bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0858_orf2
Length=265
Score E
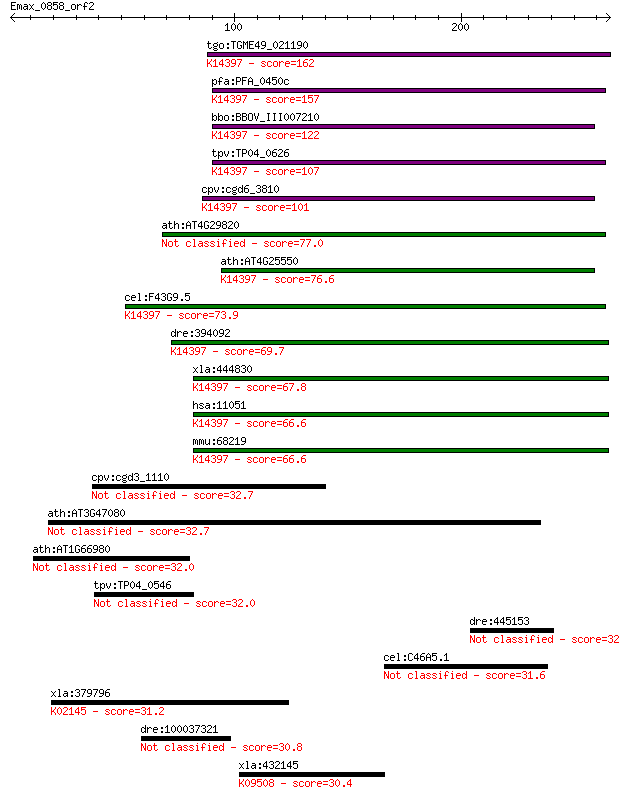
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_021190 mRNA cleavage factor-like protein, putative ... 162 1e-39
pfa:PFA_0450c mRNA cleavage factor-like protein, putative; K14... 157 5e-38
bbo:BBOV_III007210 17.m07632; hypothetical protein; K14397 cle... 122 2e-27
tpv:TP04_0626 mRNA cleavage factor protein; K14397 cleavage an... 107 7e-23
cpv:cgd6_3810 NUDIX domain protein; mRNA cleavage factor-like ... 101 2e-21
ath:AT4G29820 CFIM-25 77.0 6e-14
ath:AT4G25550 protein binding; K14397 cleavage and polyadenyla... 76.6 9e-14
cel:F43G9.5 hypothetical protein; K14397 cleavage and polyaden... 73.9 5e-13
dre:394092 cpsf5, MGC63966, zgc:63966; cleavage and polyadenyl... 69.7 9e-12
xla:444830 nudt21, MGC84447, cpsf5; nudix (nucleoside diphosph... 67.8 4e-11
hsa:11051 NUDT21, CFIM25, CPSF5, DKFZp686H1588; nudix (nucleos... 66.6 8e-11
mmu:68219 Nudt21, 25kDa, 3110048P04Rik, 5730530J16Rik, AU01486... 66.6 8e-11
cpv:cgd3_1110 P-type ATpase fused to two adenyl cyclase domain... 32.7 1.4
ath:AT3G47080 binding 32.7 1.5
ath:AT1G66980 protein kinase family protein / glycerophosphory... 32.0 2.1
tpv:TP04_0546 hypothetical protein 32.0 2.2
dre:445153 im:7149072; si:ch211-175f11.2 32.0 2.5
cel:C46A5.1 hypothetical protein 31.6 2.7
xla:379796 atp6a1, MGC54007; ATPase, H+ transporting, lysosoma... 31.2 4.1
dre:100037321 zgc:162255 30.8 5.6
xla:432145 dnajb2, MGC78895; DnaJ (Hsp40) homolog, subfamily B... 30.4 7.0
> tgo:TGME49_021190 mRNA cleavage factor-like protein, putative
; K14397 cleavage and polyadenylation specificity factor subunit
5
Length=414
Score = 162 bits (410), Expect = 1e-39, Method: Compositional matrix adjust.
