bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0891_orf1
Length=185
Score E
Sequences producing significant alignments: (Bits) Value
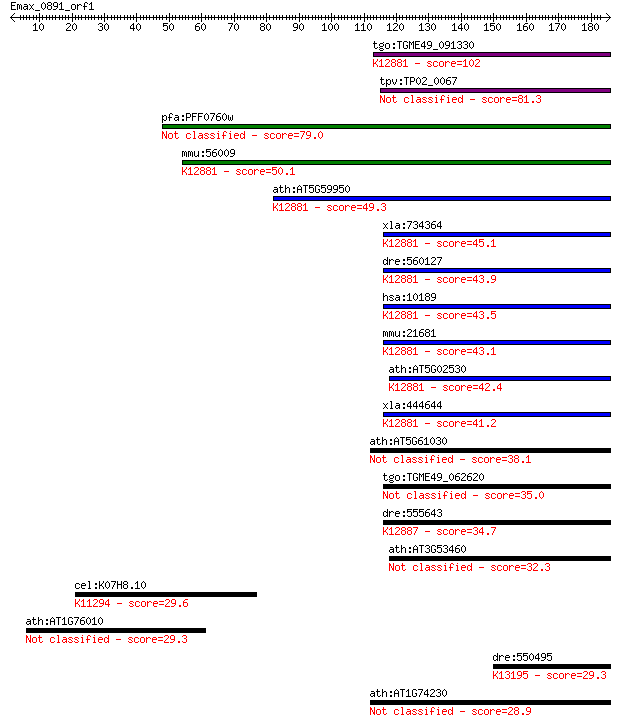
tgo:TGME49_091330 hypothetical protein ; K12881 THO complex su... 102 6e-22
tpv:TP02_0067 hypothetical protein 81.3 2e-15
pfa:PFF0760w RNA and export factor binding protein, putative 79.0 8e-15
mmu:56009 Refbp2, C130042O11Rik, MGC117570, REF2; RNA and expo... 50.1 4e-06
ath:AT5G59950 RNA and export factor-binding protein, putative;... 49.3 8e-06
xla:734364 thoc4, MGC84954, Tho4-A, thoc4-a; THO complex 4; K1... 45.1 1e-04
dre:560127 tho4, MGC171753, zgc:171753; THO complex 4; K12881 ... 43.9 3e-04
hsa:10189 THOC4, ALY, ALY/REF, BEF, REF; THO complex 4; K12881... 43.5 4e-04
mmu:21681 Thoc4, ALY, REF1, Refbp1; THO complex 4; K12881 THO ... 43.1 5e-04
ath:AT5G02530 RNA and export factor-binding protein, putative;... 42.4 0.001
xla:444644 Tho4-B, thoc4-B; MGC84169 protein; K12881 THO compl... 41.2 0.002
ath:AT5G61030 GR-RBP3 (glycine-rich RNA-binding protein 3); AT... 38.1 0.016
tgo:TGME49_062620 Gbp1p protein, putative 35.0 0.13
dre:555643 hnrnpm, fa10g07, si:ch211-197g15.1, wu:fa10g07, wu:... 34.7 0.19
ath:AT3G53460 CP29; CP29; RNA binding / poly(U) binding 32.3 0.88
cel:K07H8.10 hypothetical protein; K11294 nucleolin 29.6 6.3
ath:AT1G76010 nucleic acid binding 29.3 8.2
dre:550495 wu:fb12a12, wu:fb16h02, wu:fb55a06, wu:fc67h01, wu:... 29.3 8.5
ath:AT1G74230 GR-RBP5 (glycine-rich RNA-binding protein 5); AT... 28.9 9.5
> tgo:TGME49_091330 hypothetical protein ; K12881 THO complex
subunit 4
Length=228
Score = 102 bits (255), Expect = 6e-22, Method: Compositional matrix adjust.
