bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0848_orf1
Length=148
Score E
Sequences producing significant alignments: (Bits) Value
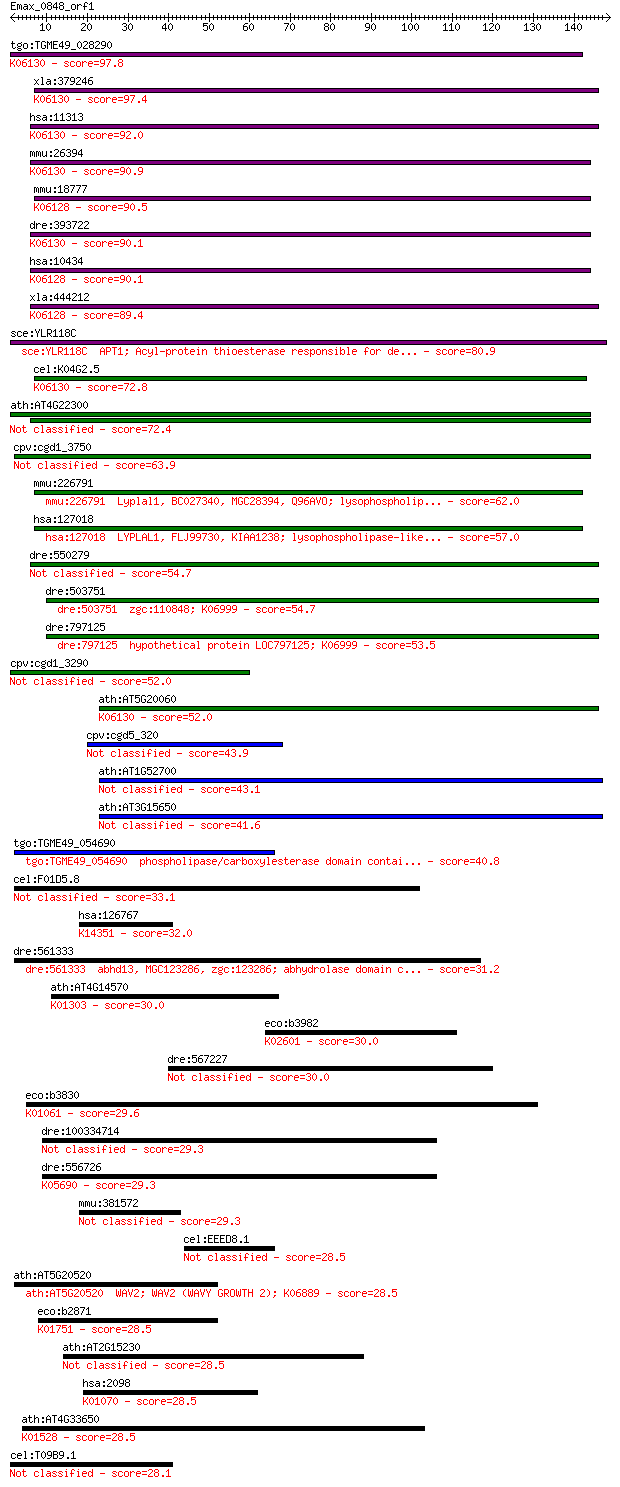
tgo:TGME49_028290 phospholipase/carboxylesterase, putative (EC... 97.8 1e-20
xla:379246 lypla2, MGC52664; lysophospholipase II (EC:3.1.1.5)... 97.4 1e-20
hsa:11313 LYPLA2, APT-2, DJ886K2.4; lysophospholipase II (EC:3... 92.0 6e-19
mmu:26394 Lypla2, LysoII; lysophospholipase 2 (EC:3.1.1.5); K0... 90.9 1e-18
mmu:18777 Lypla1, Pla1a; lysophospholipase 1 (EC:3.1.1.5); K06... 90.5 2e-18
dre:393722 MGC73210; zgc:73210 (EC:3.1.1.5); K06130 lysophosph... 90.1 2e-18
hsa:10434 LYPLA1, APT-1, LPL1, LYSOPLA, hAPT1; lysophospholipa... 90.1 2e-18
xla:444212 lypla1, MGC80756; lysophospholipase I (EC:3.1.1.5);... 89.4 4e-18
sce:YLR118C APT1; Acyl-protein thioesterase responsible for de... 80.9 1e-15
cel:K04G2.5 ath-1; Acyl protein THioesterase family member (at... 72.8 4e-13
ath:AT4G22300 SOBER1; SOBER1 (SUPPRESSOR OF AVRBST-ELICITED RE... 72.4 5e-13
cpv:cgd1_3750 hypothetical protein 63.9 2e-10
mmu:226791 Lyplal1, BC027340, MGC28394, Q96AVO; lysophospholip... 62.0 7e-10
hsa:127018 LYPLAL1, FLJ99730, KIAA1238; lysophospholipase-like... 57.0 2e-08
dre:550279 lypla1, zgc:110260; lysophospholipase I (EC:3.1.1.5) 54.7 9e-08
dre:503751 zgc:110848; K06999 54.7 1e-07
dre:797125 hypothetical protein LOC797125; K06999 53.5 2e-07
cpv:cgd1_3290 carboxylesterase 52.0 6e-07
ath:AT5G20060 phospholipase/carboxylesterase family protein; K... 52.0 8e-07
cpv:cgd5_320 carboxylesterase, lysophospholipase, signal peptide 43.9 2e-04
ath:AT1G52700 phospholipase/carboxylesterase family protein 43.1 3e-04
ath:AT3G15650 phospholipase/carboxylesterase family protein 41.6 0.001
tgo:TGME49_054690 phospholipase/carboxylesterase domain contai... 40.8 0.002
cel:F01D5.8 hypothetical protein 33.1 0.30
hsa:126767 AADACL3; arylacetamide deacetylase-like 3 (EC:3.1.1... 32.0 0.75
dre:561333 abhd13, MGC123286, zgc:123286; abhydrolase domain c... 31.2 1.3
ath:AT4G14570 acylaminoacyl-peptidase-related; K01303 acylamin... 30.0 2.5
eco:b3982 nusG, ECK3973, JW3945; transcription termination fac... 30.0 2.7
dre:567227 esyt1a, si:ch211-150a20.1; extended synaptotagmin-l... 30.0 3.1
eco:b3830 ysgA, ECK3824, JW5853; predicted hydrolase; K01061 c... 29.6 3.6
dre:100334714 hypothetical protein LOC100334714 29.3 4.6
dre:556726 ctnnd1; catenin (cadherin-associated protein), delt... 29.3 4.9
mmu:381572 9430007A20Rik, Aadacl4, Gm1034; RIKEN cDNA 9430007A... 29.3 5.2
cel:EEED8.1 mel-47; Maternal Effect Lethal family member (mel-47) 28.5 7.4
ath:AT5G20520 WAV2; WAV2 (WAVY GROWTH 2); K06889 28.5 8.1
eco:b2871 ygeX, ECK2867, JW2839; 2,3-diaminopropionate ammonia... 28.5 8.7
ath:AT2G15230 ATLIP1; ATLIP1 (Arabidopsis thaliana lipase 1); ... 28.5 8.7
hsa:2098 ESD, FGH, FLJ11763; esterase D (EC:3.1.2.12); K01070 ... 28.5 8.9
ath:AT4G33650 DRP3A; DRP3A (DYNAMIN-RELATED PROTEIN 3A); GTP b... 28.5 9.0
cel:T09B9.1 hypothetical protein 28.1 9.7
> tgo:TGME49_028290 phospholipase/carboxylesterase, putative (EC:3.1.1.1);
K06130 lysophospholipase II [EC:3.1.1.5]
Length=285
Score = 97.8 bits (242), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 57/181 (31%), Positives = 88/181 (48%), Gaps = 41/181 (22%)
