bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0871_orf1
Length=172
Score E
Sequences producing significant alignments: (Bits) Value
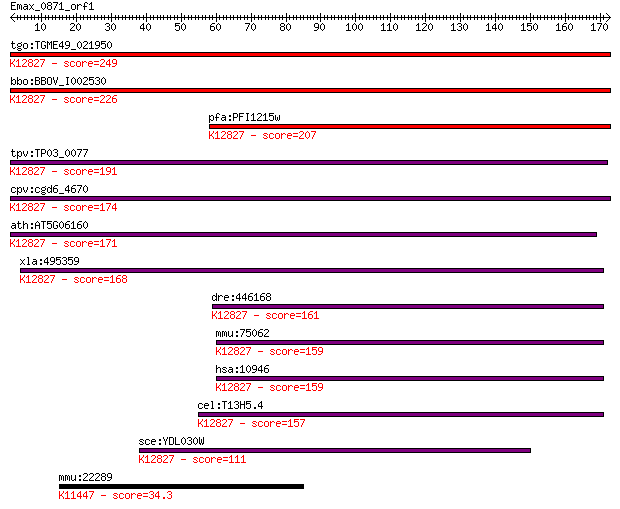
tgo:TGME49_021950 splicing factor 3a protein, putative ; K1282... 249 2e-66
bbo:BBOV_I002530 19.m02395; splicing factor 3a protein; K12827... 226 3e-59
pfa:PFI1215w splicing factor 3A; K12827 splicing factor 3A sub... 207 1e-53
tpv:TP03_0077 splicing factor 3A subunit 3; K12827 splicing fa... 191 1e-48
cpv:cgd6_4670 Prp9p-like splicing factor 3a subunit 3 snRNP. C... 174 1e-43
ath:AT5G06160 ATO; ATO (ATROPOS); nucleic acid binding / zinc ... 171 9e-43
xla:495359 sf3a3; splicing factor 3a, subunit 3, 60kDa; K12827... 168 9e-42
dre:446168 sf3a3, MGC110227, SAP61, SF3a60, wu:fi15a06, wu:fj9... 161 9e-40
mmu:75062 Sf3a3, 4930512K19Rik, 60kDa, MGC103282; splicing fac... 159 3e-39
hsa:10946 SF3A3, PRP9, PRPF9, SAP61, SF3a60; splicing factor 3... 159 4e-39
cel:T13H5.4 hypothetical protein; K12827 splicing factor 3A su... 157 1e-38
sce:YDL030W PRP9; Prp9p; K12827 splicing factor 3A subunit 3 111 1e-24
mmu:22289 Kdm6a, Utx; 4lysine (K)-specific demethylase 6A; K11... 34.3 0.19
> tgo:TGME49_021950 splicing factor 3a protein, putative ; K12827
splicing factor 3A subunit 3
Length=558
Score = 249 bits (637), Expect = 2e-66, Method: Compositional matrix adjust.
Identities = 124/179 (69%), Positives = 146/179 (81%), Gaps = 8/179 (4%)
Query 1 IKYFVDLLQPQINQTIAHYQKKQSTNAEEALEEENDEDSNEEDAAAEE-------DSEAE 53
+ + ++L+ + +T+A + KKQS AEE LEEEN +E+D A + DSE E
Sbjct 370 VNAYKEILEDVVERTVAFHHKKQSRTAEE-LEEENRISDDEDDQQAAKAVGDAERDSEDE 428
Query 54 EEEGPIYNPLNLPLGFDGRPIPYWLYKLHGLGQEFKCEICGNFSYWGRRAFERHFSEWRH 113
E++ PIYNPLNLPLGFDGRPIP+WLYKLHGLGQEFKCEICGNFSYWGRRAFERHF EWRH
Sbjct 429 EDDQPIYNPLNLPLGFDGRPIPFWLYKLHGLGQEFKCEICGNFSYWGRRAFERHFMEWRH 488
Query 114 SFGMRCLKIPNTTHFKEITKIEDAILLYEKLKQQAEGHAFKQDQELECEDAEGNVMSLR 172
+FGMRCL+IPNTTHFKEITKIEDAI LYEKLK+ AEG FK +QELECED++GNVM+LR
Sbjct 489 AFGMRCLRIPNTTHFKEITKIEDAIKLYEKLKKDAEGKTFKDEQELECEDSQGNVMNLR 547
> bbo:BBOV_I002530 19.m02395; splicing factor 3a protein; K12827
splicing factor 3A subunit 3
Length=534
Score = 226 bits (575), Expect = 3e-59, Method: Compositional matrix adjust.