Identities = 45/73 (61%), Positives = 62/73 (84%), Gaps = 1/73 (1%)
Query 113 TAIVRISGLDYSVLDSELRELMEKSCEGILKVWIDYDKTDRSRGTGGCVFRTLADAQRAV 172
TA+VR+S LDYSVL+ +L+EL E ++KVWIDYD+TDRS+GTGGC+FR++ DA+RA+
Sbjct 117 TAVVRVSNLDYSVLEEDLKELFAAVGE-VVKVWIDYDRTDRSKGTGGCIFRSVFDAKRAI 175
Query 173 ETFDGRRIEGKPL 185
E ++GRR+EG PL
Sbjct 176 EVYEGRRLEGLPL 188
> tpv:TP02_0067 hypothetical protein
Length=188
Score = 81.3 bits (199), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 36/71 (50%), Positives = 52/71 (73%), Gaps = 1/71 (1%)
Query 115 IVRISGLDYSVLDSELRELMEKSCEGILKVWIDYDKTDRSRGTGGCVFRTLADAQRAVET 174
IVR+S LD+SV+ +L EL S ++KVW+DYD TDRS+GTGGC+++ DA++A+
Sbjct 77 IVRVSNLDHSVMKEDLMELFS-SVGDVVKVWVDYDNTDRSKGTGGCIYKFTDDAKKAISK 135
Query 175 FDGRRIEGKPL 185
F+G IEGK +
Sbjct 136 FNGTVIEGKAV 146
> pfa:PFF0760w RNA and export factor binding protein, putative
Length=156
Score = 79.0 bits (193), Expect = 8e-15, Method: Compositional matrix adjust.
Identities = 44/143 (30%), Positives = 65/143 (45%), Gaps = 23/143 (16%)
Query 48 PGRTRGGGYGGPSP-----YGRSGPYSTGGGGGGMNHWSSNSSSSYGGGGGPNSSGVHWS 102
R R Y S Y + Y T +N Y G G S
Sbjct 10 EHRRRKNNYNNKSHGKYYNYNKHDDYKTNNQHNRIN--------KYRGKG---------S 52
Query 103 WSNGPRGGSSTAIVRISGLDYSVLDSELRELMEKSCEGILKVWIDYDKTDRSRGTGGCVF 162
+ + ++ V+IS LDY++ ++L EL C+ ++ WI+YD TDRS GT CVF
Sbjct 53 YGSNYEERNNEIRVKISNLDYTISKNDLMELFSNVCK-VVNAWINYDHTDRSNGTAVCVF 111
Query 163 RTLADAQRAVETFDGRRIEGKPL 185
+ DAQ+A++ +DG IEG +
Sbjct 112 ENINDAQKAIDKYDGSEIEGLSI 134
> mmu:56009 Refbp2, C130042O11Rik, MGC117570, REF2; RNA and export
factor binding protein 2; K12881 THO complex subunit 4
Length=218
Score = 50.1 bits (118), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 39/133 (29%), Positives = 62/133 (46%), Gaps = 26/133 (19%)
Query 54 GGYGGPSPYGRSGPYSTGGGGGGMNHWSSNSSSSYGGGGGPNSSGVHWSWSNGPRGGSST 113
GG P+PY R P + W + S GGG G +
Sbjct 38 GGRNRPAPYSRPKPLP--------DKWQHDLFDSGCGGG---------------EGVETG 74
Query 114 AIVRISGLDYSVLDSELRELMEKSCEGILK-VWIDYDKTDRSRGTGGCVFRTLADAQRAV 172
A + +S LD+ V D++++EL + G LK +DYD++ RS GT F ADA +A+
Sbjct 75 AKLLVSNLDFGVSDADIQELFAEF--GTLKKAAVDYDRSGRSLGTADVHFERRADALKAM 132
Query 173 ETFDGRRIEGKPL 185
+ + G ++G+P+
Sbjct 133 KQYKGVPLDGRPM 145
> ath:AT5G59950 RNA and export factor-binding protein, putative;
K12881 THO complex subunit 4
Length=211
Score = 49.3 bits (116), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 33/113 (29%), Positives = 60/113 (53%), Gaps = 10/113 (8%)