Identities = 87/192 (45%), Positives = 118/192 (61%), Gaps = 15/192 (7%)
Query 88 ELEWAVFPESNYSFEADSTLGNKW-----GSGSEE-----ELLKKRQEAYFREGTRRSVA 137
+ +WAV+ ++Y + D L KW G G ++ LL+KR +Y R+G RR+VA
Sbjct 124 DADWAVYSAASYESQVDPALEGKWEFLSPGGGEQQGGRAAALLRKRLASYQRQGLRRTVA 183
Query 138 AIFLVHRAEYPHVLLLLDQQQKKHSLLMFKYKTWQKPREVLHAKLAEYLIRPEQCSKRK- 196
+F H EY H+LLL ++ +++SL FK K+W++P VL KLA + R
Sbjct 184 PVFFCHLREYVHLLLLFHRETRRYSLFTFKAKSWERPEVVLERKLARLFTKHRSEVDRNV 243
Query 197 ---WVAQQLSNEGPDMEVGEFLGEWWRGEFDDDLVPYLPPHVTRPKERVRVHQVQLPHRR 253
WVA Q S EG EVGEFLGEWWR EFD++ P+LPPHVTRPKER+R++QVQLP +
Sbjct 244 NYTWVADQKS-EGVAAEVGEFLGEWWRVEFDEEPQPFLPPHVTRPKERIRLYQVQLPPKC 302
Query 254 SFRVPMGFCLTV 265
SFR+P F L
Sbjct 303 SFRLPPAFSLAA 314
> pfa:PFA_0450c mRNA cleavage factor-like protein, putative; K14397
cleavage and polyadenylation specificity factor subunit
5
Length=232
Score = 157 bits (396), Expect = 5e-38, Method: Compositional matrix adjust.
Identities = 79/175 (45%), Positives = 111/175 (63%), Gaps = 6/175 (3%)
Query 90 EWAVFPESNYSFEADSTLGNKWGSGSEEELLKKRQEAYFREGTRRSVAAIFLVHRAEYPH 149
EW ++P+SNY F D L NK+ E+ KKR AY + G R SV AI L HR EYPH
Sbjct 25 EWLLYPQSNYEFNIDEKLKNKFIIDKEK--CKKRINAYNKNGIRNSVLAIILCHRYEYPH 82
Query 150 VLLLLDQQQKKHSLLMFKYKTWQKPREVLHAKLAEYLIRPEQCSKRKWVAQQLSNEGPD- 208
+LLL + +K+ LL KYKTW+KP+EVL KL +Y+ + + Q++ E +
Sbjct 83 LLLLQHIESQKYYLLNGKYKTWEKPKEVLKKKLQKYI---NKIKDIHFTPAQINKEQEET 139
Query 209 MEVGEFLGEWWRGEFDDDLVPYLPPHVTRPKERVRVHQVQLPHRRSFRVPMGFCL 263
+E+G+FLGEWWR +F+ +PYLP H++RPKE +R++QV L + F +P GF L
Sbjct 140 VEIGDFLGEWWRTQFNSVFLPYLPAHISRPKEYIRLYQVILSPKCIFHLPPGFTL 194
> bbo:BBOV_III007210 17.m07632; hypothetical protein; K14397 cleavage
and polyadenylation specificity factor subunit 5
Length=357
Score = 122 bits (305), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 69/171 (40%), Positives = 99/171 (57%), Gaps = 8/171 (4%)
Query 90 EWAVFPESNYSFEADST-LGNKWGSGSEEELLKKRQEAYFREGTRRSVAAIFLVHRAEYP 148
+W V+PE +YSF DS +G + E LL KR AY + G R +V + L HR +P
Sbjct 17 DWLVYPEDSYSFRDDSAPIGQGLLFRTPESLLCKRIRAYNQNGLRITVYGLILCHRNGFP 76
Query 149 HVLLLLDQQQKKHSLLMFKYKTWQKPREVLHAKLAEYLIRPEQCSKRKWVAQQLSNEGPD 208
+L+L D LL K K+++ PREVL KLA ++ + RK V Q D
Sbjct 77 CILVLRDTSGNI-GLLGGKCKSFENPREVLKLKLARFVS-----TSRKGVHQLNVRANVD 130
Query 209 -MEVGEFLGEWWRGEFDDDLVPYLPPHVTRPKERVRVHQVQLPHRRSFRVP 258
+ VGEF+GE+WR E+D D++PYLP H+ RP+E++ ++QV L + SF P
Sbjct 131 TIIVGEFMGEFWRAEYDSDVLPYLPLHINRPREKILIYQVTLREQCSFIAP 181
> tpv:TP04_0626 mRNA cleavage factor protein; K14397 cleavage
and polyadenylation specificity factor subunit 5
Length=226
Score = 107 bits (266), Expect = 7e-23, Method: Compositional matrix adjust.
