEERNLLSVAYKNAVGARRASWRIISSVEQK-EANRNHMTNKGLAASYRQKVENE 66
G EL+ EERNLLSVAYKN VGARR+SWR+ISS+EQK E N + +A YR+K+E E
Sbjct 33 GVELSNEERNLLSVAYKNVVGARRSSWRVISSIEQKTEGNEKK---QQMAREYREKIETE 89
Query 67 LNKICQEILTLLTDKLLPRTTDSESRVFYFKMKGDYYRYISEFSNEEGK-NLVEN 120
L IC ++L LL L+ + +ES+VFY KMKGDYYRY+SE ++ + K VEN
Sbjct 90 LQDICSDVLGLLEKYLIANASQAESKVFYLKMKGDYYRYLSEVASGDSKATTVEN 144
Score = 38.9 bits (89), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 19/23 (82%), Positives = 20/23 (86%), Gaps = 0/23 (0%)
Query 133 GDELTVEERNLLSVAYKNAVGAR 155
G EL+ EERNLLSVAYKN VGAR
Sbjct 33 GVELSNEERNLLSVAYKNVVGAR 55
> hsa:2810 SFN, YWHAS; stratifin; K06644 stratifin
Length=248
Score = 121 bits (303), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 60/113 (53%), Positives = 78/113 (69%), Gaps = 2/113 (1%)
Query 7 KGDELTVEERNLLSVAYKNAVGARRASWRIISSVEQKEANRNHMTNKGLAA-SYRQKVEN 65
KG+EL+ EERNLLSVAYKN VG +RA+WR++SS+EQK +N KG YR+KVE
Sbjct 32 KGEELSCEERNLLSVAYKNVVGGQRAAWRVLSSIEQK-SNEEGSEEKGPEVREYREKVET 90
Query 66 ELNKICQEILTLLTDKLLPRTTDSESRVFYFKMKGDYYRYISEFSNEEGKNLV 118
EL +C +L LL L+ D+ESRVFY KMKGDYYRY++E + + K +
Sbjct 91 ELQGVCDTVLGLLDSHLIKEAGDAESRVFYLKMKGDYYRYLAEVATGDDKKRI 143
Score = 40.0 bits (92), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 18/24 (75%), Positives = 21/24 (87%), Gaps = 0/24 (0%)
Query 132 KGDELTVEERNLLSVAYKNAVGAR 155
KG+EL+ EERNLLSVAYKN VG +
Sbjct 32 KGEELSCEERNLLSVAYKNVVGGQ 55
> dre:335195 ywhaqb, wu:fa91b01, wu:fb02e12, wu:fb36f06, wu:fk86h02,
ywhaq1, ywhaz, zgc:55499; tyrosine 3-monooxygenase/tryptophan
5-monooxygenase activation protein, theta polypeptide
b; K06630 tyrosine 3-monooxygenase/tryptophan 5-monooxygenase
activation protein
Length=245
Score = 120 bits (301), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 62/118 (52%), Positives = 84/118 (71%), Gaps = 5/118 (4%)
Query 8 GDELTVEERNLLSVAYKNAVGARRASWRIISSVEQK-EANRNHMTNKGLAASYRQKVENE 66
G EL+ EERNLLSVAYKN VGARR++WR+ISS+EQK E N + + YR+KVE+E
Sbjct 33 GSELSNEERNLLSVAYKNVVGARRSAWRVISSIEQKTEGNDKKLQ---MVKEYREKVESE 89
Query 67 LNKICQEILTLLTDKLLPRTTDSESRVFYFKMKGDYYRYISEF-SNEEGKNLVENCLD 123
L IC ++L LL L+ +++ ES+VFY KMKGDYYRY++E + ++ K +EN D
Sbjct 90 LRDICNDVLELLNKYLIENSSNPESKVFYLKMKGDYYRYLAEVAAGDDKKATIENSQD 147
Score = 40.0 bits (92), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 19/23 (82%), Positives = 20/23 (86%), Gaps = 0/23 (0%)
Query 133 GDELTVEERNLLSVAYKNAVGAR 155
G EL+ EERNLLSVAYKN VGAR
Sbjct 33 GSELSNEERNLLSVAYKNVVGAR 55
> mmu:55948 Sfn, Er, Mme1, Ywhas; stratifin; K06644 stratifin
Length=248
Score = 119 bits (299), Expect = 3e-27, Method: Compositional matrix adjust.