Query 82 SNSSSSYGGGGGPNSSGVHWSWSN---GPRGGSSTAIVR------ISGLDYSVLDSELRE 132
S S+ Y P S+ H +S+ R G S+A + IS LDY V++ +++E
Sbjct 47 STRSAPYQSAKAPESTWGHDMFSDRSEDHRSGRSSAGIETGTKLYISNLDYGVMNEDIKE 106
Query 133 LMEKSCEGILKVWIDYDKTDRSRGTGGCVFRTLADAQRAVETFDGRRIEGKPL 185
L + E + + + +D++ RS+GT V+ DA AV+ ++ +++GKP+
Sbjct 107 LFAEVGE-LKRYTVHFDRSGRSKGTAEVVYSRRGDALAAVKKYNDVQLDGKPM 158
> xla:734364 thoc4, MGC84954, Tho4-A, thoc4-a; THO complex 4;
K12881 THO complex subunit 4
Length=256
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 22/70 (31%), Positives = 43/70 (61%), Gaps = 1/70 (1%)
Query 116 VRISGLDYSVLDSELRELMEKSCEGILKVWIDYDKTDRSRGTGGCVFRTLADAQRAVETF 175
+ +S LD+ V D++++EL + + K + YD++ RS GT F ADA +A++ +
Sbjct 108 LLVSNLDFGVSDADIQELFAE-FGTLKKAAVHYDRSGRSLGTADVHFERKADALKAMKQY 166
Query 176 DGRRIEGKPL 185
+G ++G+P+
Sbjct 167 NGVPLDGRPM 176
> dre:560127 tho4, MGC171753, zgc:171753; THO complex 4; K12881
THO complex subunit 4
Length=280
Score = 43.9 bits (102), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 24/71 (33%), Positives = 44/71 (61%), Gaps = 3/71 (4%)
Query 116 VRISGLDYSVLDSELRELMEKSCEGILK-VWIDYDKTDRSRGTGGCVFRTLADAQRAVET 174
+ +S LD+ V D++++EL + G LK + YD++ RS GT F ADA +A++
Sbjct 127 LLVSNLDFGVSDADIQELFAEF--GTLKKAAVHYDRSGRSLGTADVHFERKADALKAMKQ 184
Query 175 FDGRRIEGKPL 185
++G ++G+P+
Sbjct 185 YNGVPLDGRPM 195
> hsa:10189 THOC4, ALY, ALY/REF, BEF, REF; THO complex 4; K12881
THO complex subunit 4
Length=264
Score = 43.5 bits (101), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 24/71 (33%), Positives = 44/71 (61%), Gaps = 3/71 (4%)
Query 116 VRISGLDYSVLDSELRELMEKSCEGILK-VWIDYDKTDRSRGTGGCVFRTLADAQRAVET 174
+ +S LD+ V D++++EL + G LK + YD++ RS GT F ADA +A++
Sbjct 115 LLVSNLDFGVSDADIQELFAEF--GTLKKAAVHYDRSGRSLGTADVHFERKADALKAMKQ 172
Query 175 FDGRRIEGKPL 185
++G ++G+P+
Sbjct 173 YNGVPLDGRPM 183
> mmu:21681 Thoc4, ALY, REF1, Refbp1; THO complex 4; K12881 THO
complex subunit 4
Length=255
Score = 43.1 bits (100), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 24/71 (33%), Positives = 44/71 (61%), Gaps = 3/71 (4%)
Query 116 VRISGLDYSVLDSELRELMEKSCEGILK-VWIDYDKTDRSRGTGGCVFRTLADAQRAVET 174
+ +S LD+ V D++++EL + G LK + YD++ RS GT F ADA +A++
Sbjct 107 LLVSNLDFGVSDADIQELFAEF--GTLKKAAVHYDRSGRSLGTADVHFERKADALKAMKQ 164
Query 175 FDGRRIEGKPL 185
++G ++G+P+
Sbjct 165 YNGVPLDGRPM 175
> ath:AT5G02530 RNA and export factor-binding protein, putative;
K12881 THO complex subunit 4
Length=292
