bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0800_orf1
Length=84
Score E
Sequences producing significant alignments: (Bits) Value
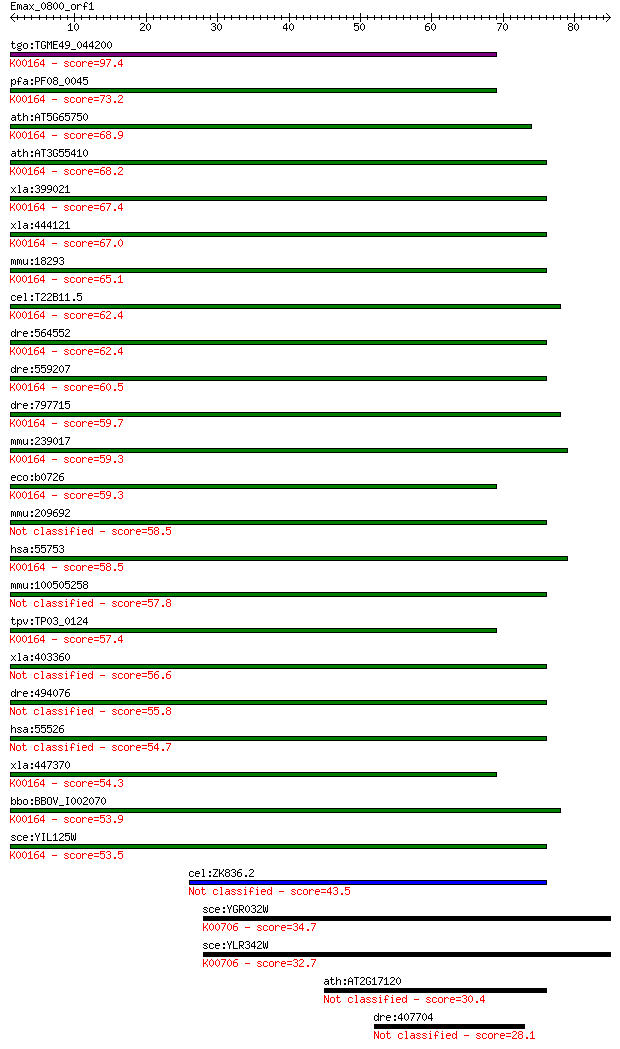
tgo:TGME49_044200 2-oxoglutarate dehydrogenase, putative (EC:1... 97.4 9e-21
pfa:PF08_0045 2-oxoglutarate dehydrogenase E1 component (EC:1.... 73.2 2e-13
ath:AT5G65750 2-oxoglutarate dehydrogenase E1 component, putat... 68.9 4e-12
ath:AT3G55410 2-oxoglutarate dehydrogenase E1 component, putat... 68.2 7e-12
xla:399021 ogdh, MGC68800, akgdh, e1k, ogdc; oxoglutarate (alp... 67.4 1e-11
xla:444121 MGC80496 protein; K00164 2-oxoglutarate dehydrogena... 67.0 1e-11
mmu:18293 Ogdh, 2210403E04Rik, 2210412K19Rik, AA409584, KIAA41... 65.1 6e-11
cel:T22B11.5 hypothetical protein; K00164 2-oxoglutarate dehyd... 62.4 3e-10
dre:564552 ogdh, MGC73296, im:7045267, wu:fa06d01, wu:fb98a04,... 62.4 4e-10
dre:559207 hypothetical LOC559207; K00164 2-oxoglutarate dehyd... 60.5 1e-09
dre:797715 si:ch211-229p19.3; K00164 2-oxoglutarate dehydrogen... 59.7 2e-09
mmu:239017 Ogdhl; oxoglutarate dehydrogenase-like (EC:1.2.4.-)... 59.3 3e-09
eco:b0726 sucA, ECK0714, JW0715, lys; 2-oxoglutarate decarboxy... 59.3 3e-09
mmu:209692 Dhtkd1, C330018I04Rik; dehydrogenase E1 and transke... 58.5 4e-09
hsa:55753 OGDHL; oxoglutarate dehydrogenase-like (EC:1.2.4.-);... 58.5 5e-09
mmu:100505258 probable 2-oxoglutarate dehydrogenase E1 compone... 57.8 9e-09
tpv:TP03_0124 2-oxoglutarate dehydrogenase e1 component (EC:1.... 57.4 1e-08