Identities = 110/179 (61%), Positives = 133/179 (74%), Gaps = 8/179 (4%)
Query 1 IKYFVDLLQPQINQTIAHYQKKQSTNAEEALEEENDEDSNEEDAAAEED-------SEAE 53
I+ + L I +TI +K++S N++E LEE ++ EE+ S E
Sbjct 346 IQQYKQFLANTIEKTIEFIEKRESRNSKE-LEESQGLALKILESVGEENNIDIGDISSEE 404
Query 54 EEEGPIYNPLNLPLGFDGRPIPYWLYKLHGLGQEFKCEICGNFSYWGRRAFERHFSEWRH 113
EEE P+YNPLNLPLG+DG+PIP+WLYKLHGLGQEFKCEICGN+SYWGR+AFE HF EWRH
Sbjct 405 EEEQPVYNPLNLPLGWDGKPIPFWLYKLHGLGQEFKCEICGNYSYWGRKAFENHFQEWRH 464
Query 114 SFGMRCLKIPNTTHFKEITKIEDAILLYEKLKQQAEGHAFKQDQELECEDAEGNVMSLR 172
SFGM+CLKIPNT HFKEITKIEDA LYEKLK Q + + FK QE+ECED+EGNVM+ R
Sbjct 465 SFGMKCLKIPNTPHFKEITKIEDAFALYEKLKNQNDKNTFKVAQEVECEDSEGNVMNAR 523
> pfa:PFI1215w splicing factor 3A; K12827 splicing factor 3A subunit
3
Length=589
Score = 207 bits (527), Expect = 1e-53, Method: Compositional matrix adjust.
Identities = 86/115 (74%), Positives = 104/115 (90%), Gaps = 0/115 (0%)
Query 58 PIYNPLNLPLGFDGRPIPYWLYKLHGLGQEFKCEICGNFSYWGRRAFERHFSEWRHSFGM 117
PIYNPLNLPLGFD +PIPYWLYKLHGL +E+KCEICGN+SY+GR FE+HF EWRHSFGM
Sbjct 464 PIYNPLNLPLGFDNKPIPYWLYKLHGLSKEYKCEICGNYSYFGRATFEKHFYEWRHSFGM 523
Query 118 RCLKIPNTTHFKEITKIEDAILLYEKLKQQAEGHAFKQDQELECEDAEGNVMSLR 172
+CLKIPNT HFKEITKIEDA+ LYEKLK++ + + FK DQE+ECED++GNVM+++
Sbjct 524 KCLKIPNTLHFKEITKIEDALNLYEKLKKETQMNIFKPDQEVECEDSKGNVMNIK 578
> tpv:TP03_0077 splicing factor 3A subunit 3; K12827 splicing
factor 3A subunit 3
Length=529
Score = 191 bits (484), Expect = 1e-48, Method: Compositional matrix adjust.
Identities = 101/177 (57%), Positives = 128/177 (72%), Gaps = 9/177 (5%)
Query 1 IKYFVDLLQPQINQTIAHYQKKQSTNAEEALEEENDEDSNEE--DA----AAEEDSEAEE 54
I+ + D+L I +T+ QK++S +E EN + +E DA AAE +SE EE
Sbjct 344 IQSYHDVLASTIEKTVEFVQKRESRTVKEL---ENSQTLAQEILDALETNAAESESEVEE 400
Query 55 EEGPIYNPLNLPLGFDGRPIPYWLYKLHGLGQEFKCEICGNFSYWGRRAFERHFSEWRHS 114
+E P+YNPLNLPLG+DG+PIP+WLYKLHGLGQEFKCEICG SYWGR+AFE HF E+RHS
Sbjct 401 DEKPVYNPLNLPLGWDGKPIPFWLYKLHGLGQEFKCEICGGSSYWGRKAFENHFQEFRHS 460
Query 115 FGMRCLKIPNTTHFKEITKIEDAILLYEKLKQQAEGHAFKQDQELECEDAEGNVMSL 171
FGM+ L IPNT HFKEIT I++A+ LY+KL +A FK E ECED EGN+++L
Sbjct 461 FGMKVLGIPNTPHFKEITNIQEALDLYKKLNNEAAAKTFKLFSEAECEDNEGNLVTL 517
> cpv:cgd6_4670 Prp9p-like splicing factor 3a subunit 3 snRNP.