Score = 42.4 bits (98), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 27/69 (39%), Positives = 42/69 (60%), Gaps = 3/69 (4%)
Query 118 ISGLDYSVLDSELRELMEKSCEGILKVW-IDYDKTDRSRGTGGCVFRTLADAQRAVETFD 176
IS LDY V + +++EL S G LK + I YD++ RS+GT VF DA AV+ ++
Sbjct 112 ISNLDYGVSNEDIKELF--SEVGDLKRYGIHYDRSGRSKGTAEVVFSRRGDALAAVKRYN 169
Query 177 GRRIEGKPL 185
+++GK +
Sbjct 170 NVQLDGKLM 178
> xla:444644 Tho4-B, thoc4-B; MGC84169 protein; K12881 THO complex
subunit 4
Length=256
Score = 41.2 bits (95), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 21/70 (30%), Positives = 42/70 (60%), Gaps = 1/70 (1%)
Query 116 VRISGLDYSVLDSELRELMEKSCEGILKVWIDYDKTDRSRGTGGCVFRTLADAQRAVETF 175
+ +S LD+ V D++++EL + + K + YD++ RS GT F ADA +A++ +
Sbjct 107 LLVSNLDFGVSDADIQELFAE-FGSLKKAAVHYDRSGRSLGTADVHFERKADALKAMKQY 165
Query 176 DGRRIEGKPL 185
+G ++G+ +
Sbjct 166 NGVPLDGRSM 175
> ath:AT5G61030 GR-RBP3 (glycine-rich RNA-binding protein 3);
ATP binding / RNA binding
Length=309
Score = 38.1 bits (87), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 25/75 (33%), Positives = 41/75 (54%), Gaps = 2/75 (2%)
Query 112 STAIVRISGLDYSVLDSELRELMEKSCEGI-LKVWIDYDKTDRSRGTGGCVFRTLADAQR 170
S++ + I G+ YS+ + LRE K E + +V +D + T RSRG G F + A
Sbjct 38 SSSKLFIGGMAYSMDEDSLREAFTKYGEVVDTRVILDRE-TGRSRGFGFVTFTSSEAASS 96
Query 171 AVETFDGRRIEGKPL 185
A++ DGR + G+ +
Sbjct 97 AIQALDGRDLHGRVV 111
> tgo:TGME49_062620 Gbp1p protein, putative
Length=293
Score = 35.0 bits (79), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 22/70 (31%), Positives = 38/70 (54%), Gaps = 1/70 (1%)
Query 116 VRISGLDYSVLDSELRELMEKSCEGILKVWIDYDKTDRSRGTGGCVFRTLADAQRAVETF 175
V IS L + +L++L + C +++ + RS+G G +F T AQRAVE F
Sbjct 217 VFISNLPWRTSWQDLKDLF-RECGDVVRADVMTMPDGRSKGVGTVLFSTPEGAQRAVEMF 275
Query 176 DGRRIEGKPL 185
+ ++G+P+
Sbjct 276 NEYMLDGRPI 285
> dre:555643 hnrnpm, fa10g07, si:ch211-197g15.1, wu:fa10g07, wu:fa98b02;
heterogeneous nuclear ribonucleoprotein M; K12887
heterogeneous nuclear ribonucleoprotein M
Length=685
Score = 34.7 bits (78), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 21/70 (30%), Positives = 34/70 (48%), Gaps = 0/70 (0%)
Query 116 VRISGLDYSVLDSELRELMEKSCEGILKVWIDYDKTDRSRGTGGCVFRTLADAQRAVETF 175
V +S + Y V L++LM++ + V D +SRG FRT ++AVE
Sbjct 49 VFVSNIPYDVKWQTLKDLMKEKVGEVTYVEHLMDGEGKSRGCAVVEFRTEELMKKAVEKV 108
Query 176 DGRRIEGKPL 185
+ + G+PL
Sbjct 109 NKHVMNGRPL 118
> ath:AT3G53460 CP29; CP29; RNA binding / poly(U) binding
Length=342
Score = 32.3 bits (72), Expect = 0.88, Method: Compositional matrix adjust.