Identities = 61/178 (34%), Positives = 97/178 (54%), Gaps = 12/178 (6%)
Query 90 EWAVFPESNYSFEADSTLGNKWGSGS----EEELLKKRQEAYFREGTRRSVAAIFLVHRA 145
EW V+P++ + FE ++T K G G +++L+KR + Y G R +V + L HR
Sbjct 22 EWDVYPQTMFKFEHNTT---KVGRGLIFRLSDDILEKRVKTYAVSGMRITVCGVILSHRK 78
Query 146 EYPHVLLLLDQQQKKHSLLMFKYKTWQKPREVLHAKLAEYLIRPEQCSKRKWVAQQLSNE 205
+P VLLL K LL K K+++ P+EVL +KLA ++ S + +
Sbjct 79 GFPFVLLLKRDLDKSVGLLGGKCKSFENPKEVLSSKLARFI-----TSTKHKHQLNIKET 133
Query 206 GPDMEVGEFLGEWWRGEFDDDLVPYLPPHVTRPKERVRVHQVQLPHRRSFRVPMGFCL 263
++VGE L ++WR +F+ + +PYLP H RPKE++ ++QV L VP G+ L
Sbjct 134 IETIQVGELLADFWRCDFNTEPLPYLPLHTNRPKEKISLYQVVLQESCKISVPKGYSL 191
> cpv:cgd6_3810 NUDIX domain protein; mRNA cleavage factor-like
protein Im like, plant+animal group ; K14397 cleavage and
polyadenylation specificity factor subunit 5
Length=277
Score = 101 bits (252), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 60/190 (31%), Positives = 87/190 (45%), Gaps = 17/190 (8%)
Query 86 DVELEWAVFPESNYSFEADSTLGNKWGSGSEEEL--LKKRQEAYFREGTRRSVAAIFLVH 143
D E W ++P NY S E+ + + + ++G RSVAA+ L H
Sbjct 46 DHEPSWLIYPLKNYGIRVQDNSDEIQSSIPINEMNGFNVKVDNFLKDGIGRSVAALMLTH 105
Query 144 RAEYPHVLLLLDQQQKKHSLLMFKYKTWQKPREVLHAKLAEYLIRPEQCSKRKWVAQQLS 203
R PHV+LL + + L YK W+ PR VL L + + + +
Sbjct 106 RYLCPHVVLLQNDLTSEWMLPNCTYKAWENPRIVLANFLKSMFLTSSSINSNTNNSGSNN 165
Query 204 N---------------EGPDMEVGEFLGEWWRGEFDDDLVPYLPPHVTRPKERVRVHQVQ 248
+EVGE+LG WWR EF+ +PYLPPH TRPKE +R++QV
Sbjct 166 GNNNSSTNNNNINNACSDNAVEVGEYLGTWWRTEFNYSPLPYLPPHSTRPKETIRIYQVI 225
Query 249 LPHRRSFRVP 258
LP + F++P
Sbjct 226 LPPKLLFKLP 235
> ath:AT4G29820 CFIM-25
Length=222
Score = 77.0 bits (188), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 64/209 (30%), Positives = 98/209 (46%), Gaps = 39/209 (18%)
Query 68 EHRQGNFKTISKTTSAPRDV--ELEWAVFPESNYSFEADSTLGNKWGSGSEEELLKKR-- 123
E R + + IS T+ DV +L ++P S+Y F G+K ++E++ R
Sbjct 4 EARALDMEEISDNTTRRNDVVHDLMVDLYPLSSYYF------GSKEALRVKDEIISDRVI 57
Query 124 --QEAYFREGTRRSVAAIFLVHRAEYPHVLLLLDQQQKKHSLLMFKYKTWQKPREVLHAK 181
