bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0765_orf3
Length=233
Score E
Sequences producing significant alignments: (Bits) Value
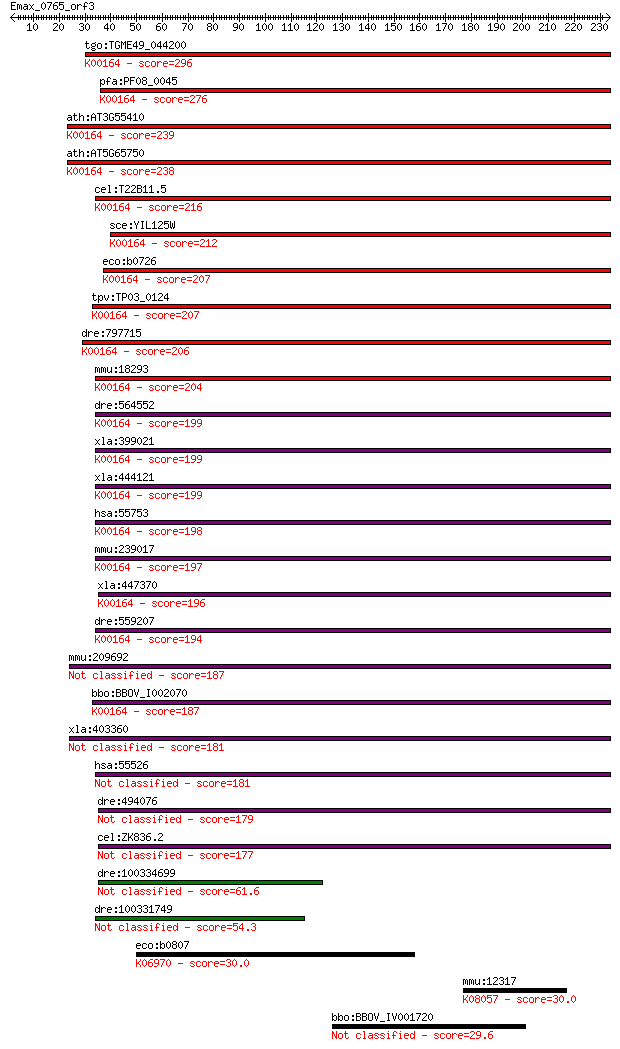
tgo:TGME49_044200 2-oxoglutarate dehydrogenase, putative (EC:1... 296 3e-80
pfa:PF08_0045 2-oxoglutarate dehydrogenase E1 component (EC:1.... 276 7e-74
ath:AT3G55410 2-oxoglutarate dehydrogenase E1 component, putat... 239 7e-63
ath:AT5G65750 2-oxoglutarate dehydrogenase E1 component, putat... 238 1e-62
cel:T22B11.5 hypothetical protein; K00164 2-oxoglutarate dehyd... 216 6e-56
sce:YIL125W KGD1, OGD1; Component of the mitochondrial alpha-k... 212 1e-54
eco:b0726 sucA, ECK0714, JW0715, lys; 2-oxoglutarate decarboxy... 207 2e-53
tpv:TP03_0124 2-oxoglutarate dehydrogenase e1 component (EC:1.... 207 3e-53
dre:797715 si:ch211-229p19.3; K00164 2-oxoglutarate dehydrogen... 206 4e-53
mmu:18293 Ogdh, 2210403E04Rik, 2210412K19Rik, AA409584, KIAA41... 204 2e-52
dre:564552 ogdh, MGC73296, im:7045267, wu:fa06d01, wu:fb98a04,... 199 5e-51
xla:399021 ogdh, MGC68800, akgdh, e1k, ogdc; oxoglutarate (alp... 199 6e-51
xla:444121 MGC80496 protein; K00164 2-oxoglutarate dehydrogena... 199 6e-51
hsa:55753 OGDHL; oxoglutarate dehydrogenase-like (EC:1.2.4.-);... 198 1e-50
mmu:239017 Ogdhl; oxoglutarate dehydrogenase-like (EC:1.2.4.-)... 197 2e-50
xla:447370 ogdhl, MGC84242; oxoglutarate dehydrogenase-like; K... 196 5e-50
dre:559207 hypothetical LOC559207; K00164 2-oxoglutarate dehyd... 194 2e-49
mmu:209692 Dhtkd1, C330018I04Rik; dehydrogenase E1 and transke... 187 3e-47
bbo:BBOV_I002070 19.m02351; 2-oxoglutarate dehydrogenase E1 co... 187 4e-47
xla:403360 dhtkd1, MGC68840; dehydrogenase E1 and transketolas... 181 2e-45
hsa:55526 DHTKD1, DKFZp762M115, KIAA1630, MGC3090; dehydrogena... 181 3e-45
dre:494076 dhtkd1, zgc:101818; dehydrogenase E1 and transketol... 179 5e-45
cel:ZK836.2 hypothetical protein 177 3e-44
dre:100334699 probable 2-oxoglutarate dehydrogenase E1 compone... 61.6 2e-09
dre:100331749 oxoglutarate (alpha-ketoglutarate) dehydrogenase... 54.3 4e-07
eco:b0807 rlmF, ECK0796, JW5107, ybiN; 23S rRNA mA1618 methylt... 30.0 8.1
mmu:12317 Calr, CRT, Calregulin; calreticulin; K08057 calretic... 30.0 8.3
bbo:BBOV_IV001720 21.m02935; hypothetical protein 29.6 8.5
> tgo:TGME49_044200 2-oxoglutarate dehydrogenase, putative (EC:1.2.4.2);
K00164 2-oxoglutarate dehydrogenase E1 component
[EC:1.2.4.2]
Length=1116
Score = 296 bits (759), Expect = 3e-80, Method: Compositional matrix adjust.