xla:403360 dhtkd1, MGC68840; dehydrogenase E1 and transketolas... 56.6 2e-08
dre:494076 dhtkd1, zgc:101818; dehydrogenase E1 and transketol... 55.8 3e-08
hsa:55526 DHTKD1, DKFZp762M115, KIAA1630, MGC3090; dehydrogena... 54.7 8e-08
xla:447370 ogdhl, MGC84242; oxoglutarate dehydrogenase-like; K... 54.3 1e-07
bbo:BBOV_I002070 19.m02351; 2-oxoglutarate dehydrogenase E1 co... 53.9 1e-07
sce:YIL125W KGD1, OGD1; Component of the mitochondrial alpha-k... 53.5 2e-07
cel:ZK836.2 hypothetical protein 43.5 2e-04
sce:YGR032W GSC2, FKS2; Catalytic subunit of 1,3-beta-glucan s... 34.7 0.069
sce:YLR342W FKS1, CND1, CWH53, ETG1, GSC1, PBR1; Catalytic sub... 32.7 0.31
ath:AT2G17120 LYM2; LYM2 (LYSM DOMAIN GPI-ANCHORED PROTEIN 2 P... 30.4 1.3
dre:407704 hypothetical protein LOC407704 28.1 6.7
> tgo:TGME49_044200 2-oxoglutarate dehydrogenase, putative (EC:1.2.4.2);
K00164 2-oxoglutarate dehydrogenase E1 component
[EC:1.2.4.2]
Length=1116
Score = 97.4 bits (241), Expect = 9e-21, Method: Composition-based stats.
Identities = 41/68 (60%), Positives = 59/68 (86%), Gaps = 2/68 (2%)
Query 1 GQIYYDLLSERERIQSENNDPDNPGNRVALARVEQLSPFPFDLAIRDIQRYPNLRSVVWA 60
GQ+YYDL++ +++++ N D + G+++A+AR+EQLSPFPFDL I D++R+PNL+SVVWA
Sbjct 975 GQVYYDLIAGKDKMK--NGDENGDGDKIAIARMEQLSPFPFDLFIEDLKRFPNLKSVVWA 1032
Query 61 QEEPMNQG 68
QEEPMNQG
Sbjct 1033 QEEPMNQG 1040
> pfa:PF08_0045 2-oxoglutarate dehydrogenase E1 component (EC:1.2.4.2);
K00164 2-oxoglutarate dehydrogenase E1 component [EC:1.2.4.2]
Length=1038
Score = 73.2 bits (178), Expect = 2e-13, Method: Composition-based stats.