Query 1 MDSKARIDKIIDSQIEAGIDPSRILVGGFSQGGAVAYLVCLTSPHKLGGILAMSSWCPLG 60
+ SK RID I+ ++ AG+ P RI++ GFSQGGA+AY L + +LGGI+A+S+W PL
Sbjct 99 LASKQRIDAILAGELAAGVAPERIILAGFSQGGALAYFTGLQASVRLGGIVALSTWTPLA 158
Query 61 KVIEVSP----------------------------------------HYVEAVPRIMHCH 80
+ + VS VE ++HCH
Sbjct 159 QELRVSAGCLGKRDTQSRKEALQTREEEKTEEEKEEEKKEEKKEEKEKRVEGPTPVLHCH 218
Query 81 GAVDDMVRPVYGQTSVESVRNRLIDAGARPEKCQEDITFKLYPGLGHFANQQELNDIKAF 140
G D++V +GQ S VR + +A + ++ + F + GLGH AN QEL+ ++ F
Sbjct 219 GEQDELVLIEFGQESAAIVRRQYAEAWGE-DVAKKAVKFLSFQGLGHSANAQELDQVRRF 277
Query 141 L 141
+
Sbjct 278 I 278
> xla:379246 lypla2, MGC52664; lysophospholipase II (EC:3.1.1.5);
K06130 lysophospholipase II [EC:3.1.1.5]
Length=231
Score = 97.4 bits (241), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 51/139 (36%), Positives = 76/139 (54%), Gaps = 9/139 (6%)
Query 7 IDKIIDSQIEAGIDPSRILVGGFSQGGAVAYLVCLTSPHKLGGILAMSSWCPLGKVIEVS 66
I II+ +++ GI +RI++GGFSQGGA++ LT HKL G++ +S W PL K +
Sbjct 99 IKTIIEHEVKNGIPANRIVLGGFSQGGALSMYTALTCQHKLAGVVGLSCWLPLHKTFPQA 158
Query 67 PHYVEAVPRIMHCHGAVDDMVRPVYGQTSVESVRNRLIDAGARPEKCQEDITFKLYPGLG 126
V +M CHG D M+ +G + E +++ L P K Q FK YPG+
Sbjct 159 ASGVNKEISVMQCHGEADPMIPVRFGNLTSEKLKSVL-----NPSKVQ----FKSYPGVM 209
Query 127 HFANQQELNDIKAFLSRTF 145
H NQ+E+ +K FL +
Sbjct 210 HSTNQEEMMAVKDFLQKVL 228
> hsa:11313 LYPLA2, APT-2, DJ886K2.4; lysophospholipase II (EC:3.1.1.5);
K06130 lysophospholipase II [EC:3.1.1.5]
Length=231
Score = 92.0 bits (227), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 48/140 (34%), Positives = 76/140 (54%), Gaps = 9/140 (6%)
Query 6 RIDKIIDSQIEAGIDPSRILVGGFSQGGAVAYLVCLTSPHKLGGILAMSSWCPLGKVIEV 65
I +I+ +++ GI +RI++GGFSQGGA++ LT PH L GI+A+S W PL +
Sbjct 98 NIKALIEHEMKNGIPANRIVLGGFSQGGALSLYTALTCPHPLAGIVALSCWLPLHRAFPQ 157
Query 66 SPHYVEAVPRIMHCHGAVDDMVRPVYGQTSVESVRNRLIDAGARPEKCQEDITFKLYPGL 125
+ + I+ CHG +D MV +G + E +R+ + A + FK YPG+
Sbjct 158 AANGSAKDLAILQCHGELDPMVPVRFGALTAEKLRSVVTPA---------RVQFKTYPGV 208
Query 126 GHFANQQELNDIKAFLSRTF 145
H + QE+ +K FL +
Sbjct 209 MHSSCPQEMAAVKEFLEKLL 228
> mmu:26394 Lypla2, LysoII; lysophospholipase 2 (EC:3.1.1.5);
K06130 lysophospholipase II [EC:3.1.1.5]
Length=231
Score = 90.9 bits (224), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 48/138 (34%), Positives = 75/138 (54%), Gaps = 9/138 (6%)
Query 6 RIDKIIDSQIEAGIDPSRILVGGFSQGGAVAYLVCLTSPHKLGGILAMSSWCPLGKVIEV 65
I +I+ +++ GI +RI++GGFSQGGA++ LT PH L GI+A+S W PL +
Sbjct 98 NIKALIEHEMKNGIPANRIVLGGFSQGGALSLYTALTCPHPLAGIVALSCWLPLHRNFPQ 157
Query 66 SPHYVEAVPRIMHCHGAVDDMVRPVYGQTSVESVRNRLIDAGARPEKCQEDITFKLYPGL 125
+ + I+ CHG +D MV +G + E +R + A + FK YPG+
Sbjct 158 AANGSAKDLAILQCHGELDPMVPVRFGALTAEKLRTVVTPA---------RVQFKTYPGV 208
Query 126 GHFANQQELNDIKAFLSR 143
H + QE+ +K FL +
Sbjct 209 MHSSCPQEMAAVKEFLEK 226
> mmu:18777 Lypla1, Pla1a; lysophospholipase 1 (EC:3.1.1.5); K06128
lysophospholipase I [EC:3.1.1.5]
Length=230
Score = 90.5 bits (223), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 47/140 (33%), Positives = 80/140 (57%), Gaps = 14/140 (10%)
Query 7 IDKIIDSQIEAGIDPSRILVGGFSQGGAVAYLVCLTSPHKLGGILAMSSWCPLGKVIEVS 66
+ +ID +++ GI +RI++GGFSQGGA++ LT+ KL G+ A+S W PL