Identities = 138/204 (67%), Positives = 157/204 (76%), Gaps = 0/204 (0%)
Query 30 AAVVAAGVRLSALRELGRRIFTIPEGFVPHATIAKIMKQRLAAVEGPQHEKSLDFGAAEN 89
AA GV L LRELG +IFT+P F H T+ KI K+RL A++ E +DFG AEN
Sbjct 670 AAPQLTGVPLDRLRELGTKIFTLPPDFNVHPTVGKIYKERLNAIQAAPDENLIDFGTAEN 729
Query 90 LAYATLLSDGFHVRLAGQDAQRGTFSHRHAVLHDQAVEAQHCIFDGLKDLNLPHCITVCN 149
L YATLLSDGFHVR+AGQD QRGTFSHRHAVLHDQ + IFD LK PH I N
Sbjct 730 LCYATLLSDGFHVRIAGQDVQRGTFSHRHAVLHDQTTFEPYSIFDSLKCYGFPHKIQTVN 789
Query 150 SPLSEYAALGYEFGYSMEHPDTVAIWEAQFGDFSNGAQIIIDQFVVSAEVKWNRQNGLVM 209
SPLSEYAA+GYE GYS+EHPD++ IWEAQFGDF+NGAQIIIDQF+ S EVKWN+Q G+V+
Sbjct 790 SPLSEYAAMGYELGYSLEHPDSLCIWEAQFGDFANGAQIIIDQFIASGEVKWNKQTGIVV 849
Query 210 LLPHGYDGQGPEHSSARIERFLQL 233
+LPHGYDGQGPEHSS RIER LQL
Sbjct 850 MLPHGYDGQGPEHSSGRIERILQL 873
> pfa:PF08_0045 2-oxoglutarate dehydrogenase E1 component (EC:1.2.4.2);
K00164 2-oxoglutarate dehydrogenase E1 component [EC:1.2.4.2]
Length=1038
Score = 276 bits (705), Expect = 7e-74, Method: Composition-based stats.
Identities = 129/198 (65%), Positives = 150/198 (75%), Gaps = 6/198 (3%)
Query 36 GVRLSALRELGRRIFTIPEGFVPHATIAKIMKQRLAAVEGPQHEKSLDFGAAENLAYATL 95
GV L LG++IFT+ E F H I K+ K R+ ++E K++DFG AE LAYATL
Sbjct 607 GVEKDVLINLGKKIFTLRENFTAHPIITKLFKSRIDSLET---GKNIDFGTAELLAYATL 663
Query 96 LSDGFHVRLAGQDAQRGTFSHRHAVLHDQAVEAQHCIFDGLKDLNLPHCITVCNSPLSEY 155
LSDGFH RL+GQD+QRGTFSHRHAVLHDQ + IFD LK PH I V NS LSEY
Sbjct 664 LSDGFHARLSGQDSQRGTFSHRHAVLHDQITYESYNIFDSLKT---PHTIEVNNSLLSEY 720
Query 156 AALGYEFGYSMEHPDTVAIWEAQFGDFSNGAQIIIDQFVVSAEVKWNRQNGLVMLLPHGY 215
A LGYE GYS EHPD + IWEAQFGDF+NGAQ++ID ++ S E KWN+Q+G+VMLLPHGY
Sbjct 721 ACLGYEIGYSYEHPDALVIWEAQFGDFANGAQVMIDNYIASGETKWNKQSGIVMLLPHGY 780
Query 216 DGQGPEHSSARIERFLQL 233
DGQGPEHSSARIERFLQL
Sbjct 781 DGQGPEHSSARIERFLQL 798
> ath:AT3G55410 2-oxoglutarate dehydrogenase E1 component, putative
/ oxoglutarate decarboxylase, putative / alpha-ketoglutaric
dehydrogenase, putative; K00164 2-oxoglutarate dehydrogenase
E1 component [EC:1.2.4.2]
Length=1017
Score = 239 bits (609), Expect = 7e-63, Method: Compositional matrix adjust.