Identities = 60/113 (53%), Positives = 78/113 (69%), Gaps = 2/113 (1%)
Query 7 KGDELTVEERNLLSVAYKNAVGARRASWRIISSVEQKEANRNHMTNKGL-AASYRQKVEN 65
KG+EL+ EERNLLSVAYKN VG +RA+WR++SS+EQK +N KG YR+KVE
Sbjct 32 KGEELSCEERNLLSVAYKNVVGGQRAAWRVLSSIEQK-SNEEGSEEKGPEVKEYREKVET 90
Query 66 ELNKICQEILTLLTDKLLPRTTDSESRVFYFKMKGDYYRYISEFSNEEGKNLV 118
EL +C +L LL L+ D+ESRVFY KMKGDYYRY++E + + K +
Sbjct 91 ELRGVCDTVLGLLDSHLIKGAGDAESRVFYLKMKGDYYRYLAEVATGDDKKRI 143
Score = 40.0 bits (92), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 18/24 (75%), Positives = 21/24 (87%), Gaps = 0/24 (0%)
Query 132 KGDELTVEERNLLSVAYKNAVGAR 155
KG+EL+ EERNLLSVAYKN VG +
Sbjct 32 KGEELSCEERNLLSVAYKNVVGGQ 55
> xla:379809 ywhaz-a, 14-3-3z, 14-3-3zeta, MGC52760, ywhaz, ywhaza;
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation
protein, zeta polypeptide; K06630 tyrosine 3-monooxygenase/tryptophan
5-monooxygenase activation protein
Length=245
Score = 117 bits (294), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 57/110 (51%), Positives = 78/110 (70%), Gaps = 2/110 (1%)
Query 7 KGDELTVEERNLLSVAYKNAVGARRASWRIISSVEQKEANRNHMTNKGLAASYRQKVENE 66
+G EL+ EERNLLSVAYKN VGARR+SWR++SS+EQK + ++ YR+K+E E
Sbjct 32 EGGELSNEERNLLSVAYKNVVGARRSSWRVVSSIEQK--TEGAEKKQEMSREYREKIEAE 89
Query 67 LNKICQEILTLLTDKLLPRTTDSESRVFYFKMKGDYYRYISEFSNEEGKN 116
L +IC ++L LL L+ T ES+VFY KMKGDYYRY++E + + K+
Sbjct 90 LREICNDVLNLLDKFLIANATQPESKVFYLKMKGDYYRYLAEVAAGDAKS 139
Score = 39.3 bits (90), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 19/24 (79%), Positives = 21/24 (87%), Gaps = 0/24 (0%)
Query 132 KGDELTVEERNLLSVAYKNAVGAR 155
+G EL+ EERNLLSVAYKN VGAR
Sbjct 32 EGGELSNEERNLLSVAYKNVVGAR 55
> xla:379608 ywhaz-b, 14-3-3z, 14-3-3zeta, MGC64423, ywhaz, ywhazb;
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation
protein, zeta polypeptide; K06630 tyrosine 3-monooxygenase/tryptophan
5-monooxygenase activation protein
Length=245
Score = 117 bits (293), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 56/104 (53%), Positives = 75/104 (72%), Gaps = 2/104 (1%)
Query 7 KGDELTVEERNLLSVAYKNAVGARRASWRIISSVEQKEANRNHMTNKGLAASYRQKVENE 66
+G EL+ EERNLLSVAYKN VGARR+SWR++SS+EQK + ++ YR+K+E E
Sbjct 32 EGGELSNEERNLLSVAYKNVVGARRSSWRVVSSIEQK--TEGAEKKQEMSREYREKIEAE 89
Query 67 LNKICQEILTLLTDKLLPRTTDSESRVFYFKMKGDYYRYISEFS 110
L +IC ++L LL L+ T ES+VFY KMKGDYYRY++E +
Sbjct 90 LREICNDVLNLLDKFLIANATQPESKVFYLKMKGDYYRYLAEVA 133
Score = 39.3 bits (90), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 19/24 (79%), Positives = 21/24 (87%), Gaps = 0/24 (0%)
Query 132 KGDELTVEERNLLSVAYKNAVGAR 155
+G EL+ EERNLLSVAYKN VGAR
Sbjct 32 EGGELSNEERNLLSVAYKNVVGAR 55