Sbjct 96 VKALIDQEVKNGIPSNRIILGGFSQGGALSLYTALTTQQKLAGVTALSCWLPLRASFSQG 155
Query 67 PHYVEAVPR---IMHCHGAVDDMVRPVYGQTSVESVRNRLIDAGARPEKCQEDITFKLYP 123
P + + R ++ CHG D +V ++G +VE ++ + A ++TFK+Y
Sbjct 156 P--INSANRDISVLQCHGDCDPLVPLMFGSLTVERLKALINPA---------NVTFKIYE 204
Query 124 GLGHFANQQELNDIKAFLSR 143
G+ H + QQE+ D+K F+ +
Sbjct 205 GMMHSSCQQEMMDVKHFIDK 224
> dre:393722 MGC73210; zgc:73210 (EC:3.1.1.5); K06130 lysophospholipase
II [EC:3.1.1.5]
Length=232
Score = 90.1 bits (222), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 47/139 (33%), Positives = 77/139 (55%), Gaps = 10/139 (7%)
Query 6 RIDKIIDSQIEAGIDPSRILVGGFSQGGAVAYLVCLTSPHKLGGILAMSSWCPLGKVI-E 64
I IID +++ GI +RI++GGFSQGGA++ LTS +L G++ +S W PL K +
Sbjct 98 NIKAIIDHEVKNGIPSNRIVLGGFSQGGALSLYTALTSQQQLAGVVGLSCWLPLHKTFPQ 157
Query 65 VSPHYVEAVPRIMHCHGAVDDMVRPVYGQTSVESVRNRLIDAGARPEKCQEDITFKLYPG 124
+ I+ CHG +D M+ +G + E ++ + E+ITF+ YPG
Sbjct 158 AAGASANKDTPILQCHGEMDPMIPVQFGAMTAEKLKTIV---------SPENITFRTYPG 208
Query 125 LGHFANQQELNDIKAFLSR 143
L H + QE++ +K F+ +
Sbjct 209 LMHSSCPQEMSAVKDFIEK 227
> hsa:10434 LYPLA1, APT-1, LPL1, LYSOPLA, hAPT1; lysophospholipase
I (EC:3.1.1.5 3.1.2.-); K06128 lysophospholipase I [EC:3.1.1.5]
Length=230
Score = 90.1 bits (222), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 49/141 (34%), Positives = 78/141 (55%), Gaps = 14/141 (9%)
Query 6 RIDKIIDSQIEAGIDPSRILVGGFSQGGAVAYLVCLTSPHKLGGILAMSSWCPLGKVIEV 65
I +ID +++ GI +RI++GGFSQGGA++ LT+ KL G+ A+S W PL
Sbjct 95 NIKALIDQEVKNGIPSNRIILGGFSQGGALSLYTALTTQQKLAGVTALSCWLPLRASFPQ 154
Query 66 SPHYVEAVPR---IMHCHGAVDDMVRPVYGQTSVESVRNRLIDAGARPEKCQEDITFKLY 122
P + R I+ CHG D +V ++G +VE ++ + A ++TFK Y
Sbjct 155 GP--IGGANRDISILQCHGDCDPLVPLMFGSLTVEKLKTLVNPA---------NVTFKTY 203
Query 123 PGLGHFANQQELNDIKAFLSR 143
G+ H + QQE+ D+K F+ +
Sbjct 204 EGMMHSSCQQEMMDVKQFIDK 224
> xla:444212 lypla1, MGC80756; lysophospholipase I (EC:3.1.1.5);
K06128 lysophospholipase I [EC:3.1.1.5]
Length=230
Score = 89.4 bits (220), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 45/141 (31%), Positives = 77/141 (54%), Gaps = 10/141 (7%)
Query 6 RIDKIIDSQIEAGIDPSRILVGGFSQGGAVAYLVCLTSPHKLGGILAMSSWCPL-GKVIE 64
+ +ID +++ GI +RI++GGFSQGGA++ LT+ KLGG++A+S W PL +
Sbjct 95 NVKALIDQEVKNGIPSNRIILGGFSQGGALSLYTALTTQQKLGGVVALSCWLPLRSSFPQ 154
Query 65 VSPHYVEAVPRIMHCHGAVDDMVRPVYGQTSVESVRNRLIDAGARPEKCQEDITFKLYPG 124
+ + ++ CHG D +V ++G + E ++ + A ++ FK Y G
Sbjct 155 AAANSANKDVAVLQCHGESDPLVPLMFGTITSEKLKTIISPA---------NVKFKTYSG 205
Query 125 LGHFANQQELNDIKAFLSRTF 145
L H + QE+ DIK F+ +
Sbjct 206 LMHSSCNQEMTDIKQFIDKQL 226
> sce:YLR118C APT1; Acyl-protein thioesterase responsible for
depalmitoylation of Gpa1p; green fluorescent protein (GFP)-fusion
protein localizes to both the cytoplasm and nucleus and
is induced in response to the DNA-damaging agent MMS (EC:3.1.2.-);
K06999
Length=227
Score = 80.9 bits (198), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 44/148 (29%), Positives = 80/148 (54%), Gaps = 11/148 (7%)
Query 1 MDSKARIDKIIDSQIEAGIDPSRILVGGFSQGGAVAYLVCLTSPHKLGGILAMSSWCPLG 60
M+S I+K + +I+ GI P +I++GGFSQG A+A +T P K+GGI+A+S +C +
Sbjct 90 MNSLNSIEKTVKQEIDKGIKPEQIIIGGFSQGAALALATSVTLPWKIGGIVALSGFCSIP 149
Query 61 KVIEVSPHYVEAVPRIMHCHGAVDDMVRPVYGQTSVESVRNRLIDAGARPEKCQ-EDITF 119