Identities = 112/211 (53%), Positives = 147/211 (69%), Gaps = 3/211 (1%)
Query 23 YISATATAAVVAAGVRLSALRELGRRIFTIPEGFVPHATIAKIMKQRLAAVEGPQHEKSL 82
+ S + V GV+ L+ +G+ I ++PE F PH + K+ +QR +E + +
Sbjct 579 FKSPEQISRVRNTGVKPEILKTVGKAISSLPENFKPHRAVKKVYEQRAQMIESGE---GV 635
Query 83 DFGAAENLAYATLLSDGFHVRLAGQDAQRGTFSHRHAVLHDQAVEAQHCIFDGLKDLNLP 142
D+ AE LA+ATL+ +G HVRL+GQD +RGTFSHRH+VLHDQ ++C D L P
Sbjct 636 DWALAEALAFATLVVEGNHVRLSGQDVERGTFSHRHSVLHDQETGEEYCPLDHLIMNQDP 695
Query 143 HCITVCNSPLSEYAALGYEFGYSMEHPDTVAIWEAQFGDFSNGAQIIIDQFVVSAEVKWN 202
TV NS LSE+ LG+E GYSME P+++ +WEAQFGDF+NGAQ+I DQF+ S E KW
Sbjct 696 EMFTVSNSSLSEFGVLGFELGYSMESPNSLVLWEAQFGDFANGAQVIFDQFISSGEAKWL 755
Query 203 RQNGLVMLLPHGYDGQGPEHSSARIERFLQL 233
RQ GLVMLLPHGYDGQGPEHSSAR+ER+LQ+
Sbjct 756 RQTGLVMLLPHGYDGQGPEHSSARLERYLQM 786
> ath:AT5G65750 2-oxoglutarate dehydrogenase E1 component, putative
/ oxoglutarate decarboxylase, putative / alpha-ketoglutaric
dehydrogenase, putative; K00164 2-oxoglutarate dehydrogenase
E1 component [EC:1.2.4.2]
Length=1025
Score = 238 bits (607), Expect = 1e-62, Method: Compositional matrix adjust.
Identities = 110/211 (52%), Positives = 146/211 (69%), Gaps = 3/211 (1%)
Query 23 YISATATAAVVAAGVRLSALRELGRRIFTIPEGFVPHATIAKIMKQRLAAVEGPQHEKSL 82
+ S + + GV+ L+ +G+ I T PE F PH + ++ +QR +E + +
Sbjct 583 FKSPEQISRIRNTGVKPEILKNVGKAISTFPENFKPHRGVKRVYEQRAQMIESGE---GI 639
Query 83 DFGAAENLAYATLLSDGFHVRLAGQDAQRGTFSHRHAVLHDQAVEAQHCIFDGLKDLNLP 142
D+G E LA+ATL+ +G HVRL+GQD +RGTFSHRH+VLHDQ ++C D L P
Sbjct 640 DWGLGEALAFATLVVEGNHVRLSGQDVERGTFSHRHSVLHDQETGEEYCPLDHLIKNQDP 699
Query 143 HCITVCNSPLSEYAALGYEFGYSMEHPDTVAIWEAQFGDFSNGAQIIIDQFVVSAEVKWN 202
TV NS LSE+ LG+E GYSME+P+++ IWEAQFGDF+NGAQ++ DQF+ S E KW
Sbjct 700 EMFTVSNSSLSEFGVLGFELGYSMENPNSLVIWEAQFGDFANGAQVMFDQFISSGEAKWL 759
Query 203 RQNGLVMLLPHGYDGQGPEHSSARIERFLQL 233
RQ GLV+LLPHGYDGQGPEHSS R+ERFLQ+
Sbjct 760 RQTGLVVLLPHGYDGQGPEHSSGRLERFLQM 790
> cel:T22B11.5 hypothetical protein; K00164 2-oxoglutarate dehydrogenase
E1 component [EC:1.2.4.2]
Length=1029
Score = 216 bits (550), Expect = 6e-56, Method: Compositional matrix adjust.
Identities = 108/201 (53%), Positives = 139/201 (69%), Gaps = 7/201 (3%)
Query 34 AAGVRLSALRELGRRIFTIPEGFVPHATIAKIMKQRLAAVEGPQHEKSLDFGAAENLAYA 93
+ G+ + ++ + PEGF H + + +K R ++ + SLD+ E LA+
Sbjct 607 STGIEQENIEQIIGKFSQYPEGFNLHRGLERTLKGRQQMLK----DNSLDWACGEALAFG 662
Query 94 TLLSDGFHVRLAGQDAQRGTFSHRHAVLHDQAVEAQHCIFDGLKDLNLPHC-ITVCNSPL 152
+LL +G HVRL+GQD QRGTFSHRH VLHDQ V+ + I++ L DL+ TVCNS L
Sbjct 663 SLLKEGIHVRLSGQDVQRGTFSHRHHVLHDQKVDQK--IYNPLNDLSEGQGEYTVCNSSL 720
Query 153 SEYAALGYEFGYSMEHPDTVAIWEAQFGDFSNGAQIIIDQFVVSAEVKWNRQNGLVMLLP 212
SEYA LG+E GYSM P+++ IWEAQFGDFSN AQ IIDQF+ S + KW RQ+GLVMLLP
Sbjct 721 SEYAVLGFELGYSMVDPNSLVIWEAQFGDFSNTAQCIIDQFISSGQSKWIRQSGLVMLLP 780
Query 213 HGYDGQGPEHSSARIERFLQL 233
HGY+G GPEHSSAR ERFLQ+
Sbjct 781 HGYEGMGPEHSSARPERFLQM 801
> sce:YIL125W KGD1, OGD1; Component of the mitochondrial alpha-ketoglutarate
dehydrogenase complex, which catalyzes a key
step in the tricarboxylic acid (TCA) cycle, the oxidative decarboxylation
of alpha-ketoglutarate to form succinyl-CoA (EC:1.2.4.2);
K00164 2-oxoglutarate dehydrogenase E1 component
[EC:1.2.4.2]
Length=1014
Score = 212 bits (539), Expect = 1e-54, Method: Compositional matrix adjust.
