bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0789_orf2
Length=186
Score E
Sequences producing significant alignments: (Bits) Value
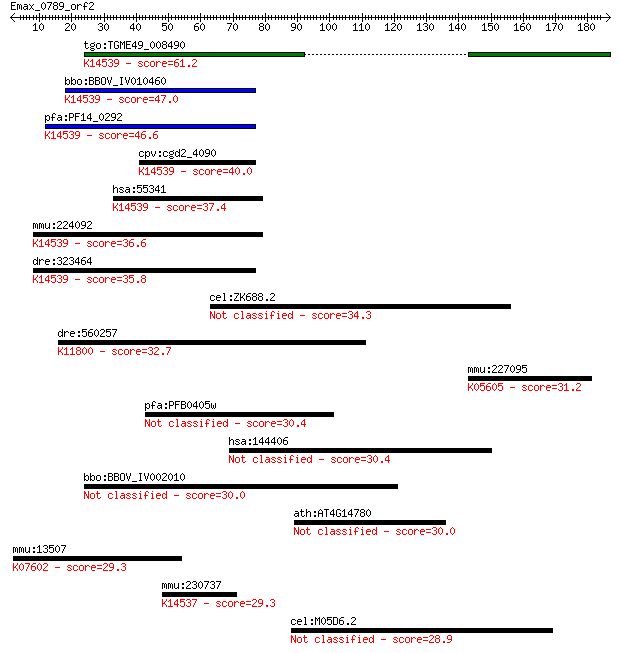
tgo:TGME49_008490 hypothetical protein ; K14539 large subunit ... 61.2 2e-09
bbo:BBOV_IV010460 23.m06487; GTPase subfamily protein; K14539 ... 47.0 4e-05
pfa:PF14_0292 cytosolic preribosomal GTPase, putative; K14539 ... 46.6 6e-05
cpv:cgd2_4090 YawG/Kre35p-like, Yjeq GTpase ; K14539 large sub... 40.0 0.005
hsa:55341 LSG1, FLJ11301, FLJ27294; large subunit GTPase 1 hom... 37.4 0.027
mmu:224092 Lsg1, 5830465I20, AA409273, D16Bwg1547e; large subu... 36.6 0.050
dre:323464 fb99b06, lsg1, wu:fb99b06; zgc:76988 (EC:3.6.1.-); ... 35.8 0.090
cel:ZK688.2 hypothetical protein 34.3 0.24
dre:560257 dcaf5, MGC153934, wdr22, zgc:153934; ddb1 and cul4 ... 32.7 0.69
mmu:227095 Hibch, 2610509I15Rik, AI648812, MGC31364; 3-hydroxy... 31.2 2.3
pfa:PFB0405w s230; transmission-blocking target antigen S230 30.4 3.3
hsa:144406 WDR66, FLJ39783, MGC33630; WD repeat domain 66 30.4
bbo:BBOV_IV002010 21.m02762; hypothetical protein 30.0 5.0
ath:AT4G14780 protein kinase, putative 30.0 5.3
mmu:13507 Dsc3; desmocollin 3; K07602 desmocollin 3 29.3
mmu:230737 Gnl2, BC003262, HUMAUANTIG, MGC7863, Ngp-1; guanine... 29.3 7.4
cel:M05D6.2 hypothetical protein 28.9 9.9
> tgo:TGME49_008490 hypothetical protein ; K14539 large subunit
GTPase 1 [EC:3.6.1.-]
Length=1064
Score = 61.2 bits (147), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 31/71 (43%), Positives = 41/71 (57%), Gaps = 3/71 (4%)
Query 24 PLEHIEAVHVMAAIATHRHFISGGKGGQLDMYRTAKMILRDFTTGRLQRCHLPKGSVY-G 82
P + A + ++A R F +GGKGGQ D+YR AKM+L+D +GR+ C P G Y G
Sbjct 808 PELQLHAPAFLESLAQKRRFTAGGKGGQWDLYRVAKMVLKDHASGRVTACRGPDGRYYDG 867
Query 83 EDE--ETANIN 91
E E ET N
Sbjct 868 EQERRETEQTN 878
Score = 31.2 bits (69), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 21/52 (40%), Positives = 31/52 (59%), Gaps = 9/52 (17%)
Query 143 LRELEQDEDLLEVLGSGGNSQAVG---GNVQCV-----MTKRKARQLQKQML 186
L +L DED +++G+ + A+G G++ C MTKRK R LQKQ+L
Sbjct 1007 LLDLTIDEDFAQIVGADKDVLALGEKPGHI-CREGNNGMTKRKMRHLQKQIL 1057
> bbo:BBOV_IV010460 23.m06487; GTPase subfamily protein; K14539
large subunit GTPase 1 [EC:3.6.1.-]
Length=826
Score = 47.0 bits (110), Expect = 4e-05, Method: Composition-based stats.