C-terminal C2H2 ; K12827 splicing factor 3A subunit 3
Length=493
Score = 174 bits (441), Expect = 1e-43, Method: Compositional matrix adjust.
Identities = 96/198 (48%), Positives = 119/198 (60%), Gaps = 26/198 (13%)
Query 1 IKYFVDLLQPQINQTIAHYQKKQSTNAEEA-----LEEEN-------------------- 35
+ F LL Q + I H K QS+ EE LE EN
Sbjct 285 VSKFSQLLSVQRREAIDHVNKLQSSTREELEIDKQLEMENSGLETLITELNDCLNKNKSK 344
Query 36 -DEDSNEEDAAAEEDSEAEEEEGPIYNPLNLPLGFDGRPIPYWLYKLHGLGQEFKCEICG 94
D DSN+ + + D + +E + +YNPL LPLG DGRP+PYWLYKL+GLG EFKCEICG
Sbjct 345 GDRDSNKMEFDTDSDDDFDELQEKVYNPLKLPLGPDGRPMPYWLYKLNGLGIEFKCEICG 404
Query 95 NFSYWGRRAFERHFSEWRHSFGMRCLKIPNTTHFKEITKIEDAILLYEKLKQQAEGHAFK 154
N SYWGRRAFERHFSE RH+ G+ L IPNT HFKEITKI DA LY L +QA+ +F
Sbjct 405 NCSYWGRRAFERHFSETRHANGLSALGIPNTCHFKEITKISDAQELYSALCKQAKDLSFD 464
Query 155 QDQELECEDAEGNVMSLR 172
+E ED++GN++ L+
Sbjct 465 DQNYVEMEDSQGNILPLK 482
> ath:AT5G06160 ATO; ATO (ATROPOS); nucleic acid binding / zinc
ion binding; K12827 splicing factor 3A subunit 3
Length=504
Score = 171 bits (434), Expect = 9e-43, Method: Compositional matrix adjust.
Identities = 91/168 (54%), Positives = 119/168 (70%), Gaps = 7/168 (4%)
Query 1 IKYFVDLLQPQINQTIAHYQKKQSTNAEEALEEENDEDSNEEDAAAEEDSEAEEEEGPIY 60
+K +LL I +T + KKQS EE E E++N E E+++E+G IY
Sbjct 329 VKKLCNLLDETIERTKQNIVKKQSLTYEEMEGEREGEEANTE-------LESDDEDGLIY 381
Query 61 NPLNLPLGFDGRPIPYWLYKLHGLGQEFKCEICGNFSYWGRRAFERHFSEWRHSFGMRCL 120
NPL LP+G+DG+PIPYWLYKLHGLGQEFKCEICGN+SYWGRRAFERHF EWRH GMRCL
Sbjct 382 NPLKLPIGWDGKPIPYWLYKLHGLGQEFKCEICGNYSYWGRRAFERHFKEWRHQHGMRCL 441
Query 121 KIPNTTHFKEITKIEDAILLYEKLKQQAEGHAFKQDQELECEDAEGNV 168
IPNT +F EIT IE+A L+++++++ + ++ + E E ED EGN+
Sbjct 442 GIPNTKNFNEITSIEEAKELWKRIQERQGVNKWRPELEEEYEDREGNI 489
> xla:495359 sf3a3; splicing factor 3a, subunit 3, 60kDa; K12827
splicing factor 3A subunit 3
Length=501
Score = 168 bits (425), Expect = 9e-42, Method: Compositional matrix adjust.