Identities = 102/195 (52%), Positives = 137/195 (70%), Gaps = 7/195 (3%)
Query 40 SALRELGRRIFTIPEGFVPHATIAKIMKQRLAAVEGPQHEKSLDFGAAENLAYATLLSDG 99
S L+ELG+ + + PEGF H + +I+K R ++E + +D+ E LA+ TL+ DG
Sbjct 607 STLKELGKVLSSWPEGFEVHKNLKRILKNRGKSIETGE---GIDWATGEALAFGTLVLDG 663
Query 100 FHVRLAGQDAQRGTFSHRHAVLHDQAVEAQHCIFDGLKDLNLPHC-ITVCNSPLSEYAAL 158
+VR++G+D +RGTFS RHAVLHDQ EA I+ L LN T+ NS LSEY +
Sbjct 664 QNVRVSGEDVERGTFSQRHAVLHDQQSEA---IYTPLSTLNNEKADFTIANSSLSEYGVM 720
Query 159 GYEFGYSMEHPDTVAIWEAQFGDFSNGAQIIIDQFVVSAEVKWNRQNGLVMLLPHGYDGQ 218
G+E+GYS+ PD + +WEAQFGDF+N AQ+IIDQF+ E KW +++GLV+ LPHGYDGQ
Sbjct 721 GFEYGYSLTSPDYLVMWEAQFGDFANTAQVIIDQFIAGGEQKWKQRSGLVLSLPHGYDGQ 780
Query 219 GPEHSSARIERFLQL 233
GPEHSS R+ERFLQL
Sbjct 781 GPEHSSGRLERFLQL 795
> eco:b0726 sucA, ECK0714, JW0715, lys; 2-oxoglutarate decarboxylase,
thiamin-requiring (EC:1.2.4.2); K00164 2-oxoglutarate
dehydrogenase E1 component [EC:1.2.4.2]
Length=933
Score = 207 bits (528), Expect = 2e-53, Method: Compositional matrix adjust.
Identities = 105/197 (53%), Positives = 135/197 (68%), Gaps = 5/197 (2%)
Query 37 VRLSALRELGRRIFTIPEGFVPHATIAKIMKQRLAAVEGPQHEKSLDFGAAENLAYATLL 96
V + L+EL +RI T+PE + +AKI R A G EK D+G AENLAYATL+
Sbjct 549 VEMKRLQELAKRISTVPEAVEMQSRVAKIYGDRQAMAAG---EKLFDWGGAENLAYATLV 605
Query 97 SDGFHVRLAGQDAQRGTFSHRHAVLHDQAVEAQHCIFDGLKDLNLPHCITVCNSPLSEYA 156
+G VRL+G+D+ RGTF HRHAV+H+Q+ + + + N V +S LSE A
Sbjct 606 DEGIPVRLSGEDSGRGTFFHRHAVIHNQSNGSTYTPLQHIH--NGQGAFRVWDSVLSEEA 663
Query 157 ALGYEFGYSMEHPDTVAIWEAQFGDFSNGAQIIIDQFVVSAEVKWNRQNGLVMLLPHGYD 216
L +E+GY+ P T+ IWEAQFGDF+NGAQ++IDQF+ S E KW R GLVMLLPHGY+
Sbjct 664 VLAFEYGYATAEPRTLTIWEAQFGDFANGAQVVIDQFISSGEQKWGRMCGLVMLLPHGYE 723
Query 217 GQGPEHSSARIERFLQL 233
GQGPEHSSAR+ER+LQL
Sbjct 724 GQGPEHSSARLERYLQL 740
> tpv:TP03_0124 2-oxoglutarate dehydrogenase e1 component (EC:1.2.4.2);
K00164 2-oxoglutarate dehydrogenase E1 component [EC:1.2.4.2]
Length=1030
Score = 207 bits (527), Expect = 3e-53, Method: Composition-based stats.