Identities = 25/69 (36%), Positives = 38/69 (55%), Gaps = 2/69 (2%)
Query 118 ISGLDYSVLDSELRELMEKSCEGILKVWIDYDK-TDRSRGTGGCVFRTLADAQRAVETFD 176
+ L ++V ++L +L E S + V + YDK T RSRG G T A+ + A + F+
Sbjct 103 VGNLSFNVDSAQLAQLFE-SAGNVEMVEVIYDKVTGRSRGFGFVTMSTAAEVEAAAQQFN 161
Query 177 GRRIEGKPL 185
G EG+PL
Sbjct 162 GYEFEGRPL 170
> cel:K07H8.10 hypothetical protein; K11294 nucleolin
Length=798
Score = 29.6 bits (65), Expect = 6.3, Method: Composition-based stats.
Identities = 24/65 (36%), Positives = 28/65 (43%), Gaps = 9/65 (13%)
Query 21 GGYRGGRGGG--WNGPYRSYGREG-------TWGPTPGRTRGGGYGGPSPYGRSGPYSTG 71
G +RGG GG W G R G + G RGGG GG +P GR G +
Sbjct 110 GNFRGGDRGGSPWRGGDRGVANRGRGDFSGGSRGGNKFSPRGGGRGGFTPRGRGGNDFSP 169
Query 72 GGGGG 76
GG G
Sbjct 170 RGGRG 174
> ath:AT1G76010 nucleic acid binding
Length=350
Score = 29.3 bits (64), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 31/58 (53%), Positives = 35/58 (60%), Gaps = 9/58 (15%)
Query 6 DGPIRRGGYMQERGEGGYRG---GRGGGWNGPYRSYGREGTWGPTPGRTRGGGYGGPS 60
DGP RGGY +G GY G GR GG++GP S GR G GP+ GR GGY GPS
Sbjct 251 DGPQGRGGYDGPQGRRGYDGPPQGR-GGYDGP--SQGRGGYDGPSQGR---GGYDGPS 302
> dre:550495 wu:fb12a12, wu:fb16h02, wu:fb55a06, wu:fc67h01, wu:fj10g10,
zgc:109887; zgc:112425; K13195 cold-inducible RNA-binding
protein
Length=185
Score = 29.3 bits (64), Expect = 8.5, Method: Compositional matrix adjust.
Identities = 13/36 (36%), Positives = 23/36 (63%), Gaps = 0/36 (0%)
Query 150 KTDRSRGTGGCVFRTLADAQRAVETFDGRRIEGKPL 185
+T+RSRG G F DA+ A+E +G+ ++G+ +
Sbjct 41 ETNRSRGFGFVTFENPDDAKDALEGMNGKSVDGRTI 76
> ath:AT1G74230 GR-RBP5 (glycine-rich RNA-binding protein 5);
ATP binding / RNA binding
Length=289
Score = 28.9 bits (63), Expect = 9.5, Method: Compositional matrix adjust.
Identities = 22/75 (29%), Positives = 40/75 (53%), Gaps = 3/75 (4%)
Query 112 STAIVRISGLDYSVLDSELRELMEKSCEGI-LKVWIDYDKTDRSRGTGGCVFRTLADAQR 170
S++ + + G+ YS + LRE K E + K+ +D +T RSRG F + +A
Sbjct 32 SSSKIFVGGISYSTDEFGLREAFSKYGEVVDAKIIVDR-ETGRSRGFAFVTFTSTEEASN 90
Query 171 AVETFDGRRIEGKPL 185
A++ DG+ + G+ +
Sbjct 91 AMQ-LDGQDLHGRRI 104
Lambda K H
0.313 0.140 0.454
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5106150148
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40