Identities = 36/68 (52%), Positives = 45/68 (66%), Gaps = 9/68 (13%)
Query 1 GQIYYDLLSERERIQSENNDPDNPGNRVALARVEQLSPFPFDLAIRDIQRYPNLRSVVWA 60
GQ+YYDLL+ R N + VA+AR+EQLSPFPF + D+Q YPNLR ++WA
Sbjct 902 GQVYYDLLNYRY---------TNKIDDVAIARIEQLSPFPFKQIMNDLQTYPNLRDIIWA 952
Query 61 QEEPMNQG 68
QEE MN G
Sbjct 953 QEEHMNMG 960
> ath:AT5G65750 2-oxoglutarate dehydrogenase E1 component, putative
/ oxoglutarate decarboxylase, putative / alpha-ketoglutaric
dehydrogenase, putative; K00164 2-oxoglutarate dehydrogenase
E1 component [EC:1.2.4.2]
Length=1025
Score = 68.9 bits (167), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 36/73 (49%), Positives = 48/73 (65%), Gaps = 10/73 (13%)
Query 1 GQIYYDLLSERERIQSENNDPDNPGNRVALARVEQLSPFPFDLAIRDIQRYPNLRSVVWA 60
G++YY+L + ER +SE D VA+ RVEQL PFP+DL R+++RYPN +VW
Sbjct 903 GKVYYEL--DEERKKSETKD-------VAICRVEQLCPFPYDLIQRELKRYPNAE-IVWC 952
Query 61 QEEPMNQGKERRI 73
QEEPMN G + I
Sbjct 953 QEEPMNMGGYQYI 965
> ath:AT3G55410 2-oxoglutarate dehydrogenase E1 component, putative
/ oxoglutarate decarboxylase, putative / alpha-ketoglutaric
dehydrogenase, putative; K00164 2-oxoglutarate dehydrogenase
E1 component [EC:1.2.4.2]
Length=1017
Score = 68.2 bits (165), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 33/75 (44%), Positives = 48/75 (64%), Gaps = 10/75 (13%)
Query 1 GQIYYDLLSERERIQSENNDPDNPGNRVALARVEQLSPFPFDLAIRDIQRYPNLRSVVWA 60
G++YY+L ER+++ + + VA+ RVEQL PFP+DL R+++RYPN +VW
Sbjct 899 GKVYYELDDERKKVGATD---------VAICRVEQLCPFPYDLIQRELKRYPNAE-IVWC 948
Query 61 QEEPMNQGKERRINP 75
QEE MN G I+P
Sbjct 949 QEEAMNMGAFSYISP 963
> xla:399021 ogdh, MGC68800, akgdh, e1k, ogdc; oxoglutarate (alpha-ketoglutarate)
dehydrogenase (lipoamide) (EC:1.2.4.2);
K00164 2-oxoglutarate dehydrogenase E1 component [EC:1.2.4.2]
Length=1021
Score = 67.4 bits (163), Expect = 1e-11, Method: Composition-based stats.
Identities = 35/75 (46%), Positives = 46/75 (61%), Gaps = 9/75 (12%)
Query 1 GQIYYDLLSERERIQSENNDPDNPGNRVALARVEQLSPFPFDLAIRDIQRYPNLRSVVWA 60
G++YY+L ER E + VA+ARVEQLSPFPFDL +++Q+YPN +VW
Sbjct 904 GKVYYELTKERSGRDMEGD--------VAIARVEQLSPFPFDLVEKEVQKYPN-ADLVWC 954
Query 61 QEEPMNQGKERRINP 75
QEE NQG + P
Sbjct 955 QEEHKNQGYYDYVKP 969
> xla:444121 MGC80496 protein; K00164 2-oxoglutarate dehydrogenase
E1 component [EC:1.2.4.2]
Length=1018
Score = 67.0 bits (162), Expect = 1e-11, Method: Composition-based stats.
Identities = 35/75 (46%), Positives = 45/75 (60%), Gaps = 9/75 (12%)
Query 1 GQIYYDLLSERERIQSENNDPDNPGNRVALARVEQLSPFPFDLAIRDIQRYPNLRSVVWA 60
G++YYDL ER E + VA+ RVEQLSPFPFDL +++Q+YPN +VW
Sbjct 901 GKVYYDLTKERSGRGMEGD--------VAITRVEQLSPFPFDLVEKEVQKYPN-ADLVWC 951
Query 61 QEEPMNQGKERRINP 75
QEE NQG + P
Sbjct 952 QEEHKNQGYYDYVKP 966
> mmu:18293 Ogdh, 2210403E04Rik, 2210412K19Rik, AA409584, KIAA4192,
d1401, mKIAA4192; oxoglutarate dehydrogenase (lipoamide)
(EC:1.2.4.2); K00164 2-oxoglutarate dehydrogenase E1 component
[EC:1.2.4.2]
Length=1023
Score = 65.1 bits (157), Expect = 6e-11, Method: Composition-based stats.