Identities = 102/204 (50%), Positives = 138/204 (67%), Gaps = 6/204 (2%)
Query 33 VAAGVRLSALRELGRRIFTIPEGFVPHATIAKIMKQRLAAVEGPQHEKSLDFGAAENLAY 92
V G+ + L ELG + T+P H ++ KI RL + + D +E LA+
Sbjct 597 VETGLDKNLLLELGTKCVTVPSDIKMHNSVKKIFDARLQCLSTGS---NFDTAMSEILAF 653
Query 93 ATLLSDGFHVRLAGQDAQRGTFSHRHAVLHDQAVEAQHCIFDGLK---DLNLPHCITVCN 149
++L ++GFHVRL+GQ+++RGTFSHRH+ + Q H IF G++ ++ +++ N
Sbjct 654 SSLANEGFHVRLSGQESKRGTFSHRHSHVQCQTTFKYHNIFKGIECFINVFSGFDVSIYN 713
Query 150 SPLSEYAALGYEFGYSMEHPDTVAIWEAQFGDFSNGAQIIIDQFVVSAEVKWNRQNGLVM 209
S LSE AALG+E+GYS+ P T+ IWEAQFGDF NGAQ+IID FV SAE KWN +GLV+
Sbjct 714 SYLSELAALGFEYGYSLYSPKTLNIWEAQFGDFMNGAQVIIDAFVTSAETKWNYFSGLVL 773
Query 210 LLPHGYDGQGPEHSSARIERFLQL 233
LPHGYDGQGP+HSS+RIERFLQL
Sbjct 774 FLPHGYDGQGPDHSSSRIERFLQL 797
> dre:797715 si:ch211-229p19.3; K00164 2-oxoglutarate dehydrogenase
E1 component [EC:1.2.4.2]
Length=1023
Score = 206 bits (525), Expect = 4e-53, Method: Compositional matrix adjust.
Identities = 102/207 (49%), Positives = 138/207 (66%), Gaps = 8/207 (3%)
Query 29 TAAVVAAGVRLSALRELGRRIFTIP-EGFVPHATIAKIMKQRLAAVEGPQHEKSLDFGAA 87
T + + G+ L +G+ ++P E F H +++I+K R V+ +S+D+
Sbjct 602 TMSCPSTGLSEETLAHIGQTASSVPVEDFTIHGGLSRILKSRSLMVQN----RSVDWALG 657
Query 88 ENLAYATLLSDGFHVRLAGQDAQRGTFSHRHAVLHDQAVEAQHCIFDGLKDLN-LPHCIT 146
E +A+ +LL +G HVRL+GQD +RGTFSHRH VLHDQ V+ + CI D N P+ T
Sbjct 658 EYMAFGSLLKEGIHVRLSGQDVERGTFSHRHHVLHDQNVDKRTCIPMNYMDPNQAPY--T 715
Query 147 VCNSPLSEYAALGYEFGYSMEHPDTVAIWEAQFGDFSNGAQIIIDQFVVSAEVKWNRQNG 206
VCNS LSEY LG+E G++M P+ + +WEAQFGDF N AQ IIDQF+ + KW RQNG
Sbjct 716 VCNSSLSEYGVLGFELGFAMASPNALVLWEAQFGDFHNTAQCIIDQFICPGQAKWVRQNG 775
Query 207 LVMLLPHGYDGQGPEHSSARIERFLQL 233
+V+LLPHG +G GPEHSSAR ERFLQ+
Sbjct 776 IVLLLPHGMEGMGPEHSSARPERFLQM 802
> mmu:18293 Ogdh, 2210403E04Rik, 2210412K19Rik, AA409584, KIAA4192,
d1401, mKIAA4192; oxoglutarate dehydrogenase (lipoamide)
(EC:1.2.4.2); K00164 2-oxoglutarate dehydrogenase E1 component
[EC:1.2.4.2]
Length=1023
Score = 204 bits (519), Expect = 2e-52, Method: Compositional matrix adjust.
Identities = 100/202 (49%), Positives = 137/202 (67%), Gaps = 8/202 (3%)
Query 34 AAGVRLSALRELGRRIFTIP-EGFVPHATIAKIMKQRLAAVEGPQHEKSLDFGAAENLAY 92
+ G+ L +G+ ++P E F H +++I+K R V +++D+ AE +A+
Sbjct 606 STGLEEDVLFHIGKVASSVPVENFTIHGGLSRILKTRRELVTN----RTVDWALAEYMAF 661
Query 93 ATLLSDGFHVRLAGQDAQRGTFSHRHAVLHDQAVEAQHCI-FDGLKDLNLPHCITVCNSP 151
+LL +G HVRL+GQD +RGTFSHRH VLHDQ V+ + CI + L P+ TVCNS
Sbjct 662 GSLLKEGIHVRLSGQDVERGTFSHRHHVLHDQNVDKRTCIPMNHLWPNQAPY--TVCNSS 719
Query 152 LSEYAALGYEFGYSMEHPDTVAIWEAQFGDFSNGAQIIIDQFVVSAEVKWNRQNGLVMLL 211
LSEY LG+E G++M P+ + +WEAQFGDF+N AQ IIDQF+ + KW RQNG+V+LL
Sbjct 720 LSEYGVLGFELGFAMASPNALVLWEAQFGDFNNMAQCIIDQFICPGQAKWVRQNGIVLLL 779
Query 212 PHGYDGQGPEHSSARIERFLQL 233
PHG +G GPEHSSAR ERFLQ+
Sbjct 780 PHGMEGMGPEHSSARPERFLQM 801
> dre:564552 ogdh, MGC73296, im:7045267, wu:fa06d01, wu:fb98a04,
zgc:73296; oxoglutarate (alpha-ketoglutarate) dehydrogenase
(lipoamide) (EC:1.2.4.2); K00164 2-oxoglutarate dehydrogenase
E1 component [EC:1.2.4.2]
Length=1022
Score = 199 bits (507), Expect = 5e-51, Method: Compositional matrix adjust.