> mmu:100039786 Gm2423; predicted gene 2423; K06630 tyrosine 3-monooxygenase/tryptophan
5-monooxygenase activation protein
Length=319
Score = 117 bits (292), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 59/115 (51%), Positives = 79/115 (68%), Gaps = 3/115 (2%)
Query 7 KGDELTVEERNLLSVAYKNAVGARRASWRIISSVEQKEANRNHMTNKGLAASYRQKVENE 66
+G EL+ EERNLLSVAYKN VG RR++WR+ISS+EQK + L YR+KVE+E
Sbjct 106 QGAELSNEERNLLSVAYKNVVGGRRSAWRVISSIEQKTDTSDKKLQ--LIKDYREKVESE 163
Query 67 LNKICQEILTLLTDKLLPRTTDSESRVFYFKMKGDYYRYISEFS-NEEGKNLVEN 120
L IC +L LL L+ T+ ES+VFY KMKGDY+RY++E + ++ K +EN
Sbjct 164 LRSICTTVLELLDKYLIANATNPESKVFYLKMKGDYFRYLAEVACGDDQKQTIEN 218
Score = 38.1 bits (87), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 18/24 (75%), Positives = 20/24 (83%), Gaps = 0/24 (0%)
Query 132 KGDELTVEERNLLSVAYKNAVGAR 155
+G EL+ EERNLLSVAYKN VG R
Sbjct 106 QGAELSNEERNLLSVAYKNVVGGR 129
> dre:560560 ywhag2, ywhagl; 3-monooxygenase/tryptophan 5-monooxygenase
activation protein, gamma polypeptide 2; K06630 tyrosine
3-monooxygenase/tryptophan 5-monooxygenase activation
protein
Length=247
Score = 116 bits (291), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 57/110 (51%), Positives = 78/110 (70%), Gaps = 2/110 (1%)
Query 11 LTVEERNLLSVAYKNAVGARRASWRIISSVEQKEANRNHMTNKGLAASYRQKVENELNKI 70
L+ EERNLLSVAYKN VGARR+SWR+ISS+EQK + + + +YR+K+E EL +
Sbjct 37 LSNEERNLLSVAYKNVVGARRSSWRVISSIEQKTSADGNEKKIEMVRAYREKIEKELEAV 96
Query 71 CQEILTLLTDKLLPRT--TDSESRVFYFKMKGDYYRYISEFSNEEGKNLV 118
CQ++L LL + L+ T ES+VFY KMKGDYYRY++E + E ++ V
Sbjct 97 CQDVLNLLDNFLIKNCSETQHESKVFYLKMKGDYYRYLAEVATGEKRSTV 146
Score = 36.2 bits (82), Expect = 0.042, Method: Compositional matrix adjust.
Identities = 17/20 (85%), Positives = 18/20 (90%), Gaps = 0/20 (0%)
Query 136 LTVEERNLLSVAYKNAVGAR 155
L+ EERNLLSVAYKN VGAR
Sbjct 37 LSNEERNLLSVAYKNVVGAR 56
> mmu:22628 Ywhag, 14-3-3gamma, D7Bwg1348e; tyrosine 3-monooxygenase/tryptophan
5-monooxygenase activation protein, gamma
polypeptide; K06630 tyrosine 3-monooxygenase/tryptophan 5-monooxygenase
activation protein
Length=247
Score = 116 bits (290), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 56/110 (50%), Positives = 80/110 (72%), Gaps = 2/110 (1%)
Query 11 LTVEERNLLSVAYKNAVGARRASWRIISSVEQKEANRNHMTNKGLAASYRQKVENELNKI 70
L+ EERNLLSVAYKN VGARR+SWR+ISS+EQK + + + +YR+K+E EL +
Sbjct 37 LSNEERNLLSVAYKNVVGARRSSWRVISSIEQKTSADGNEKKIEMVRAYREKIEKELEAV 96
Query 71 CQEILTLLTDKLLPRTTDS--ESRVFYFKMKGDYYRYISEFSNEEGKNLV 118
CQ++L+LL + L+ +++ ES+VFY KMKGDYYRY++E + E + V
Sbjct 97 CQDVLSLLDNYLIKNCSETQYESKVFYLKMKGDYYRYLAEVATGEKRATV 146
Score = 36.2 bits (82), Expect = 0.047, Method: Compositional matrix adjust.