Identities = 87/167 (52%), Positives = 117/167 (70%), Gaps = 8/167 (4%)
Query 4 FVDLLQPQINQTIAHYQKKQSTNAEEALEEENDEDSNEEDAAAEEDSEAEEEEGPIYNPL 63
+V++L Q + T + Q+KQ+ EE EEE ++ S+ E + +ED+E IYNP
Sbjct 330 YVEILGEQRHLTHENVQRKQARTGEEREEEEEEQISDSE--SDDEDNEI------IYNPK 381
Query 64 NLPLGFDGRPIPYWLYKLHGLGQEFKCEICGNFSYWGRRAFERHFSEWRHSFGMRCLKIP 123
NLPLG+DG+PIPYWLYKLHGL + CEICGNF+Y G +AF+RHF+EWRH+ GMRCL IP
Sbjct 382 NLPLGWDGKPIPYWLYKLHGLNINYNCEICGNFTYRGPKAFQRHFAEWRHAHGMRCLGIP 441
Query 124 NTTHFKEITKIEDAILLYEKLKQQAEGHAFKQDQELECEDAEGNVMS 170
NT HF +T+IEDA+ L+ KLK Q ++ D E E ED+ GNV++
Sbjct 442 NTAHFANVTQIEDAVSLWSKLKLQKSSERWQPDTEEEYEDSSGNVVN 488
> dre:446168 sf3a3, MGC110227, SAP61, SF3a60, wu:fi15a06, wu:fj94f01,
zgc:110227; splicing factor 3a, subunit 3; K12827 splicing
factor 3A subunit 3
Length=501
Score = 161 bits (408), Expect = 9e-40, Method: Compositional matrix adjust.
Identities = 69/112 (61%), Positives = 87/112 (77%), Gaps = 0/112 (0%)
Query 59 IYNPLNLPLGFDGRPIPYWLYKLHGLGQEFKCEICGNFSYWGRRAFERHFSEWRHSFGMR 118
IYNP NLPLG+DG+PIPYWLYKLHGL + CEICGN++Y G +AF+RHF+EWRH+ GMR
Sbjct 377 IYNPKNLPLGWDGKPIPYWLYKLHGLNINYNCEICGNYTYRGPKAFQRHFAEWRHAHGMR 436
Query 119 CLKIPNTTHFKEITKIEDAILLYEKLKQQAEGHAFKQDQELECEDAEGNVMS 170
CL IPNT HF +T+IEDA+ L+ KLK Q ++ D E E ED+ GNV++
Sbjct 437 CLGIPNTAHFANVTQIEDAVSLWSKLKSQKALERWQPDTEEEYEDSSGNVVN 488
> mmu:75062 Sf3a3, 4930512K19Rik, 60kDa, MGC103282; splicing factor
3a, subunit 3; K12827 splicing factor 3A subunit 3
Length=501
Score = 159 bits (403), Expect = 3e-39, Method: Compositional matrix adjust.
Identities = 68/111 (61%), Positives = 86/111 (77%), Gaps = 0/111 (0%)
Query 60 YNPLNLPLGFDGRPIPYWLYKLHGLGQEFKCEICGNFSYWGRRAFERHFSEWRHSFGMRC 119
YNP NLPLG+DG+PIPYWLYKLHGL + CEICGN++Y G +AF+RHF+EWRH+ GMRC
Sbjct 378 YNPKNLPLGWDGKPIPYWLYKLHGLNINYNCEICGNYTYRGPKAFQRHFAEWRHAHGMRC 437
Query 120 LKIPNTTHFKEITKIEDAILLYEKLKQQAEGHAFKQDQELECEDAEGNVMS 170
L IPNT HF +T+IEDA+ L+ KLK Q ++ D E E ED+ GNV++
Sbjct 438 LGIPNTAHFANVTQIEDAVSLWAKLKLQKASERWQPDTEEEYEDSSGNVVN 488
> hsa:10946 SF3A3, PRP9, PRPF9, SAP61, SF3a60; splicing factor
3a, subunit 3, 60kDa; K12827 splicing factor 3A subunit 3
Length=501
Score = 159 bits (403), Expect = 4e-39, Method: Compositional matrix adjust.