Identities = 97/202 (48%), Positives = 137/202 (67%), Gaps = 8/202 (3%)
Query 34 AAGVRLSALRELGRRIFTIP-EGFVPHATIAKIMKQRLAAVEGPQHEKSLDFGAAENLAY 92
+ G+ L ++G+ ++P E F H +++I+K R ++ +++D+ E +A+
Sbjct 606 STGLPEEELAQIGQVASSVPVEDFTIHGGLSRILKGRGDMIKN----RTVDWALGEYMAF 661
Query 93 ATLLSDGFHVRLAGQDAQRGTFSHRHAVLHDQAVEAQHCI-FDGLKDLNLPHCITVCNSP 151
+LL +G HVRL+GQD +RGTFSHRH VLHDQ V+ + CI + + P+ TVCNS
Sbjct 662 GSLLKEGIHVRLSGQDVERGTFSHRHHVLHDQNVDKRICIPMNHMSPNQAPY--TVCNSS 719
Query 152 LSEYAALGYEFGYSMEHPDTVAIWEAQFGDFSNGAQIIIDQFVVSAEVKWNRQNGLVMLL 211
LSEY LG+E G++M P+ + +WEAQFGDF N AQ IIDQF+ + KW RQNG+V+LL
Sbjct 720 LSEYGVLGFELGFAMASPNALVLWEAQFGDFHNTAQCIIDQFICPGQAKWVRQNGIVLLL 779
Query 212 PHGYDGQGPEHSSARIERFLQL 233
PHG +G GPEHSSAR ERFLQ+
Sbjct 780 PHGMEGMGPEHSSARPERFLQM 801
> xla:399021 ogdh, MGC68800, akgdh, e1k, ogdc; oxoglutarate (alpha-ketoglutarate)
dehydrogenase (lipoamide) (EC:1.2.4.2);
K00164 2-oxoglutarate dehydrogenase E1 component [EC:1.2.4.2]
Length=1021
Score = 199 bits (507), Expect = 6e-51, Method: Compositional matrix adjust.
Identities = 100/202 (49%), Positives = 136/202 (67%), Gaps = 8/202 (3%)
Query 34 AAGVRLSALRELGRRIFTIP-EGFVPHATIAKIMKQRLAAVEGPQHEKSLDFGAAENLAY 92
+ G+ L +G ++P E F+ H +++I+K R V+ +++D+ AE +A
Sbjct 605 STGLTEEDLTHIGNVASSVPVEDFMIHGGLSRILKGRGEMVKN----RTVDWALAEYMAL 660
Query 93 ATLLSDGFHVRLAGQDAQRGTFSHRHAVLHDQAVEAQHCI-FDGLKDLNLPHCITVCNSP 151
+LL +G H+RL+GQD +RGTFSHRH VLHDQ V+ + CI + L P+ TVCNS
Sbjct 661 GSLLKEGIHIRLSGQDVERGTFSHRHHVLHDQNVDKRTCIPMNHLWPNQAPY--TVCNSS 718
Query 152 LSEYAALGYEFGYSMEHPDTVAIWEAQFGDFSNGAQIIIDQFVVSAEVKWNRQNGLVMLL 211
LSEY LG+E G++M P+ + +WEAQFGDF N AQ IIDQFV + KW RQNG+V+LL
Sbjct 719 LSEYGVLGFELGFAMASPNALVLWEAQFGDFHNTAQCIIDQFVCPGQAKWVRQNGIVLLL 778
Query 212 PHGYDGQGPEHSSARIERFLQL 233
PHG +G GPEHSSAR ERFLQ+
Sbjct 779 PHGMEGMGPEHSSARPERFLQM 800
> xla:444121 MGC80496 protein; K00164 2-oxoglutarate dehydrogenase
E1 component [EC:1.2.4.2]
Length=1018
Score = 199 bits (506), Expect = 6e-51, Method: Compositional matrix adjust.