Identities = 17/20 (85%), Positives = 18/20 (90%), Gaps = 0/20 (0%)
Query 136 LTVEERNLLSVAYKNAVGAR 155
L+ EERNLLSVAYKN VGAR
Sbjct 37 LSNEERNLLSVAYKNVVGAR 56
> hsa:7532 YWHAG, 14-3-3GAMMA; tyrosine 3-monooxygenase/tryptophan
5-monooxygenase activation protein, gamma polypeptide;
K06630 tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation
protein
Length=247
Score = 116 bits (290), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 56/110 (50%), Positives = 80/110 (72%), Gaps = 2/110 (1%)
Query 11 LTVEERNLLSVAYKNAVGARRASWRIISSVEQKEANRNHMTNKGLAASYRQKVENELNKI 70
L+ EERNLLSVAYKN VGARR+SWR+ISS+EQK + + + +YR+K+E EL +
Sbjct 37 LSNEERNLLSVAYKNVVGARRSSWRVISSIEQKTSADGNEKKIEMVRAYREKIEKELEAV 96
Query 71 CQEILTLLTDKLLPRTTDS--ESRVFYFKMKGDYYRYISEFSNEEGKNLV 118
CQ++L+LL + L+ +++ ES+VFY KMKGDYYRY++E + E + V
Sbjct 97 CQDVLSLLDNYLIKNCSETQYESKVFYLKMKGDYYRYLAEVATGEKRATV 146
Score = 36.2 bits (82), Expect = 0.047, Method: Compositional matrix adjust.
Identities = 17/20 (85%), Positives = 18/20 (90%), Gaps = 0/20 (0%)
Query 136 LTVEERNLLSVAYKNAVGAR 155
L+ EERNLLSVAYKN VGAR
Sbjct 37 LSNEERNLLSVAYKNVVGAR 56
> mmu:22630 Ywhaq, 2700028P07Rik, AA409740, AU021156, MGC118161,
R74690; tyrosine 3-monooxygenase/tryptophan 5-monooxygenase
activation protein, theta polypeptide; K06630 tyrosine 3-monooxygenase/tryptophan
5-monooxygenase activation protein
Length=245
Score = 116 bits (290), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 59/115 (51%), Positives = 79/115 (68%), Gaps = 3/115 (2%)
Query 7 KGDELTVEERNLLSVAYKNAVGARRASWRIISSVEQKEANRNHMTNKGLAASYRQKVENE 66
+G EL+ EERNLLSVAYKN VG RR++WR+ISS+EQK + L YR+KVE+E
Sbjct 32 QGAELSNEERNLLSVAYKNVVGGRRSAWRVISSIEQKTDTSDKKLQ--LIKDYREKVESE 89
Query 67 LNKICQEILTLLTDKLLPRTTDSESRVFYFKMKGDYYRYISEFS-NEEGKNLVEN 120
L IC +L LL L+ T+ ES+VFY KMKGDY+RY++E + ++ K +EN
Sbjct 90 LRSICTTVLELLDKYLIANATNPESKVFYLKMKGDYFRYLAEVACGDDRKQTIEN 144
Score = 38.1 bits (87), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 18/24 (75%), Positives = 20/24 (83%), Gaps = 0/24 (0%)
Query 132 KGDELTVEERNLLSVAYKNAVGAR 155
+G EL+ EERNLLSVAYKN VG R
Sbjct 32 QGAELSNEERNLLSVAYKNVVGGR 55
> mmu:100503129 14-3-3 protein theta-like; K06630 tyrosine 3-monooxygenase/tryptophan
5-monooxygenase activation protein
Length=245
Score = 116 bits (290), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 59/115 (51%), Positives = 79/115 (68%), Gaps = 3/115 (2%)
Query 7 KGDELTVEERNLLSVAYKNAVGARRASWRIISSVEQKEANRNHMTNKGLAASYRQKVENE 66
+G EL+ EERNLLSVAYKN VG RR++WR+ISS+EQK + L YR+KVE+E
Sbjct 32 QGAELSNEERNLLSVAYKNVVGGRRSAWRVISSIEQKTDTSDKKLQ--LIKDYREKVESE 89
Query 67 LNKICQEILTLLTDKLLPRTTDSESRVFYFKMKGDYYRYISEFS-NEEGKNLVEN 120
L IC +L LL L+ T+ ES+VFY KMKGDY+RY++E + ++ K +EN
Sbjct 90 LRSICTTVLELLDKYLIANATNPESKVFYLKMKGDYFRYLAEVACGDDRKQTIEN 144
Lambda K H
0.310 0.128 0.351
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3386671600
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40