Identities = 21/59 (35%), Positives = 33/59 (55%), Gaps = 2/59 (3%)
Query 18 VNAAARPLEHIEAVHVMAAIATHRHFISGGKGGQLDMYRTAKMILRDFTTGRLQRCHLP 76
+N +P+ + + + I R F SGGKGGQ D+ R AK++++D+ G L C P
Sbjct 673 INKNKKPI--LLSTKFLECICNSRKFFSGGKGGQPDLGRAAKLVVKDYVNGNLLYCAWP 729
> pfa:PF14_0292 cytosolic preribosomal GTPase, putative; K14539
large subunit GTPase 1 [EC:3.6.1.-]
Length=833
Score = 46.6 bits (109), Expect = 6e-05, Method: Composition-based stats.
Identities = 22/65 (33%), Positives = 34/65 (52%), Gaps = 0/65 (0%)
Query 12 NAGHRKVNAAARPLEHIEAVHVMAAIATHRHFISGGKGGQLDMYRTAKMILRDFTTGRLQ 71
N H+ +N ++A + T R F+SGGKGGQL+ ++I+ DF +G+L
Sbjct 705 NIIHQYLNEKGHISYFLDASEFLKKFCTFRKFVSGGKGGQLNFSHATRIIIHDFISGKLL 764
Query 72 RCHLP 76
LP
Sbjct 765 YNFLP 769
> cpv:cgd2_4090 YawG/Kre35p-like, Yjeq GTpase ; K14539 large subunit
GTPase 1 [EC:3.6.1.-]
Length=666
Score = 40.0 bits (92), Expect = 0.005, Method: Composition-based stats.
Identities = 16/36 (44%), Positives = 21/36 (58%), Gaps = 0/36 (0%)
Query 41 RHFISGGKGGQLDMYRTAKMILRDFTTGRLQRCHLP 76
RH GKG D + +MILRD+ +G+L CH P
Sbjct 553 RHLFQQGKGAIPDWSKAGRMILRDYWSGKLLYCHTP 588
> hsa:55341 LSG1, FLJ11301, FLJ27294; large subunit GTPase 1 homolog
(S. cerevisiae); K14539 large subunit GTPase 1 [EC:3.6.1.-]
Length=658
Score = 37.4 bits (85), Expect = 0.027, Method: Compositional matrix adjust.
Identities = 17/46 (36%), Positives = 27/46 (58%), Gaps = 2/46 (4%)
Query 33 VMAAIATHRHFISGGKGGQLDMYRTAKMILRDFTTGRLQRCHLPKG 78
++ A R F++ GQ D R+A+ IL+D+ +G+L CH P G
Sbjct 509 LLTAYGYMRGFMTAH--GQPDQPRSARYILKDYVSGKLLYCHPPPG 552
> mmu:224092 Lsg1, 5830465I20, AA409273, D16Bwg1547e; large subunit
GTPase 1 homolog (S. cerevisiae); K14539 large subunit
GTPase 1 [EC:3.6.1.-]
Length=644
Score = 36.6 bits (83), Expect = 0.050, Method: Compositional matrix adjust.
Identities = 23/71 (32%), Positives = 33/71 (46%), Gaps = 5/71 (7%)
Query 8 YGAGNAGHRKVNAAARPLEHIEAVHVMAAIATHRHFISGGKGGQLDMYRTAKMILRDFTT 67
YG R+ RP E ++ A R F++ GQ D R+A+ IL+D+
Sbjct 473 YGINIIKPREDEDPYRPPTSEE---LLTAYGCMRGFMTAH--GQPDQPRSARYILKDYVG 527
Query 68 GRLQRCHLPKG 78
G+L CH P G
Sbjct 528 GKLLYCHPPPG 538
> dre:323464 fb99b06, lsg1, wu:fb99b06; zgc:76988 (EC:3.6.1.-);
K14539 large subunit GTPase 1 [EC:3.6.1.-]
Length=640
Score = 35.8 bits (81), Expect = 0.090, Method: Compositional matrix adjust.