+++ + + I H HG +D +V G + + + + C+ ++ F
Sbjct 150 GILKQHKNGINVKTPIFHGHGDMDPVVPIGLGIKAKQFYQ----------DSCEIQNYEF 199
Query 120 KLYPGLGHFANQQELNDIKAFLSRTFGS 147
K+Y G+ H EL D+ +F+ ++ S
Sbjct 200 KVYKGMAHSTVPDELEDLASFIKKSLSS 227
> cel:K04G2.5 ath-1; Acyl protein THioesterase family member (ath-1);
K06130 lysophospholipase II [EC:3.1.1.5]
Length=223
Score = 72.8 bits (177), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 48/136 (35%), Positives = 69/136 (50%), Gaps = 12/136 (8%)
Query 7 IDKIIDSQIEAGIDPSRILVGGFSQGGAVAYLVCLTSPHKLGGILAMSSWCPLGKVIEVS 66
+ ++ID+++ AGI SRI VGGFS GGA+A LT P KLGGI+ +SS+ S
Sbjct 95 VHQLIDAEVAAGIPASRIAVGGFSMGGALAIYAGLTYPQKLGGIVGLSSFFLQRTKFPGS 154
Query 67 PHYVEAVPRIMHCHGAVDDMVRPVYGQTSVESVRNRLIDAGARPEKCQEDITFKLYPGLG 126
A P I HG D +V +GQ S + ++ K + Y G+
Sbjct 155 FTANNATP-IFLGHGTDDFLVPLQFGQMSEQYIK-----------KFNPKVELHTYRGMQ 202
Query 127 HFANQQELNDIKAFLS 142
H + +E+ D+K FLS
Sbjct 203 HSSCGEEMRDVKTFLS 218
> ath:AT4G22300 SOBER1; SOBER1 (SUPPRESSOR OF AVRBST-ELICITED
RESISTANCE 1); carboxylesterase
Length=502
Score = 72.4 bits (176), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 47/152 (30%), Positives = 71/152 (46%), Gaps = 20/152 (13%)
Query 1 MDSKARIDKIIDSQIEAGIDPSRILVGGFSQGGAVAYLVCLTSPHKLGGILAMSSWCPL- 59
+++ + IID +I G +P + + G SQGGA+ L P LGG +S W P
Sbjct 77 LEAVKNVHAIIDQEIAEGTNPENVFICGLSQGGALTLASVLLYPKTLGGGAVLSGWVPFT 136
Query 60 GKVIEVSPHYVEAVPR--------IMHCHGAVDDMVRPVYGQTSVESVRNRLIDAGARPE 111
+I P + VP I+ HG D MV GQ ++ ++ +AG E
Sbjct 137 SSIISQFPEEAKKVPHLCFLINTPILWSHGTDDRMVLFEAGQAALPFLK----EAGVTCE 192
Query 112 KCQEDITFKLYPGLGHFANQQELNDIKAFLSR 143
FK YPGLGH + +EL I++++ R
Sbjct 193 -------FKAYPGLGHSISNKELKYIESWIKR 217
Score = 67.0 bits (162), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 44/140 (31%), Positives = 66/140 (47%), Gaps = 15/140 (10%)
Query 6 RIDKIIDSQIEAGIDPSRILVGGFSQGGAVAYLVCLTSPHKLGGILAMSSWCPLGKVIEV 65
+ IID +I GI+P + + GFSQGGA+ L P +GG S W P I
Sbjct 367 NVHAIIDKEIAGGINPENVYICGFSQGGALTLASVLLYPKTIGGGAVFSGWIPFNSSI-- 424
Query 66 SPHYVEAVPR--IMHCHGAVDDMVRPVYGQTSVESVRNRLIDAGARPEKCQEDITFKLYP 123
+ + E + I+ HG D V GQ ++ ++ AG E FK YP
Sbjct 425 TNQFTEDAKKTPILWSHGIDDKTVLFEAGQAALPFLQ----QAGVTCE-------FKAYP 473
Query 124 GLGHFANQQELNDIKAFLSR 143
GLGH + +EL ++++L +
Sbjct 474 GLGHSISNEELQYLESWLKQ 493
> cpv:cgd1_3750 hypothetical protein
Length=244
Score = 63.9 bits (154), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 47/159 (29%), Positives = 75/159 (47%), Gaps = 27/159 (16%)
Query 2 DSKARIDKIIDSQIEAGIDPSRILVGGFSQGGAVAYLVCLTS-PHKLGGILAMSSWCPLG 60
+S +RI ++I +IE GIDP +I +GGFSQG A+ +L+ + S + LG + + W PL
Sbjct 94 NSVSRITRLISLEIEKGIDPKKISLGGFSQGSAIVFLISMASRKYTLGSCIVVGGWLPLT 153
Query 61 K----------------VIEVSPHYVEAVPRIMHCHGAVDDMVRPVYGQTSVESVRNRLI 104
+ +V E V I+ HG D +V + + + V +
Sbjct 154 ERGFKEGKESKIATEELTFDVRESVKEHVDFIV-LHGEADPVVLYQWSLMNKDFVLEFI- 211
Query 105 DAGARPEKCQEDITFKLYPGLGHFANQQELNDIKAFLSR 143
+P+K +K YPG+ H Q + DI FLS+
Sbjct 212 ----KPKK----FIYKSYPGVVHTITSQMMVDIFNFLSK 242
> mmu:226791 Lyplal1, BC027340, MGC28394, Q96AVO; lysophospholipase-like
1 (EC:3.1.2.-); K06999
Length=239
Score = 62.0 bits (149), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 34/136 (25%), Positives = 66/136 (48%), Gaps = 12/136 (8%)