Identities = 33/75 (44%), Positives = 44/75 (58%), Gaps = 9/75 (12%)
Query 1 GQIYYDLLSERERIQSENNDPDNPGNRVALARVEQLSPFPFDLAIRDIQRYPNLRSVVWA 60
G++YYDL ER+ E VA+ R+EQLSPFPFDL +++ Q+YPN + W
Sbjct 906 GKVYYDLTRERKARNMEE--------EVAITRIEQLSPFPFDLLLKEAQKYPN-AELAWC 956
Query 61 QEEPMNQGKERRINP 75
QEE NQG + P
Sbjct 957 QEEHKNQGYYDYVKP 971
> cel:T22B11.5 hypothetical protein; K00164 2-oxoglutarate dehydrogenase
E1 component [EC:1.2.4.2]
Length=1029
Score = 62.4 bits (150), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 31/77 (40%), Positives = 47/77 (61%), Gaps = 9/77 (11%)
Query 1 GQIYYDLLSERERIQSENNDPDNPGNRVALARVEQLSPFPFDLAIRDIQRYPNLRSVVWA 60
G++YYD+++ R+ + EN+ VAL RVEQLSPFP+DL ++ ++Y ++WA
Sbjct 908 GKVYYDMVAARKHVGKEND--------VALVRVEQLSPFPYDLVQQECRKYQGAE-ILWA 958
Query 61 QEEPMNQGKERRINPII 77
QEE N G + P I
Sbjct 959 QEEHKNMGAWSFVQPRI 975
> dre:564552 ogdh, MGC73296, im:7045267, wu:fa06d01, wu:fb98a04,
zgc:73296; oxoglutarate (alpha-ketoglutarate) dehydrogenase
(lipoamide) (EC:1.2.4.2); K00164 2-oxoglutarate dehydrogenase
E1 component [EC:1.2.4.2]
Length=1022
Score = 62.4 bits (150), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 33/75 (44%), Positives = 45/75 (60%), Gaps = 9/75 (12%)
Query 1 GQIYYDLLSERERIQSENNDPDNPGNRVALARVEQLSPFPFDLAIRDIQRYPNLRSVVWA 60
G+IYY+L ER+ EN+ VA+ R+EQLSPFPFDL + +++PN +VW
Sbjct 905 GKIYYELTRERKARNMENS--------VAITRIEQLSPFPFDLVRAETEKFPN-ADLVWC 955
Query 61 QEEPMNQGKERRINP 75
QEE NQG + P
Sbjct 956 QEEHKNQGYYDYVKP 970
> dre:559207 hypothetical LOC559207; K00164 2-oxoglutarate dehydrogenase
E1 component [EC:1.2.4.2]
Length=1008
Score = 60.5 bits (145), Expect = 1e-09, Method: Composition-based stats.
Identities = 30/75 (40%), Positives = 46/75 (61%), Gaps = 9/75 (12%)
Query 1 GQIYYDLLSERERIQSENNDPDNPGNRVALARVEQLSPFPFDLAIRDIQRYPNLRSVVWA 60
G++YY+L ER++++ E N VA+ R+EQ+SPFPFDL ++++Y N ++W
Sbjct 891 GKVYYELAKERKQLKLEEN--------VAIVRLEQISPFPFDLIKAEVEKYSN-AELIWC 941
Query 61 QEEPMNQGKERRINP 75
QEE N G I P
Sbjct 942 QEEHKNMGYYDYIRP 956
> dre:797715 si:ch211-229p19.3; K00164 2-oxoglutarate dehydrogenase
E1 component [EC:1.2.4.2]
Length=1023
Score = 59.7 bits (143), Expect = 2e-09, Method: Composition-based stats.