+ Y G R V A+ LV ++PHVLLL Q ++S+ K
Sbjct 58 RLKSNYAAHGLRTCVEAVLLVELFKHPHVLLL----QYRNSIF----------------K 97
Query 182 LAEYLIRPEQCS----KRKWVAQQLSNEGPDM---EVGEFLGEWWRGEFDDDLVPYLPPH 234
L +RP + KRK ++ NE + EVGE +G WWR F+ + P+LPP+
Sbjct 98 LPGGRLRPGESDIEGLKRKLASKLSVNENVGVSGYEVGECIGMWWRPNFETLMYPFLPPN 157
Query 235 VTRPKERVRVHQVQLPHRRSFRVPMGFCL 263
+ PKE ++ V+LP + F VP F L
Sbjct 158 IKHPKECTKLFLVRLPVHQQFVVPKNFKL 186
> ath:AT4G25550 protein binding; K14397 cleavage and polyadenylation
specificity factor subunit 5
Length=200
Score = 76.6 bits (187), Expect = 9e-14, Method: Compositional matrix adjust.
Identities = 55/179 (30%), Positives = 81/179 (45%), Gaps = 43/179 (24%)
Query 94 FPESNYSF-------EADSTLGNKWGSGSEEELLKKRQEAYFREGTRRSVAAIFLVHRAE 146
+P SNYSF E D+++ ++ L + + Y +EG R SV I LV
Sbjct 10 YPLSNYSFGTKEPKLEKDTSVADR---------LARMKINYMKEGMRTSVEGILLVQEHN 60
Query 147 YPHVLLLLDQQQKKHSLLMFKYKTWQKPREVLHAKLAEYLIRPEQCS----KRKWVAQQL 202
+PH+LLL Q ++ KL ++P + KRK ++
Sbjct 61 HPHILLL----QIGNTF----------------CKLPGGRLKPGENEADGLKRKLTSKLG 100
Query 203 SNEG---PDMEVGEFLGEWWRGEFDDDLVPYLPPHVTRPKERVRVHQVQLPHRRSFRVP 258
N PD VGE + WWR F+ + PY PPH+T+PKE R++ V L + F VP
Sbjct 101 GNSAALVPDWTVGECVATWWRPNFETMMYPYCPPHITKPKECKRLYIVHLSEKEYFAVP 159
> cel:F43G9.5 hypothetical protein; K14397 cleavage and polyadenylation
specificity factor subunit 5
Length=227
Score = 73.9 bits (180), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 61/216 (28%), Positives = 94/216 (43%), Gaps = 37/216 (17%)
Query 52 FEREGLSASM-DATSSIEHRQGNFKTISKTTSAPRDVELEWAVFPESNYSFEADSTLGNK 110
ER +SAS+ +A ++ + + +TI+ V+P +NY+F K
Sbjct 8 IERTTISASVPEAPANFDEKPPFNRTIN--------------VYPLTNYTFGTKDAQAEK 53
Query 111 WGSGSEEELLKKRQEAYFREGTRRSVAAIFLVHRAEYPHVLLLLDQQQKKHSLLMFKYKT 170
S E K+ ++ Y G RRSV A+ +VH PH+LLL
Sbjct 54 --DKSVPERFKRMKDEYEVMGMRRSVEAVLIVHEHSLPHILLL-----------QIGTTF 100
Query 171 WQKPREVLHAKLAEYLIRPEQCSKRKWVAQQL---SNEGPDMEVGEFLGEWWRGEFDDDL 227
++ P L L E + + + L E + + + +G WWR FD
Sbjct 101 YKLPGGELE------LGEDEISGVTRLLNETLGRTDGETNEWTIEDEIGNWWRPNFDPPR 154