Query 7 IDKIIDSQIEAGIDPSRILVGGFSQGGAVAYLVCLTSPHKLGGILAMSSWCPLGKVI-EV 65
+ ++D +++ GI SRIL+GGFS GG +A + S + G+ +S + V+ +
Sbjct 102 LSGLVDEEVKTGIQKSRILIGGFSMGGCMAMHLAYRSHPDVAGVFVLSGFLNKASVVYQD 161
Query 66 SPHYVEAVPRIMHCHGAVDDMVRPVYGQTSVESVRNRLIDAGARPEKCQEDITFKLYPGL 125
+P + CHG+ D++V +G+ + +++ + TF P L
Sbjct 162 LQQGGRMLPELFQCHGSADNLVLHAWGKETNSKLKSLGVST-----------TFHSLPNL 210
Query 126 GHFANQQELNDIKAFL 141
H N+ EL +K+++
Sbjct 211 NHELNKTELEKLKSWI 226
> hsa:127018 LYPLAL1, FLJ99730, KIAA1238; lysophospholipase-like
1 (EC:3.1.2.-); K06999
Length=237
Score = 57.0 bits (136), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 33/136 (24%), Positives = 67/136 (49%), Gaps = 12/136 (8%)
Query 7 IDKIIDSQIEAGIDPSRILVGGFSQGGAVAYLVCLTSPHKLGGILAMSSWCPLGKVIEVS 66
+ +ID ++++GI +RIL+GGFS GG +A + + + G+ A+SS+ + +
Sbjct 101 LTDLIDEEVKSGIKKNRILIGGFSMGGCMAMHLAYRNHQDVAGVFALSSFLNKASAVYQA 160
Query 67 PHYVEAV-PRIMHCHGAVDDMVRPVYGQTSVESVRNRLIDAGARPEKCQEDITFKLYPGL 125
V P + CHG D++V + + + N ++ + K F +P +
Sbjct 161 LQKSNGVLPELFQCHGTADELVLHSWAEET-----NSMLKSLGVTTK------FHSFPNV 209
Query 126 GHFANQQELNDIKAFL 141
H ++ EL+ +K ++
Sbjct 210 YHELSKTELDILKLWI 225
> dre:550279 lypla1, zgc:110260; lysophospholipase I (EC:3.1.1.5)
Length=196
Score = 54.7 bits (130), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 36/140 (25%), Positives = 63/140 (45%), Gaps = 43/140 (30%)
Query 6 RIDKIIDSQIEAGIDPSRILVGGFSQGGAVAYLVCLTSPHKLGGILAMSSWCPLGKVIEV 65
+ +ID +++ GI RI++GGFSQ + K ++
Sbjct 95 NVKALIDQEVKNGIPSHRIVLGGFSQSV-------------------------ISKNKDI 129
Query 66 SPHYVEAVPRIMHCHGAVDDMVRPVYGQTSVESVRNRLIDAGARPEKCQEDITFKLYPGL 125
S ++ CHG D +V ++GQ +VE +++ L +P ++TFK Y G+
Sbjct 130 S---------VLQCHGEADPLVPLIFGQLTVEKLKSML-----KPS----NVTFKTYSGM 171
Query 126 GHFANQQELNDIKAFLSRTF 145
H A +E+ DIK F+ +
Sbjct 172 THSACPEEMMDIKQFIEKQL 191
> dre:503751 zgc:110848; K06999
Length=228
Score = 54.7 bits (130), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 36/139 (25%), Positives = 69/139 (49%), Gaps = 16/139 (11%)
Query 10 IIDSQIEAGIDPSRILVGGFSQGGAVAYLVCLTSPHKLGGILAMSSWCPLGK---VIEVS 66
I+ ++ AGI R+++GGF GGA+A + + GI +SS+ L K V +
Sbjct 101 IVQDELRAGIPKHRMVIGGFPMGGAMALHLVCRHHQDIAGIFCLSSF--LNKDSAVYQAV 158
Query 67 PHYVEAVPRIMHCHGAVDDMVRPVYGQTSVESVRNRLIDAGARPEKCQEDITFKLYPGLG 126
+ +P ++ CHG D++V +G+ + N L+ +K + +F +P L
Sbjct 159 ENAQRPLPELLQCHGTSDELVFHDWGEKT-----NTLL------KKAGLNASFHSFPDLN 207
Query 127 HFANQQELNDIKAFLSRTF 145
H +QEL +++++ +
Sbjct 208 HQLCRQELELLRSWILKKL 226
> dre:797125 hypothetical protein LOC797125; K06999
Length=228
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 34/139 (24%), Positives = 68/139 (48%), Gaps = 16/139 (11%)
Query 10 IIDSQIEAGIDPSRILVGGFSQGGAVAYLVCLTSPHKLGGILAMSSWCPLGK---VIEVS 66
I+ +++ AGI R+++GGFS GGA+A + + GI +SS+ L K V +
Sbjct 101 IVQNELRAGIPKQRMVIGGFSMGGAMALHLVCRHHQDIAGIFCLSSF--LNKDSAVYQAV 158
Query 67 PHYVEAVPRIMHCHGAVDDMVRPVYGQTSVESVRNRLIDAGARPEKCQEDITFKLYPGLG 126
+ +P ++ CHG ++V +G+ + +R ++A +F +P L
Sbjct 159 ENAQRPLPELLQCHGTSGELVFHDWGEKTNTLLRKAGLNA-----------SFHSFPDLN 207
Query 127 HFANQQELNDIKAFLSRTF 145
H + EL +++++ +
Sbjct 208 HQLCRHELELLRSWILKKL 226
> cpv:cgd1_3290 carboxylesterase
Length=729
Score = 52.0 bits (123), Expect = 6e-07, Method: Composition-based stats.