Identities = 32/77 (41%), Positives = 46/77 (59%), Gaps = 9/77 (11%)
Query 1 GQIYYDLLSERERIQSENNDPDNPGNRVALARVEQLSPFPFDLAIRDIQRYPNLRSVVWA 60
G+++YDL ER+ E RVA++R+EQLSPFPFDL + ++YP+ ++W
Sbjct 906 GKVFYDLQRERKSRGLEE--------RVAISRIEQLSPFPFDLVKAEAEKYPHAH-LLWC 956
Query 61 QEEPMNQGKERRINPII 77
QEE NQG + P I
Sbjct 957 QEEHKNQGYYDYVKPRI 973
> mmu:239017 Ogdhl; oxoglutarate dehydrogenase-like (EC:1.2.4.-);
K00164 2-oxoglutarate dehydrogenase E1 component [EC:1.2.4.2]
Length=1029
Score = 59.3 bits (142), Expect = 3e-09, Method: Composition-based stats.
Identities = 32/78 (41%), Positives = 45/78 (57%), Gaps = 9/78 (11%)
Query 1 GQIYYDLLSERERIQSENNDPDNPGNRVALARVEQLSPFPFDLAIRDIQRYPNLRSVVWA 60
G++YYDL+ ER E +VA+ R+EQ+SPFPFDL +R+ ++Y +VW
Sbjct 911 GKVYYDLVKERSSQGLEQ--------QVAITRLEQISPFPFDLIMREAEKYSG-AELVWC 961
Query 61 QEEPMNQGKERRINPIIM 78
QEE N G I+P M
Sbjct 962 QEEHKNMGYYDYISPRFM 979
> eco:b0726 sucA, ECK0714, JW0715, lys; 2-oxoglutarate decarboxylase,
thiamin-requiring (EC:1.2.4.2); K00164 2-oxoglutarate
dehydrogenase E1 component [EC:1.2.4.2]
Length=933
Score = 59.3 bits (142), Expect = 3e-09, Method: Composition-based stats.
Identities = 29/68 (42%), Positives = 42/68 (61%), Gaps = 9/68 (13%)
Query 1 GQIYYDLLSERERIQSENNDPDNPGNRVALARVEQLSPFPFDLAIRDIQRYPNLRSVVWA 60
G++YYDLL +R + NN D VA+ R+EQL PFP +Q++ +++ VW
Sbjct 822 GKVYYDLLEQRRK----NNQHD-----VAIVRIEQLYPFPHKAMQEVLQQFAHVKDFVWC 872
Query 61 QEEPMNQG 68
QEEP+NQG
Sbjct 873 QEEPLNQG 880
> mmu:209692 Dhtkd1, C330018I04Rik; dehydrogenase E1 and transketolase
domain containing 1 (EC:1.2.4.2)
Length=921
Score = 58.5 bits (140), Expect = 4e-09, Method: Composition-based stats.
Identities = 26/75 (34%), Positives = 46/75 (61%), Gaps = 7/75 (9%)
Query 1 GQIYYDLLSERERIQSENNDPDNPGNRVALARVEQLSPFPFDLAIRDIQRYPNLRSVVWA 60
G+ +Y LL +RE + ++ +D A+ R+E+L PFP D +++ +Y ++R V+W+
Sbjct 811 GKHFYALLKQRESLGTKKHD-------FAIIRLEELCPFPLDALQQEMSKYKHVRDVIWS 863
Query 61 QEEPMNQGKERRINP 75
QEEP N G ++P
Sbjct 864 QEEPQNMGPWSFVSP 878
> hsa:55753 OGDHL; oxoglutarate dehydrogenase-like (EC:1.2.4.-);
K00164 2-oxoglutarate dehydrogenase E1 component [EC:1.2.4.2]
Length=953
Score = 58.5 bits (140), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 31/78 (39%), Positives = 44/78 (56%), Gaps = 9/78 (11%)
Query 1 GQIYYDLLSERERIQSENNDPDNPGNRVALARVEQLSPFPFDLAIRDIQRYPNLRSVVWA 60
G++YYDL+ ER E +VA+ R+EQ+SPFPFDL ++ ++YP + W
Sbjct 835 GKVYYDLVKERSSQDLEE--------KVAITRLEQISPFPFDLIKQEAEKYPG-AELAWC 885
Query 61 QEEPMNQGKERRINPIIM 78
QEE N G I+P M
Sbjct 886 QEEHKNMGYYDYISPRFM 903
> mmu:100505258 probable 2-oxoglutarate dehydrogenase E1 component
DHKTD1, mitochondrial-like
Length=532
Score = 57.8 bits (138), Expect = 9e-09, Method: Composition-based stats.