Query 228 VPYLPPHVTRPKERVRVHQVQLPHRRSFRVPMGFCL 263
PY+P HVT+PKE ++ VQLP + +F VP F L
Sbjct 155 YPYIPAHVTKPKEHTKLLLVQLPSKSTFCVPKNFKL 190
> dre:394092 cpsf5, MGC63966, zgc:63966; cleavage and polyadenylation
specific factor 5; K14397 cleavage and polyadenylation
specificity factor subunit 5
Length=228
Score = 69.7 bits (169), Expect = 9e-12, Method: Compositional matrix adjust.
Identities = 56/193 (29%), Positives = 87/193 (45%), Gaps = 18/193 (9%)
Query 72 GNFKTISKTTSAPRDVELEWAVFPESNYSFEADSTLGNKWGSGSEEELLKKRQEAYFREG 131
GN K I++ T P +E ++P +NY+F L K S ++ +E + + G
Sbjct 21 GN-KYITQATK-PLTLERTINLYPLTNYTFGTKEPLYEK--DSSVAARFQRMREEFEKIG 76
Query 132 TRRSVAAIFLVHRAEYPHVLLLLDQQQKKHSLLMFKYKTWQKPREVLHAKLAEYLIRPEQ 191
RR+V + +VH PHVLLL L + + E L + E L R +
Sbjct 77 MRRTVEGVLIVHEHRLPHVLLL-QLGTTFFKLPGGELNPGEDEVEGLKRLMTEILGR-QD 134
Query 192 CSKRKWVAQQLSNEGPDMEVGEFLGEWWRGEFDDDLVPYLPPHVTRPKERVRVHQVQLPH 251
K+ WV + + +G WWR F+ PY+P H+T+PKE ++ VQL
Sbjct 135 GVKQDWV------------IDDCIGNWWRPNFEPPQYPYIPAHITKPKEHKKLFLVQLQE 182
Query 252 RRSFRVPMGFCLT 264
+ F VP + L
Sbjct 183 KALFAVPKNYKLV 195
> xla:444830 nudt21, MGC84447, cpsf5; nudix (nucleoside diphosphate
linked moiety X)-type motif 21; K14397 cleavage and polyadenylation
specificity factor subunit 5
Length=227
Score = 67.8 bits (164), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 51/188 (27%), Positives = 82/188 (43%), Gaps = 26/188 (13%)
Query 82 SAPRDVELEWAVFPESNYSFEADSTLGNKWGSGSEEELLKKRQEAYFREGTRRSVAAIFL 141
+ P +E ++P +NY+F L K S ++ +E + + G RR+V + +
Sbjct 28 TKPLTLERTINLYPLTNYTFGTKEPLYEK--DSSVAARFQRMREEFDKIGMRRTVEGVLI 85
Query 142 VHRAEYPHVLLLLDQQQKKHSLLMFKY-----KTWQKPREVLHAKLAEYLIRPEQCSKRK 196
VH PHVLLL + FK + E L + E L R + ++
Sbjct 86 VHEHRLPHVLLL------QLGTTFFKLPGGELNPGEDEVEGLKRLMTEILGR-QDGVQQD 138
Query 197 WVAQQLSNEGPDMEVGEFLGEWWRGEFDDDLVPYLPPHVTRPKERVRVHQVQLPHRRSFR 256
WV + + +G WWR F+ PY+P H+T+PKE ++ VQL + F
Sbjct 139 WV------------IDDCIGNWWRPNFEPPQYPYIPAHITKPKEHKKLFLVQLQEKALFA 186
Query 257 VPMGFCLT 264
VP + L
Sbjct 187 VPKNYKLV 194
> hsa:11051 NUDT21, CFIM25, CPSF5, DKFZp686H1588; nudix (nucleoside