Identities = 25/60 (41%), Positives = 38/60 (63%), Gaps = 1/60 (1%)
Query 1 MDSKARIDKIIDSQIEAGIDPSRILVGGFSQGGAVAYLVCLTSPH-KLGGILAMSSWCPL 59
++S RI II S+I+ GID SRI + GFSQG A+A + + +GG++ +S W P+
Sbjct 133 LESVKRIRNIIKSEIDLGIDQSRIFLIGFSQGSAMALITSMIMRDITIGGVIGVSGWIPM 192
> ath:AT5G20060 phospholipase/carboxylesterase family protein;
K06130 lysophospholipase II [EC:3.1.1.5]
Length=252
Score = 52.0 bits (123), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 41/140 (29%), Positives = 64/140 (45%), Gaps = 28/140 (20%)
Query 23 RILVGGFSQGGAVAYL--VCLT---------SPHKLGGILAMSSWCPLGKVI------EV 65
++ VGGFS G A + C P L I+ +S W P K + E
Sbjct 120 KLGVGGFSMGAATSLYSATCFALGKYGNGNPYPINLSAIIGLSGWLPCAKTLAGKLEEEQ 179
Query 66 SPHYVEAVPRIMHCHGAVDDMVRPVYGQTSVESVRNRLIDAGARPEKCQEDITFKLYPGL 125
+ ++P I+ CHG DD+V +G+ S ++ L+ G + +TFK Y L
Sbjct 180 IKNRAASLP-IVVCHGKADDVVPFKFGEKSSQA----LLSNGFK------KVTFKPYSAL 228
Query 126 GHFANQQELNDIKAFLSRTF 145
GH QEL+++ A+L+ T
Sbjct 229 GHHTIPQELDELCAWLTSTL 248
> cpv:cgd5_320 carboxylesterase, lysophospholipase, signal peptide
Length=473
Score = 43.9 bits (102), Expect = 2e-04, Method: Composition-based stats.
Identities = 20/51 (39%), Positives = 34/51 (66%), Gaps = 3/51 (5%)
Query 20 DPSRILVGGFSQGGAVAYLVCLTSPHKLGGILAMSSWCP---LGKVIEVSP 67
DP RI + G+SQGGA++ V L + + LGG+++ +S+ P + K+I + P
Sbjct 312 DPKRIFIYGYSQGGALSLSVTLRTKYVLGGLVSTASFLPERAMKKLISMDP 362
> ath:AT1G52700 phospholipase/carboxylesterase family protein
Length=255
Score = 43.1 bits (100), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 42/144 (29%), Positives = 63/144 (43%), Gaps = 33/144 (22%)
Query 23 RILVGGFSQGGAVA-----------YLVCLTSPHKLGGILAMSSWCP----LGKVIEVSP 67
++ +GGFS G A++ Y P L ++ +S W P L IE S
Sbjct 120 KVGIGGFSMGAAISLYSATCYALGRYGTGHAYPINLQAVVGLSGWLPGWKSLRSKIECS- 178
Query 68 HYVEAVPR-----IMHCHGAVDDMVRPVYGQTSVESVRNRLIDAGARPEKCQEDITFKLY 122
EA R I+ HG DD+V +G+ S +S L AG R FK Y
Sbjct 179 --FEAARRAASLPIILTHGTSDDVVPYRFGEKSAQS----LGMAGFRLA------MFKPY 226
Query 123 PGLGHFANQQELNDIKAFLSRTFG 146
GLGH+ +E++++ +L+ G
Sbjct 227 EGLGHYTVPREMDEVVHWLTTMLG 250
> ath:AT3G15650 phospholipase/carboxylesterase family protein
Length=255
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 38/143 (26%), Positives = 60/143 (41%), Gaps = 31/143 (21%)
Query 23 RILVGGFSQGGAVAYLVCLTSPHKLG-------------GILAMSSWCPLGKVIEVSPHY 69
++ +GGFS G A+A + T+ + LG + +S W P + +
Sbjct 120 KVGIGGFSMGAAIA--LYSTTCYALGRYGTGHAYTINLRATVGLSGWLPGWRSLRSKIES 177
Query 70 VEAVPR------IMHCHGAVDDMVRPVYGQTSVESVRNRLIDAGARPEKCQEDITFKLYP 123
V R I+ HG DD+V +G+ S S L AG R FK Y
Sbjct 178 SNEVARRAASIPILLAHGTSDDVVPYRFGEKSAHS----LAMAGFR------QTMFKPYE 227
Query 124 GLGHFANQQELNDIKAFLSRTFG 146
GLGH+ +E++++ +L G
Sbjct 228 GLGHYTVPKEMDEVVHWLVSRLG 250
> tgo:TGME49_054690 phospholipase/carboxylesterase domain containing
protein (EC:3.1.-.-); K06889
Length=497
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 20/64 (31%), Positives = 39/64 (60%), Gaps = 0/64 (0%)
Query 2 DSKARIDKIIDSQIEAGIDPSRILVGGFSQGGAVAYLVCLTSPHKLGGILAMSSWCPLGK 61
D +A +D +++ Q E ID ++I + G S GGAVA + + PH++ G++ +++ L
Sbjct 164 DGEAALDMLVERQNELHIDANKIFLFGRSLGGAVAIDLAVQRPHQVRGVIVENTFTSLLD 223
Query 62 VIEV 65
++ V
Sbjct 224 MVWV 227
> cel:F01D5.8 hypothetical protein
Length=305
Score = 33.1 bits (74), Expect = 0.30, Method: Compositional matrix adjust.
Identities = 24/115 (20%), Positives = 52/115 (45%), Gaps = 20/115 (17%)
Query 2 DSKARIDKIIDSQIEAGIDPSRILVGGFSQGGAVAYLVCLTSPHKLGGILAMSSWCPLGK 61
D +A +KI++ + + +I+V G+S G A + T+P +L G++ ++ + +
Sbjct 135 DVRAVYEKILEMRPD-----KKIVVMGYSIGTTAAVDLAATNPDRLAGVVLIAPFTSGLR 189
Query 62 VIEVSPHYVEAV---------------PRIMHCHGAVDDMVRPVYGQTSVESVRN 101
+ P + R++ CHG VD+++ +G E ++N
Sbjct 190 LFSSKPDKPDTCWADSFKSFDKINNIDTRVLICHGDVDEVIPLSHGLALYEKLKN 244
> hsa:126767 AADACL3; arylacetamide deacetylase-like 3 (EC:3.1.1.-);
K14351 arylacetamide deacetylase-like 3/4 [EC:3.1.1.-]
Length=280
Score = 32.0 bits (71), Expect = 0.75, Method: Compositional matrix adjust.