Identities = 26/75 (34%), Positives = 46/75 (61%), Gaps = 7/75 (9%)
Query 1 GQIYYDLLSERERIQSENNDPDNPGNRVALARVEQLSPFPFDLAIRDIQRYPNLRSVVWA 60
G+ +Y LL +RE + ++ +D A+ R+E+L PFP D +++ +Y ++R V+W+
Sbjct 422 GKHFYALLKQRESLGTKKHD-------FAIIRLEELCPFPLDALQQEMSKYKHVRDVIWS 474
Query 61 QEEPMNQGKERRINP 75
QEEP N G ++P
Sbjct 475 QEEPQNMGPWSFVSP 489
> tpv:TP03_0124 2-oxoglutarate dehydrogenase e1 component (EC:1.2.4.2);
K00164 2-oxoglutarate dehydrogenase E1 component [EC:1.2.4.2]
Length=1030
Score = 57.4 bits (137), Expect = 1e-08, Method: Composition-based stats.
Identities = 27/68 (39%), Positives = 41/68 (60%), Gaps = 8/68 (11%)
Query 1 GQIYYDLLSERERIQSENNDPDNPGNRVALARVEQLSPFPFDLAIRDIQRYPNLRSVVWA 60
GQIYYDLL R+ + N P +AR+E+++PFP + D++++ NL ++VW
Sbjct 910 GQIYYDLLEYRDSNEEWKNIP--------VARIEEITPFPAQNILDDLKKFKNLETLVWC 961
Query 61 QEEPMNQG 68
QEE N G
Sbjct 962 QEEHENSG 969
> xla:403360 dhtkd1, MGC68840; dehydrogenase E1 and transketolase
domain containing 1 (EC:1.2.4.2)
Length=927
Score = 56.6 bits (135), Expect = 2e-08, Method: Composition-based stats.
Identities = 27/75 (36%), Positives = 40/75 (53%), Gaps = 7/75 (9%)
Query 1 GQIYYDLLSERERIQSENNDPDNPGNRVALARVEQLSPFPFDLAIRDIQRYPNLRSVVWA 60
G+ YY L +RE + + G A+ RVE+L PFP + ++I RYP + +W+
Sbjct 818 GKHYYALHKQREALGEQ-------GRSSAIIRVEELCPFPLEALQQEIHRYPKAKDFIWS 870
Query 61 QEEPMNQGKERRINP 75
QEEP N G + P
Sbjct 871 QEEPQNMGAWTFVAP 885
> dre:494076 dhtkd1, zgc:101818; dehydrogenase E1 and transketolase
domain containing 1 (EC:1.2.4.2)
Length=925
Score = 55.8 bits (133), Expect = 3e-08, Method: Composition-based stats.
Identities = 29/76 (38%), Positives = 43/76 (56%), Gaps = 9/76 (11%)
Query 1 GQIYYDLLSERERI-QSENNDPDNPGNRVALARVEQLSPFPFDLAIRDIQRYPNLRSVVW 59
G+ YY LL RE I ++E N AL RVE+L PFP + +++ +Y N + +W
Sbjct 816 GKHYYALLKHRETIPEAEKN--------TALVRVEELCPFPTEALQQELNKYTNAKEFIW 867
Query 60 AQEEPMNQGKERRINP 75
+QEEP N G ++P
Sbjct 868 SQEEPQNMGCWSFVSP 883
> hsa:55526 DHTKD1, DKFZp762M115, KIAA1630, MGC3090; dehydrogenase
E1 and transketolase domain containing 1 (EC:1.2.4.2)
Length=919
Score = 54.7 bits (130), Expect = 8e-08, Method: Composition-based stats.