Identities = 99/202 (49%), Positives = 135/202 (66%), Gaps = 8/202 (3%)
Query 34 AAGVRLSALRELGRRIFTIP-EGFVPHATIAKIMKQRLAAVEGPQHEKSLDFGAAENLAY 92
+ G+ L +G ++P E F H +++I+K R V+ +++D+ AE ++
Sbjct 602 STGLSEEELTHIGNVASSVPVEDFTIHGGLSRILKGRGEMVKN----RTVDWALAEYMSL 657
Query 93 ATLLSDGFHVRLAGQDAQRGTFSHRHAVLHDQAVEAQHCI-FDGLKDLNLPHCITVCNSP 151
+LL +G H+RL+GQD +RGTFSHRH VLHDQ V+ + CI + L P+ TVCNS
Sbjct 658 GSLLKEGIHIRLSGQDVERGTFSHRHHVLHDQNVDKRTCIPMNHLWPNQAPY--TVCNSS 715
Query 152 LSEYAALGYEFGYSMEHPDTVAIWEAQFGDFSNGAQIIIDQFVVSAEVKWNRQNGLVMLL 211
LSEY LG+E G++M P+ + +WEAQFGDF N AQ IIDQFV + KW RQNG+V+LL
Sbjct 716 LSEYGVLGFELGFAMASPNALVLWEAQFGDFHNTAQCIIDQFVCPGQAKWVRQNGIVLLL 775
Query 212 PHGYDGQGPEHSSARIERFLQL 233
PHG +G GPEHSSAR ERFLQ+
Sbjct 776 PHGMEGMGPEHSSARPERFLQM 797
> hsa:55753 OGDHL; oxoglutarate dehydrogenase-like (EC:1.2.4.-);
K00164 2-oxoglutarate dehydrogenase E1 component [EC:1.2.4.2]
Length=953
Score = 198 bits (504), Expect = 1e-50, Method: Compositional matrix adjust.
Identities = 98/202 (48%), Positives = 135/202 (66%), Gaps = 8/202 (3%)
Query 34 AAGVRLSALRELGRRIFTIP-EGFVPHATIAKIMKQRLAAVEGPQHEKSLDFGAAENLAY 92
A G+ L +G ++P E F H +++I++ R + +++D+ AE +A+
Sbjct 536 ATGIPEDMLTHIGSVASSVPLEDFKIHTGLSRILRGRADMTKN----RTVDWALAEYMAF 591
Query 93 ATLLSDGFHVRLAGQDAQRGTFSHRHAVLHDQAVEAQHCI-FDGLKDLNLPHCITVCNSP 151
+LL +G HVRL+GQD +RGTFSHRH VLHDQ V+ + C+ + L P+ TVCNS
Sbjct 592 GSLLKEGIHVRLSGQDVERGTFSHRHHVLHDQEVDRRTCVPMNHLWPDQAPY--TVCNSS 649
Query 152 LSEYAALGYEFGYSMEHPDTVAIWEAQFGDFSNGAQIIIDQFVVSAEVKWNRQNGLVMLL 211
LSEY LG+E GY+M P+ + +WEAQFGDF N AQ IIDQF+ + + KW R NG+V+LL
Sbjct 650 LSEYGVLGFELGYAMASPNALVLWEAQFGDFHNTAQCIIDQFISTGQAKWVRHNGIVLLL 709
Query 212 PHGYDGQGPEHSSARIERFLQL 233
PHG +G GPEHSSAR ERFLQ+
Sbjct 710 PHGMEGMGPEHSSARPERFLQM 731
> mmu:239017 Ogdhl; oxoglutarate dehydrogenase-like (EC:1.2.4.-);
K00164 2-oxoglutarate dehydrogenase E1 component [EC:1.2.4.2]
Length=1029
Score = 197 bits (502), Expect = 2e-50, Method: Compositional matrix adjust.
Identities = 97/202 (48%), Positives = 135/202 (66%), Gaps = 8/202 (3%)
Query 34 AAGVRLSALRELGRRIFTIP-EGFVPHATIAKIMKQRLAAVEGPQHEKSLDFGAAENLAY 92
G+ L +G ++P E F H +++I++ R + ++++D+ AE +A+
Sbjct 612 TTGIPEEMLTHIGSVASSVPLEDFKIHTGLSRILRGRADMTK----KRTVDWALAEYMAF 667
Query 93 ATLLSDGFHVRLAGQDAQRGTFSHRHAVLHDQAVEAQHCI-FDGLKDLNLPHCITVCNSP 151
+LL +G HVRL+GQD +RGTFSHRH VLHDQ V+ + C+ + L P+ TVCNS
Sbjct 668 GSLLKEGIHVRLSGQDVERGTFSHRHHVLHDQEVDRRTCVPMNHLWPDQAPY--TVCNSS 725
Query 152 LSEYAALGYEFGYSMEHPDTVAIWEAQFGDFSNGAQIIIDQFVVSAEVKWNRQNGLVMLL 211
LSEY LG+E GY+M P+ + +WEAQFGDF N AQ IIDQF+ + + KW R NG+V+LL
Sbjct 726 LSEYGVLGFELGYAMASPNALVLWEAQFGDFHNTAQCIIDQFISTGQAKWVRHNGIVLLL 785
Query 212 PHGYDGQGPEHSSARIERFLQL 233
PHG +G GPEHSSAR ERFLQ+
Sbjct 786 PHGMEGMGPEHSSARPERFLQM 807
> xla:447370 ogdhl, MGC84242; oxoglutarate dehydrogenase-like;
K00164 2-oxoglutarate dehydrogenase E1 component [EC:1.2.4.2]
Length=1018
Score = 196 bits (498), Expect = 5e-50, Method: Compositional matrix adjust.