diphosphate linked moiety X)-type motif 21; K14397 cleavage
and polyadenylation specificity factor subunit 5
Length=227
Score = 66.6 bits (161), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 51/184 (27%), Positives = 81/184 (44%), Gaps = 18/184 (9%)
Query 82 SAPRDVELEWAVFPESNYSFEADSTLGNKWGSGSEEELLKKRQEAYFREGTRRSVAAIFL 141
+ P +E ++P +NY+F L K S ++ +E + + G RR+V + +
Sbjct 28 TKPLTLERTINLYPLTNYTFGTKEPLYEK--DSSVAARFQRMREEFDKIGMRRTVEGVLI 85
Query 142 VHRAEYPHVLLLLDQQQKKHSLLMFKYKTWQ-KPREVLHAKLAEYLIRPEQCSKRKWVAQ 200
VH PHVLLL + FK + P E L + E ++ V Q
Sbjct 86 VHEHRLPHVLLL------QLGTTFFKLPGGELNPGEDEVEGLKRLMT--EILGRQDGVLQ 137
Query 201 QLSNEGPDMEVGEFLGEWWRGEFDDDLVPYLPPHVTRPKERVRVHQVQLPHRRSFRVPMG 260
D + + +G WWR F+ PY+P H+T+PKE ++ VQL + F VP
Sbjct 138 -------DWVIDDCIGNWWRPNFEPPQYPYIPAHITKPKEHKKLFLVQLQEKALFAVPKN 190
Query 261 FCLT 264
+ L
Sbjct 191 YKLV 194
> mmu:68219 Nudt21, 25kDa, 3110048P04Rik, 5730530J16Rik, AU014860,
AW549947, Cpsf5; nudix (nucleoside diphosphate linked moiety
X)-type motif 21; K14397 cleavage and polyadenylation
specificity factor subunit 5
Length=227
Score = 66.6 bits (161), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 51/184 (27%), Positives = 81/184 (44%), Gaps = 18/184 (9%)
Query 82 SAPRDVELEWAVFPESNYSFEADSTLGNKWGSGSEEELLKKRQEAYFREGTRRSVAAIFL 141
+ P +E ++P +NY+F L K S ++ +E + + G RR+V + +
Sbjct 28 TKPLTLERTINLYPLTNYTFGTKEPLYEK--DSSVAARFQRMREEFDKIGMRRTVEGVLI 85
Query 142 VHRAEYPHVLLLLDQQQKKHSLLMFKYKTWQ-KPREVLHAKLAEYLIRPEQCSKRKWVAQ 200
VH PHVLLL + FK + P E L + E ++ V Q
Sbjct 86 VHEHRLPHVLLL------QLGTTFFKLPGGELNPGEDEVEGLKRLMT--EILGRQDGVLQ 137
Query 201 QLSNEGPDMEVGEFLGEWWRGEFDDDLVPYLPPHVTRPKERVRVHQVQLPHRRSFRVPMG 260
D + + +G WWR F+ PY+P H+T+PKE ++ VQL + F VP
Sbjct 138 -------DWVIDDCIGNWWRPNFEPPQYPYIPAHITKPKEHKKLFLVQLQEKALFAVPKN 190
Query 261 FCLT 264
+ L
Sbjct 191 YKLV 194
> cpv:cgd3_1110 P-type ATpase fused to two adenyl cyclase domains
and 21 predicted transmembrane regions
Length=3848
Score = 32.7 bits (73), Expect = 1.4, Method: Composition-based stats.