Identities = 24/75 (32%), Positives = 45/75 (60%), Gaps = 7/75 (9%)
Query 1 GQIYYDLLSERERIQSENNDPDNPGNRVALARVEQLSPFPFDLAIRDIQRYPNLRSVVWA 60
G+ +Y L+ +RE + ++ +D A+ RVE+L PFP D +++ +Y +++ +W+
Sbjct 810 GKHFYSLVKQRESLGAKKHD-------FAIIRVEELCPFPLDSLQQEMSKYKHVKDHIWS 862
Query 61 QEEPMNQGKERRINP 75
QEEP N G ++P
Sbjct 863 QEEPQNMGPWSFVSP 877
> xla:447370 ogdhl, MGC84242; oxoglutarate dehydrogenase-like;
K00164 2-oxoglutarate dehydrogenase E1 component [EC:1.2.4.2]
Length=1018
Score = 54.3 bits (129), Expect = 1e-07, Method: Composition-based stats.
Identities = 27/68 (39%), Positives = 41/68 (60%), Gaps = 9/68 (13%)
Query 1 GQIYYDLLSERERIQSENNDPDNPGNRVALARVEQLSPFPFDLAIRDIQRYPNLRSVVWA 60
G++YY+L+ ER R ++VA+ R+EQ+SPFPFDL ++ ++Y +VW
Sbjct 900 GKVYYELVKERHR--------KGLDSQVAITRLEQISPFPFDLVKQEAEKYAT-SELVWC 950
Query 61 QEEPMNQG 68
QEE N G
Sbjct 951 QEEHKNMG 958
> bbo:BBOV_I002070 19.m02351; 2-oxoglutarate dehydrogenase E1
component (EC:1.2.4.2); K00164 2-oxoglutarate dehydrogenase
E1 component [EC:1.2.4.2]
Length=891
Score = 53.9 bits (128), Expect = 1e-07, Method: Composition-based stats.
Identities = 31/77 (40%), Positives = 43/77 (55%), Gaps = 9/77 (11%)
Query 1 GQIYYDLLSERERIQSENNDPDNPGNRVALARVEQLSPFPFDLAIRDIQRYPNLRSVVWA 60
GQ+Y+D+ S+R S N VA+ VEQL PFP +++RYPNL+ +VW
Sbjct 777 GQVYFDI-SDRVSELSVGN--------VAVTTVEQLCPFPAGALKSELERYPNLKRLVWC 827
Query 61 QEEPMNQGKERRINPII 77
QEE N G ++P I
Sbjct 828 QEEHANAGGWSYVSPRI 844
> sce:YIL125W KGD1, OGD1; Component of the mitochondrial alpha-ketoglutarate
dehydrogenase complex, which catalyzes a key
step in the tricarboxylic acid (TCA) cycle, the oxidative decarboxylation
of alpha-ketoglutarate to form succinyl-CoA (EC:1.2.4.2);
K00164 2-oxoglutarate dehydrogenase E1 component
[EC:1.2.4.2]
Length=1014
Score = 53.5 bits (127), Expect = 2e-07, Method: Composition-based stats.
Identities = 27/75 (36%), Positives = 35/75 (46%), Gaps = 9/75 (12%)
Query 1 GQIYYDLLSERERIQSENNDPDNPGNRVALARVEQLSPFPFDLAIRDIQRYPNLRSVVWA 60
GQ+Y L RE + + A ++EQL PFPF + YPNL +VW
Sbjct 898 GQVYTALHKRRESLGDKTT---------AFLKIEQLHPFPFAQLRDSLNSYPNLEEIVWC 948
Query 61 QEEPMNQGKERRINP 75
QEEP+N G P
Sbjct 949 QEEPLNMGSWAYTEP 963
> cel:ZK836.2 hypothetical protein
Length=911
Score = 43.5 bits (101), Expect = 2e-04, Method: Composition-based stats.