Identities = 13/23 (56%), Positives = 19/23 (82%), Gaps = 0/23 (0%)
Query 18 GIDPSRILVGGFSQGGAVAYLVC 40
G+DP+R++V G S GGA+A +VC
Sbjct 54 GVDPARVVVCGDSFGGAIAAVVC 76
> dre:561333 abhd13, MGC123286, zgc:123286; abhydrolase domain
containing 13; K06889
Length=337
Score = 31.2 bits (69), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 29/137 (21%), Positives = 52/137 (37%), Gaps = 26/137 (18%)
Query 2 DSKARIDKIIDSQIEAGIDPSRILVGGFSQGGAVAYLVCLTSPHKLGGILAMSS------ 55
D++A +D ++ ID +++++ G S GGAVA + +PH++ I+ ++
Sbjct 168 DAEATLDYVMT---RPDIDKTKVVLFGRSLGGAVAIRLASCNPHRVAAIMVENTFLSIPH 224
Query 56 ----------------WCPLGKVIEVSPHYVEAVPRIMHCHGAVDDMVRPVYGQTSVESV 99
WC K + H V + G D ++ PV + E
Sbjct 225 MAATLFSFFPMRYLPLWCYKNKFLSYR-HVVPCRMPSLFISGLSDQLIPPVMMKQLYELS 283
Query 100 RNRLIDAGARPEKCQED 116
+R PE D
Sbjct 284 PSRTKRLAIFPEGTHND 300
> ath:AT4G14570 acylaminoacyl-peptidase-related; K01303 acylaminoacyl-peptidase
[EC:3.4.19.1]
Length=764
Score = 30.0 bits (66), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 19/57 (33%), Positives = 29/57 (50%), Gaps = 1/57 (1%)
Query 11 IDSQIEAGI-DPSRILVGGFSQGGAVAYLVCLTSPHKLGGILAMSSWCPLGKVIEVS 66
+D IE GI DPSRI V G S GG + + +P K A + C + ++ ++
Sbjct 598 VDHAIEMGIADPSRITVLGGSHGGFLTTHLIGQAPDKFVAAAARNPVCNMASMVGIT 654
> eco:b3982 nusG, ECK3973, JW3945; transcription termination factor;
K02601 transcriptional antiterminator NusG
Length=181
Score = 30.0 bits (66), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 16/47 (34%), Positives = 24/47 (51%), Gaps = 1/47 (2%)
Query 64 EVSPHYVEAVPRIMHCHGAVDDMVRPVYGQTSVESVRNRLIDAGARP 110
+ S H V +VPR+M G D P+ V+++ NRL G +P
Sbjct 77 DASWHLVRSVPRVMGFIGGTSDRPAPI-SDKEVDAIMNRLQQVGDKP 122
> dre:567227 esyt1a, si:ch211-150a20.1; extended synaptotagmin-like
protein 1a
Length=1079
Score = 30.0 bits (66), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 24/86 (27%), Positives = 35/86 (40%), Gaps = 11/86 (12%)
Query 40 CLTSPHKLGGILAMSSWCPLGKVIEVSPHYVEAVPRIMHCH--GAVDDMVRPV----YGQ 93
L+SP L M W L K S Y+ A+ R++ + + V P+ YG+
Sbjct 551 LLSSPE-----LTMDQWFQLEKSGPASRIYITAMLRVLWLNEDAILTSPVSPIPGEGYGE 605
Query 94 TSVESVRNRLIDAGARPEKCQEDITF 119
T V S ++ RPE D F
Sbjct 606 TEVSSGATKVTATPKRPEHTSPDSNF 631
> eco:b3830 ysgA, ECK3824, JW5853; predicted hydrolase; K01061
carboxymethylenebutenolidase [EC:3.1.1.45]
Length=271
Score = 29.6 bits (65), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 32/135 (23%), Positives = 57/135 (42%), Gaps = 26/135 (19%)
Query 5 ARIDKIIDSQIEAGIDPSRILVGGFSQGGAVAYLVCLTSPH---------KLGGILAMSS 55
A +D + G D R+++ GF GG + +L +P KL G +++S
Sbjct 122 ADLDHVASWASRNGGDVHRLMITGFCWGGRITWLYAAHNPQLKAAVAWYGKLTGDKSLNS 181
Query 56 WCPLGKVIEVSPHYVEAVPRIMHCHGAVDDMVRPVYGQTSVESVRNRLIDAGARPEKCQE 115
+ ++++ + I+ +G D+ + Q SVE++R L A A+ E
Sbjct 182 ---PKQPVDIA---TDLNAPILGLYGGQDNSIP----QESVETMRQALRAANAKAE---- 227
Query 116 DITFKLYPGLGHFAN 130
+YP GH N
Sbjct 228 ---IIVYPDAGHAFN 239
> dre:100334714 hypothetical protein LOC100334714
Length=908
Score = 29.3 bits (64), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 25/97 (25%), Positives = 43/97 (44%), Gaps = 4/97 (4%)
Query 9 KIIDSQIEAGIDPSRILVGGFSQGGAVAYLVCLTSPHKLGGILAMSSWCPLGKVIEVSPH 68
+I+D + A D + G+ +G C P L A+++ G + VS
Sbjct 476 EIVDHALHALSDEVMVPHSGWERGNEGGEESC--KPRHLEWETALTNTT--GCLRNVSSE 531
Query 69 YVEAVPRIMHCHGAVDDMVRPVYGQTSVESVRNRLID 105
EA ++ C G VD ++ V Q + + V N+LI+
Sbjct 532 RSEARRKLRECSGLVDSLMYIVQSQINCKDVDNKLIE 568
> dre:556726 ctnnd1; catenin (cadherin-associated protein), delta
1; K05690 catenin (cadherin-associated protein), delta 1
Length=925
Score = 29.3 bits (64), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 25/97 (25%), Positives = 43/97 (44%), Gaps = 4/97 (4%)
Query 9 KIIDSQIEAGIDPSRILVGGFSQGGAVAYLVCLTSPHKLGGILAMSSWCPLGKVIEVSPH 68
+I+D + A D + G+ +G C P L A+++ G + VS
Sbjct 476 EIVDHALHALSDEVMVPHSGWERGNEGGEESC--KPRHLEWETALTNTT--GCLRNVSSE 531
Query 69 YVEAVPRIMHCHGAVDDMVRPVYGQTSVESVRNRLID 105
EA ++ C G VD ++ V Q + + V N+LI+
Sbjct 532 RSEARRKLRECSGLVDSLMYIVQSQINCKDVDNKLIE 568
> mmu:381572 9430007A20Rik, Aadacl4, Gm1034; RIKEN cDNA 9430007A20
gene
Length=407
Score = 29.3 bits (64), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 14/25 (56%), Positives = 18/25 (72%), Gaps = 0/25 (0%)
Query 18 GIDPSRILVGGFSQGGAVAYLVCLT 42
GIDPSR+++ G S GGA A +V T
Sbjct 181 GIDPSRVVICGESIGGAAAVVVTQT 205
> cel:EEED8.1 mel-47; Maternal Effect Lethal family member (mel-47)
Length=367
Score = 28.5 bits (62), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 13/22 (59%), Positives = 14/22 (63%), Gaps = 0/22 (0%)
Query 44 PHKLGGILAMSSWCPLGKVIEV 65
P K G +L SSW PL K IEV
Sbjct 113 PSKSGPVLLSSSWLPLNKDIEV 134
> ath:AT5G20520 WAV2; WAV2 (WAVY GROWTH 2); K06889
Length=308
Score = 28.5 bits (62), Expect = 8.1, Method: Compositional matrix adjust.