Identities = 24/107 (22%), Positives = 43/107 (40%), Gaps = 4/107 (3%)
Query 37 QELSSGFVECQRNGDFEREGLSASMDATSSIEHRQGNFKTISKTTSAPRDVELEWAVFPE 96
QEL G +C+ ++++E + + G + S + D+EL
Sbjct 3079 QELGKGIDDCREEAEYDKEDNTEDTPLQDGLLKEIGRQDSFSLNENIGGDIELHLKSQDS 3138
Query 97 SNYSFEADSTLGNKWGSGSEEELL----KKRQEAYFREGTRRSVAAI 139
S+ F L + + SE E + +KR + FR+ T S +I
Sbjct 3139 SSIGFFKKKELNSAFLVASEHETVAKSNRKRMKTRFRKSTEHSFISI 3185
> ath:AT3G47080 binding
Length=515
Score = 32.7 bits (73), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 56/246 (22%), Positives = 100/246 (40%), Gaps = 36/246 (14%)
Query 18 ATGGEGTADPALNVSVDTSQELSSGFVECQRNGDFEREGLSASMDA-TSSIEHRQGNFKT 76
A+ G ++D A +V + QEL+S F + E + S ++D S HR G+ K
Sbjct 103 ASSGFFSSDEAFSVKM---QELASQFRNAGDEEEEENKQKSEAVDNDNDSNNHRFGSLKL 159
Query 77 ISKTTSAPRDVELEWA-VFPESNYSFEADST--------LGNKWGSGSEEELLKKRQEAY 127
+ ++ +E WA + S+ +A+S + K +EE LKK
Sbjct 160 LQESVPGLASLEAPWAEMVNHSSIERKANSVDLPLSLRIIKRKL----QEEALKKASATT 215
Query 128 FREGTRRSVAAIFLVHR----AEYPHVLLLLDQQQKKHSLLM--FKYKTWQKPREVLHAK 181
+ R + +F++ A V +L +++ H+ L+ F+ Q P +++
Sbjct 216 YCSINRAFSSMVFMIEELHSFALQTRVGVLKQVKKEMHASLLWIFQRVFSQTPTLMVYVM 275
Query 182 --LAEYLIRPEQCSKRKWVAQQLSNEGPDM-----------EVGEFLGEWWRGEFDDDLV 228
LA Y + + + + N+GPD E + G W G + D V
Sbjct 276 ILLANYTVHSVASNLPIAASPPVVNKGPDQTQQRIDFSSLKESTKLDGSKWLGSINFDKV 335
Query 229 PYLPPH 234
+LP H
Sbjct 336 SHLPRH 341
> ath:AT1G66980 protein kinase family protein / glycerophosphoryl
diester phosphodiesterase family protein
Length=1109
Score = 32.0 bits (71), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 28/80 (35%), Positives = 39/80 (48%), Gaps = 15/80 (18%)
Query 11 RGGMKMQATGGEGTADPALNVSVDTSQELSSGFVECQRNGDFEREGL--------SASMD 62
+ G+K+ A+G D A N S D E S FV+ NG+F +G+ SAS+D
Sbjct 299 KAGLKLYASGFANDVDIAYNYSWDPVSEYLS-FVD---NGNFSVDGMLSDFPLTASASVD 354
Query 63 ATSSI---EHRQGNFKTISK 79
S I +Q +F ISK
Sbjct 355 CFSHIGRNATKQVDFLVISK 374
> tpv:TP04_0546 hypothetical protein
Length=275
Score = 32.0 bits (71), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 16/51 (31%), Positives = 27/51 (52%), Gaps = 7/51 (13%)
Query 38 ELSSGFVECQRNGD-------FEREGLSASMDATSSIEHRQGNFKTISKTT 81
++GFVE + N D F+ + L S D ++EH++GN TI+ +
Sbjct 184 NFTNGFVEKELNFDIDQVIKKFKMDDLILSPDIIENLEHKKGNIYTINSSI 234
> dre:445153 im:7149072; si:ch211-175f11.2
Length=1741