Identities = 20/50 (40%), Positives = 28/50 (56%), Gaps = 0/50 (0%)
Query 26 NRVALARVEQLSPFPFDLAIRDIQRYPNLRSVVWAQEEPMNQGKERRINP 75
+ VA+ RVE L PFP +++YP + VW+QEEP N G + P
Sbjct 820 DSVAIVRVEMLCPFPVVDLQAVLKKYPGAQDFVWSQEEPRNAGAWSFVRP 869
> sce:YGR032W GSC2, FKS2; Catalytic subunit of 1,3-beta-glucan
synthase, involved in formation of the inner layer of the spore
wall; activity positively regulated by Rho1p and negatively
by Smk1p; has similarity to an alternate catalytic subunit,
Fks1p (Gsc1p) (EC:2.4.1.34); K00706 1,3-beta-glucan synthase
[EC:2.4.1.34]
Length=1895
Score = 34.7 bits (78), Expect = 0.069, Method: Composition-based stats.
Identities = 16/57 (28%), Positives = 32/57 (56%), Gaps = 0/57 (0%)
Query 28 VALARVEQLSPFPFDLAIRDIQRYPNLRSVVWAQEEPMNQGKERRINPIIMDKEVDI 84
V++ R+ + P + A ++ YP+L+ +E P+N+G+E RI ++D +I
Sbjct 1021 VSMQRLAKFKPHELENAEFLLRAYPDLQIAYLDEEPPLNEGEEPRIYSALIDGHCEI 1077
> sce:YLR342W FKS1, CND1, CWH53, ETG1, GSC1, PBR1; Catalytic subunit
of 1,3-beta-D-glucan synthase, functionally redundant
with alternate catalytic subunit Gsc2p; binds to regulatory
subunit Rho1p; involved in cell wall synthesis and maintenance;
localizes to sites of cell wall remodeling (EC:2.4.1.34);
K00706 1,3-beta-glucan synthase [EC:2.4.1.34]
Length=1876
Score = 32.7 bits (73), Expect = 0.31, Method: Composition-based stats.
Identities = 15/57 (26%), Positives = 31/57 (54%), Gaps = 0/57 (0%)
Query 28 VALARVEQLSPFPFDLAIRDIQRYPNLRSVVWAQEEPMNQGKERRINPIIMDKEVDI 84
V++ R+ + P + A ++ YP+L+ +E P+ +G+E RI ++D +I
Sbjct 1002 VSMQRLAKFKPHELENAEFLLRAYPDLQIAYLDEEPPLTEGEEPRIYSALIDGHCEI 1058
> ath:AT2G17120 LYM2; LYM2 (LYSM DOMAIN GPI-ANCHORED PROTEIN 2
PRECURSOR)
Length=350
Score = 30.4 bits (67), Expect = 1.3, Method: Composition-based stats.
Identities = 13/34 (38%), Positives = 22/34 (64%), Gaps = 3/34 (8%)
Query 45 IRDIQRY---PNLRSVVWAQEEPMNQGKERRINP 75
+R+IQ NLRS++ A P+N +++R+NP
Sbjct 52 LRNIQTLFAVKNLRSILGANNLPLNTSRDQRVNP 85
> dre:407704 hypothetical protein LOC407704
Length=700
Score = 28.1 bits (61), Expect = 6.7, Method: Composition-based stats.
Identities = 9/21 (42%), Positives = 17/21 (80%), Gaps = 0/21 (0%)
Query 52 PNLRSVVWAQEEPMNQGKERR 72
P++ SV WA+++P++ G+E R
Sbjct 195 PSISSVKWAEQKPVHSGEELR 215
Lambda K H
0.317 0.138 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2040069136
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40