Identities = 17/50 (34%), Positives = 28/50 (56%), Gaps = 3/50 (6%)
Query 2 DSKARIDKIIDSQIEAGIDPSRILVGGFSQGGAVAYLVCLTSPHKLGGIL 51
D++A +D + ID SRI+V G S GGAV ++ +P K+ ++
Sbjct 134 DAQAALDHLSG---RTDIDTSRIVVFGRSLGGAVGAVLTKNNPDKVSALI 180
> eco:b2871 ygeX, ECK2867, JW2839; 2,3-diaminopropionate ammonia-lyase
(EC:4.3.1.15); K01751 diaminopropionate ammonia-lyase
[EC:4.3.1.15]
Length=398
Score = 28.5 bits (62), Expect = 8.7, Method: Compositional matrix adjust.
Identities = 15/47 (31%), Positives = 25/47 (53%), Gaps = 3/47 (6%)
Query 8 DKIIDSQIEAGIDPSRILVG---GFSQGGAVAYLVCLTSPHKLGGIL 51
D+ ++ E G+ P+ +L+ G GG + YLV + SP L I+
Sbjct 211 DEAVEQMREMGVTPTHVLLQAGVGAMAGGVLGYLVDVYSPQNLHSII 257
> ath:AT2G15230 ATLIP1; ATLIP1 (Arabidopsis thaliana lipase 1);
galactolipase/ hydrolase/ phospholipase/ triacylglycerol lipase
(EC:3.1.1.3)
Length=393
Score = 28.5 bits (62), Expect = 8.7, Method: Compositional matrix adjust.
Identities = 23/74 (31%), Positives = 36/74 (48%), Gaps = 6/74 (8%)
Query 14 QIEAGIDPSRILVGGFSQGGAVAYLVCLTSPHKLGGILAMSSWCPLGKVIEVSPHYVEAV 73
Q I S+I + G SQG +++ LT PH + A + CP+ + V+ VE
Sbjct 150 QYLYSISNSKIFLVGHSQGTIMSF-AALTQPHVAEMVEAAALLCPISYLDHVTAPLVE-- 206
Query 74 PRIMHCHGAVDDMV 87
R++ H +D MV
Sbjct 207 -RMVFMH--LDQMV 217
> hsa:2098 ESD, FGH, FLJ11763; esterase D (EC:3.1.2.12); K01070
S-formylglutathione hydrolase [EC:3.1.2.12]
Length=282
Score = 28.5 bits (62), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 16/48 (33%), Positives = 23/48 (47%), Gaps = 5/48 (10%)
Query 19 IDPSRILVGGFSQGGAVAYLVCLTSPHKLGGILAMSS-----WCPLGK 61
+DP R+ + G S GG A + L +P K + A + CP GK
Sbjct 138 VDPQRMSIFGHSMGGHGALICALKNPGKYKSVSAFAPICNPVLCPWGK 185
> ath:AT4G33650 DRP3A; DRP3A (DYNAMIN-RELATED PROTEIN 3A); GTP
binding / GTPase/ phosphoinositide binding; K01528 dynamin
GTPase [EC:3.6.5.5]
Length=808
Score = 28.5 bits (62), Expect = 9.0, Method: Compositional matrix adjust.
Identities = 24/99 (24%), Positives = 45/99 (45%), Gaps = 13/99 (13%)
Query 4 KARIDKIIDSQIEAGIDPSRILVGGFSQGGAVAYLVCLTSPHKLGGILAMSSWCPLGKVI 63
+ R+D++I + G++PS ++G + Y + + P+ +GG K +
Sbjct 489 RKRMDEVIGDFLREGLEPSEAMIGDIID-MEMDY-INTSHPNFIGGT----------KAV 536
Query 64 EVSPHYVEAVPRIMHCHGAVDDMVRPVYGQTSVESVRNR 102
E + H V++ RI H D V P +S V++R
Sbjct 537 EAAMHQVKS-SRIPHPVARPKDTVEPDRTSSSTSQVKSR 574
> cel:T09B9.1 hypothetical protein
Length=431
Score = 28.1 bits (61), Expect = 9.7, Method: Compositional matrix adjust.
Identities = 14/41 (34%), Positives = 26/41 (63%), Gaps = 1/41 (2%)
Query 1 MDSKARIDKIID-SQIEAGIDPSRILVGGFSQGGAVAYLVC 40
MD +A I+ + + ++ GID S+I++ G S GG +A ++
Sbjct 175 MDCEAAIEHLFEFGAVQFGIDTSKIVIMGDSAGGNMATVIA 215
Lambda K H
0.321 0.138 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3003468616
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40