Identities = 21/69 (30%), Positives = 34/69 (49%), Gaps = 5/69 (7%)
Query 8 YGAGNAGHRKVNAAARPLEHIEAVHVMAAIATHRHFISGGKGGQLDMYRTAKMILRDFTT 67
YG R+ RP + E ++ A R F++ GQ D R+A+ +L+D+ +
Sbjct 469 YGINIIRPREDEDPDRPPTYEE---LLMAYGYMRGFMTAH--GQPDQSRSARYVLKDYVS 523
Query 68 GRLQRCHLP 76
G+L CH P
Sbjct 524 GKLLYCHPP 532
> cel:ZK688.2 hypothetical protein
Length=632
Score = 34.3 bits (77), Expect = 0.24, Method: Composition-based stats.
Identities = 29/95 (30%), Positives = 42/95 (44%), Gaps = 8/95 (8%)
Query 63 RDFTTGRLQRCHLPKGSVYGEDEETANINKFLSE--VSSSVETRIAQADTSRVAPQLAGT 120
+ F+ G L+ + D AN N F + V+S + + DTS PQLA
Sbjct 449 KTFSHGMLENMEFEGSNTNIADGSHANNNSFTNSAFVNSGEDLSNKRIDTSSSQPQLATG 508
Query 121 SSGSSKPGGDVKQHLQMNPTDVLRELEQDEDLLEV 155
GS P QH ++ DVL + E DE+ L +
Sbjct 509 KRGSEHP----FQHHVLD--DVLEDDESDENQLTI 537
> dre:560257 dcaf5, MGC153934, wdr22, zgc:153934; ddb1 and cul4
associated factor 5; K11800 WD repeat-containing protein 22
Length=789
Score = 32.7 bits (73), Expect = 0.69, Method: Compositional matrix adjust.
Identities = 27/99 (27%), Positives = 43/99 (43%), Gaps = 15/99 (15%)
Query 16 RKVNAAARPL----EHIEAVHVMAAIATHRHFISGGKGGQLDMYRTAKMILRDFTTGRLQ 71
+ +++ A+P+ EH+ + +A +T++ SGG Q+ IL D G
Sbjct 83 KAIHSRAKPMKLKGEHLSNIFCLAFDSTNKRVFSGGNDEQV--------ILHDVERGETL 134
Query 72 RCHLPKGSVYGEDEETANINKFLSEVSSSVETRIAQADT 110
L +VYG N N F SSS + R+ DT
Sbjct 135 NVFLHDDAVYGLSVSPVNDNVF---ASSSDDGRVLIWDT 170
> mmu:227095 Hibch, 2610509I15Rik, AI648812, MGC31364; 3-hydroxyisobutyryl-Coenzyme
A hydrolase (EC:3.1.2.4); K05605 3-hydroxyisobutyryl-CoA
hydrolase [EC:3.1.2.4]
Length=385
Score = 31.2 bits (69), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 15/40 (37%), Positives = 25/40 (62%), Gaps = 2/40 (5%)
Query 143 LRELEQDED--LLEVLGSGGNSQAVGGNVQCVMTKRKARQ 180
L+ EQD D L+ + G+GG + GG+++ + +KARQ
Sbjct 71 LKTWEQDPDTFLIIIKGAGGKAFCAGGDIKALSEAKKARQ 110
> pfa:PFB0405w s230; transmission-blocking target antigen S230
Length=3135
Score = 30.4 bits (67), Expect = 3.3, Method: Composition-based stats.
Identities = 19/62 (30%), Positives = 30/62 (48%), Gaps = 4/62 (6%)
Query 43 FISGGKGGQLDMYRTAKMILRDFTTGRLQRCHLPKGSVYGED----EETANINKFLSEVS 98
FI GG+G + +K++L D T R+ + H + YGE E+ N+ K + V
Sbjct 485 FIEGGEGDDVYKVDGSKVLLDDDTISRVSKKHTARDGEYGEYGEAVEDGENVIKIIRSVL 544
Query 99 SS 100
S
Sbjct 545 QS 546
> hsa:144406 WDR66, FLJ39783, MGC33630; WD repeat domain 66
Length=941
Score = 30.4 bits (67), Expect = 3.9, Method: Composition-based stats.