Score = 32.0 bits (71), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 16/37 (43%), Positives = 19/37 (51%), Gaps = 1/37 (2%)
Query 204 NEGPDMEVGEFLGEWWRGEFDDDLVPYLPPHVTRPKE 240
NEG + V E GEWWRG D P + +PKE
Sbjct 1048 NEGDTILVSEREGEWWRGSIGDR-AGLFPSNYVKPKE 1083
> cel:C46A5.1 hypothetical protein
Length=497
Score = 31.6 bits (70), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 23/82 (28%), Positives = 38/82 (46%), Gaps = 20/82 (24%)
Query 166 FKYKTWQKPREVLHAKLAEYLIRPEQCSK--------RKW--VAQQLSNEGPDMEVGEFL 215
K K+ QK E +H K + R ++C+K KW VAQ+ + ++ V EF
Sbjct 166 LKGKSKQKSEEKVHQKAKDDAKRGDECNKFKISDEKLTKWKLVAQETLKDDANIAVNEF- 224
Query 216 GEWWRGEFDDDLVPYLPPHVTR 237
+ + YLPPH+++
Sbjct 225 ---------EKVSGYLPPHISK 237
> xla:379796 atp6a1, MGC54007; ATPase, H+ transporting, lysosomal
70kDa, V1 subunit A, isoform 1; K02145 V-type H+-transporting
ATPase subunit A [EC:3.6.3.14]
Length=617
Score = 31.2 bits (69), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 30/105 (28%), Positives = 43/105 (40%), Gaps = 9/105 (8%)
Query 19 TGGEGTADPALNVSVDTSQELSSGFVECQRNGDFEREGLSASMDATSSIEHRQGNFKTIS 78
T G DP L S EL G + +G +R L D T SI +G
Sbjct 70 TSGVSVGDPVLRTGKPLSVELGPGIMGNIFDG-IQRP-LKDIADLTKSIYIPRG-----I 122
Query 79 KTTSAPRDVELEWAVFPESNYSFEADSTLGNKWGSGSEEELLKKR 123
T+ RD L+W P+ N + T G+ +G+ E L+K +
Sbjct 123 NVTALSRD--LKWEFLPDKNIRAGSHVTGGDIYGTVMENSLIKHK 165
> dre:100037321 zgc:162255
Length=309
Score = 30.8 bits (68), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 15/39 (38%), Positives = 22/39 (56%), Gaps = 1/39 (2%)
Query 59 ASMDATSSIEHRQGNFKTISKTTSAPRDVELEWAVFPES 97
+ MD SS+ NF + +S PR+ L+W +FPES
Sbjct 178 SQMDQFSSVSQTPANFTLLWSVSSRPRESVLQW-LFPES 215
> xla:432145 dnajb2, MGC78895; DnaJ (Hsp40) homolog, subfamily
B, member 2; K09508 DnaJ homolog subfamily B member 2
Length=281
Score = 30.4 bits (67), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 17/69 (24%), Positives = 33/69 (47%), Gaps = 5/69 (7%)
Query 102 EADSTLGNKWGSGSEEEL-----LKKRQEAYFREGTRRSVAAIFLVHRAEYPHVLLLLDQ 156
E D L + W +G +++L L KR++A+ + R+ ++ V R P ++LD
Sbjct 193 EEDGELKSIWVNGIQDDLALAIELSKREQAFLPRASTRTDGTLYNVQRRSPPDTPVVLDS 252
Query 157 QQKKHSLLM 165
+ L +
Sbjct 253 DDEDEELQL 261
Lambda K H
0.318 0.134 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 9827099128
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40