Identities = 68/111 (61%), Positives = 86/111 (77%), Gaps = 0/111 (0%)
Query 60 YNPLNLPLGFDGRPIPYWLYKLHGLGQEFKCEICGNFSYWGRRAFERHFSEWRHSFGMRC 119
YNP NLPLG+DG+PIPYWLYKLHGL + CEICGN++Y G +AF+RHF+EWRH+ GMRC
Sbjct 378 YNPKNLPLGWDGKPIPYWLYKLHGLNINYNCEICGNYTYRGPKAFQRHFAEWRHAHGMRC 437
Query 120 LKIPNTTHFKEITKIEDAILLYEKLKQQAEGHAFKQDQELECEDAEGNVMS 170
L IPNT HF +T+IEDA+ L+ KLK Q ++ D E E ED+ GNV++
Sbjct 438 LGIPNTAHFANVTQIEDAVSLWAKLKLQKASERWQPDTEEEYEDSSGNVVN 488
> cel:T13H5.4 hypothetical protein; K12827 splicing factor 3A
subunit 3
Length=500
Score = 157 bits (398), Expect = 1e-38, Method: Compositional matrix adjust.
Identities = 69/116 (59%), Positives = 88/116 (75%), Gaps = 0/116 (0%)
Query 55 EEGPIYNPLNLPLGFDGRPIPYWLYKLHGLGQEFKCEICGNFSYWGRRAFERHFSEWRHS 114
+E YNP NLPLG+DG+PIPYWLYKLHGL + CEICGN +Y G +AF++HF+EWRHS
Sbjct 372 DESAPYNPKNLPLGWDGKPIPYWLYKLHGLNLSYSCEICGNQTYKGPKAFQKHFNEWRHS 431
Query 115 FGMRCLKIPNTTHFKEITKIEDAILLYEKLKQQAEGHAFKQDQELECEDAEGNVMS 170
GMRCL IPNT+HF ITKI+DA+ L+ KLK + E + D + E ED+ GNV++
Sbjct 432 HGMRCLGIPNTSHFANITKIKDALDLWNKLKTEKEMAKWNPDIDEEYEDSSGNVVT 487
> sce:YDL030W PRP9; Prp9p; K12827 splicing factor 3A subunit 3
Length=530
Score = 111 bits (277), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 55/116 (47%), Positives = 78/116 (67%), Gaps = 4/116 (3%)
Query 38 DSNEEDAAAEEDSEAE----EEEGPIYNPLNLPLGFDGRPIPYWLYKLHGLGQEFKCEIC 93
DS E++ A + D E +EE ++PLG DG P+PYWLYKLHGL +E++CEIC
Sbjct 367 DSTEKEGAEQVDGEQRDGQLQEEHLSGKSFDMPLGPDGLPMPYWLYKLHGLDREYRCEIC 426
Query 94 GNFSYWGRRAFERHFSEWRHSFGMRCLKIPNTTHFKEITKIEDAILLYEKLKQQAE 149
N Y GRR FERHF+E RH + +RCL I ++ FK ITKI++A L++ ++ Q++
Sbjct 427 SNKVYNGRRTFERHFNEERHIYHLRCLGIEPSSVFKGITKIKEAQELWKNMQGQSQ 482
> mmu:22289 Kdm6a, Utx; 4lysine (K)-specific demethylase 6A; K11447
histone demethylase [EC:1.14.11.-]
Length=1424
Score = 34.3 bits (77), Expect = 0.19, Method: Composition-based stats.
Identities = 21/74 (28%), Positives = 38/74 (51%), Gaps = 5/74 (6%)
Query 15 TIAHYQKKQSTNAEEALEEENDEDSNEEDAAAEE----DSEAEEEEGPIYNPLNLPLGFD 70
TIA Y + Q+++ +E+L EEN++ S+ +D + E D+ + +GP + + D
Sbjct 1031 TIAKYAQYQASSFQESLREENEKRSHHKDHSDSESTSSDNSGKRRKGP-FKTIKFGTNID 1089
Query 71 GRPIPYWLYKLHGL 84
W +LH L
Sbjct 1090 LSDDKKWKLQLHEL 1103
Lambda K H
0.315 0.134 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4341553636
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40