Identities = 21/81 (25%), Positives = 40/81 (49%), Gaps = 7/81 (8%)
Query 69 RLQRCHLPKGSVYGEDEETANINKFLSEVSSSVETRIAQADTSRVAPQLAGTSSGSSKPG 128
+ QR K S+ ED ET ++ L ++S+ +E D +++P+ SS +P
Sbjct 127 KTQRGSKSKLSLQLEDAET---DELLRDLSTQIEF----LDLDQISPEEQQISSPERQPS 179
Query 129 GDVKQHLQMNPTDVLRELEQD 149
G++++ P D L + +D
Sbjct 180 GELEEKTDRMPQDELGQERRD 200
> bbo:BBOV_IV002010 21.m02762; hypothetical protein
Length=319
Score = 30.0 bits (66), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 24/99 (24%), Positives = 48/99 (48%), Gaps = 5/99 (5%)
Query 24 PLEHIEAVHVMAAIATHRHFISGGKGGQLDMYRTAKMILRDFTTGRL--QRCHLPKGSVY 81
P H+ +++V ++A + F SG G L + +K I+ G + + K +V
Sbjct 189 PERHLASIYV--SLAQCQMF-SGQLGLALRTFTESKEIMMRLLMGNMTDEEQQRMKDTVE 245
Query 82 GEDEETANINKFLSEVSSSVETRIAQADTSRVAPQLAGT 120
D + A++ K + VS+ + + +Q + + + PQ GT
Sbjct 246 DVDIQIADLKKLIESVSTKTDNQGSQCNAADLVPQTTGT 284
> ath:AT4G14780 protein kinase, putative
Length=364
Score = 30.0 bits (66), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 16/47 (34%), Positives = 21/47 (44%), Gaps = 0/47 (0%)
Query 89 NINKFLSEVSSSVETRIAQADTSRVAPQLAGTSSGSSKPGGDVKQHL 135
N+ KF+ + I AD+ PQ A PGG +KQHL
Sbjct 124 NVTKFVGASMGTTNLNIRSADSKGSLPQQACCVVVEYLPGGTLKQHL 170
> mmu:13507 Dsc3; desmocollin 3; K07602 desmocollin 3
Length=896
Score = 29.3 bits (64), Expect = 7.4, Method: Composition-based stats.
Identities = 16/54 (29%), Positives = 25/54 (46%), Gaps = 2/54 (3%)
Query 2 AYNSSQYGAGNAGHRKVNAAARPLEHIEAVHVMAA--IATHRHFISGGKGGQLD 53
A NSSQ G G N +E ++ + + +A H H + G+GG +D
Sbjct 757 ANNSSQGFCGTMGSGMRNGGQETIEMMKGHQTLDSCRVAGHHHTLDSGRGGHMD 810
> mmu:230737 Gnl2, BC003262, HUMAUANTIG, MGC7863, Ngp-1; guanine
nucleotide binding protein-like 2 (nucleolar); K14537 nuclear
GTP-binding protein
Length=728
Score = 29.3 bits (64), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 11/23 (47%), Positives = 16/23 (69%), Gaps = 0/23 (0%)
Query 48 KGGQLDMYRTAKMILRDFTTGRL 70
KGG+ DM +KM+L D+ GR+
Sbjct 434 KGGEPDMLTVSKMVLNDWQRGRI 456
> cel:M05D6.2 hypothetical protein
Length=451
Score = 28.9 bits (63), Expect = 9.9, Method: Compositional matrix adjust.
Identities = 22/81 (27%), Positives = 32/81 (39%), Gaps = 0/81 (0%)
Query 88 ANINKFLSEVSSSVETRIAQADTSRVAPQLAGTSSGSSKPGGDVKQHLQMNPTDVLRELE 147
A IN L E + + Q D +V + S P DVK TDV+ E
Sbjct 78 AEINSMLDETALRGKLDQGQLDIKKVMKYIVDLCSRLCSPARDVKVAELRTRTDVIDIFE 137
Query 148 QDEDLLEVLGSGGNSQAVGGN 168
DLLE++ + + + N
Sbjct 138 GTMDLLELMKNDLTNYQISQN 158
Lambda K H
0.313 0.128 0.358
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5170784960
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40