Identities = 97/201 (48%), Positives = 133/201 (66%), Gaps = 8/201 (3%)
Query 35 AGVRLSALRELGRRIFTIP-EGFVPHATIAKIMKQRLAAVEGPQHEKSLDFGAAENLAYA 93
G+ L +G ++P + F H +++I+K RL + +++D+ AE + +
Sbjct 602 TGIPEDMLSHIGAIASSVPLKDFKIHGGLSRILKSRLEMT----NSRTVDWALAEYMTFG 657
Query 94 TLLSDGFHVRLAGQDAQRGTFSHRHAVLHDQAVEAQHCI-FDGLKDLNLPHCITVCNSPL 152
+LL +G HVRL+GQD +RGTFSHRH VLHDQ V+ C+ + L P+ TVCNS L
Sbjct 658 SLLKEGIHVRLSGQDVERGTFSHRHHVLHDQEVDRWTCVPMNHLWPNQAPY--TVCNSSL 715
Query 153 SEYAALGYEFGYSMEHPDTVAIWEAQFGDFSNGAQIIIDQFVVSAEVKWNRQNGLVMLLP 212
SEY LG+E G++M P+ + +WEAQFGDF N AQ IIDQF+ S + KW R NG+V+LLP
Sbjct 716 SEYGVLGFELGFAMASPNALVLWEAQFGDFYNTAQCIIDQFISSGQAKWVRHNGIVLLLP 775
Query 213 HGYDGQGPEHSSARIERFLQL 233
HG +G GPEHSSAR ERFLQ+
Sbjct 776 HGMEGMGPEHSSARPERFLQM 796
> dre:559207 hypothetical LOC559207; K00164 2-oxoglutarate dehydrogenase
E1 component [EC:1.2.4.2]
Length=1008
Score = 194 bits (494), Expect = 2e-49, Method: Compositional matrix adjust.
Identities = 95/202 (47%), Positives = 135/202 (66%), Gaps = 8/202 (3%)
Query 34 AAGVRLSALRELGRRIFTIP-EGFVPHATIAKIMKQRLAAVEGPQHEKSLDFGAAENLAY 92
+ G+ L+ +G ++P + F H+ +++I++ R + ++ D+ AE +A+
Sbjct 592 STGLSEEVLKHIGEVASSVPLKDFAIHSGLSRILRGRADMIT----KRMADWALAEYMAF 647
Query 93 ATLLSDGFHVRLAGQDAQRGTFSHRHAVLHDQAVEAQHCIFDGLKDLNLPHCI-TVCNSP 151
+LL DG HVRL+GQD +RGTFSHRH VLHDQ V+ + C+ + L + TVCNS
Sbjct 648 GSLLKDGIHVRLSGQDVERGTFSHRHHVLHDQEVDKRFCV--PMNHLWQNQALYTVCNSS 705
Query 152 LSEYAALGYEFGYSMEHPDTVAIWEAQFGDFSNGAQIIIDQFVVSAEVKWNRQNGLVMLL 211
LSEY LG+E G++M +P+ + WEAQFGDF N AQ IIDQF+ + + KW R NG+V+LL
Sbjct 706 LSEYGVLGFELGFAMANPNALVCWEAQFGDFHNTAQCIIDQFISAGQAKWVRHNGIVLLL 765
Query 212 PHGYDGQGPEHSSARIERFLQL 233
PHG +G GPEHSSAR ERFLQ+
Sbjct 766 PHGMEGMGPEHSSARPERFLQM 787
> mmu:209692 Dhtkd1, C330018I04Rik; dehydrogenase E1 and transketolase
domain containing 1 (EC:1.2.4.2)
Length=921
Score = 187 bits (475), Expect = 3e-47, Method: Compositional matrix adjust.
Identities = 99/210 (47%), Positives = 131/210 (62%), Gaps = 3/210 (1%)
Query 24 ISATATAAVVAAGVRLSALRELGRRIFTIPEGFVPHATIAKIMKQRLAAVEGPQHEKSLD 83
+ A GV L LR +G + +PE H+ + K+ Q + +E ++ LD
Sbjct 514 VQPEACVTTWDTGVPLELLRFIGVKSVEVPEELQVHSHLLKMYVQ--SRMEKVKNGSGLD 571
Query 84 FGAAENLAYATLLSDGFHVRLAGQDAQRGTFSHRHAVLHDQAVEAQHCIFDGLKDLNLPH 143
+ AE LA +LL+ GF+VRL+GQD RGTFS RHA++ Q + + + + D N
Sbjct 572 WATAETLALGSLLAQGFNVRLSGQDVGRGTFSQRHAMVVCQDTDDAYIPLNHM-DPNQKG 630
Query 144 CITVCNSPLSEYAALGYEFGYSMEHPDTVAIWEAQFGDFSNGAQIIIDQFVVSAEVKWNR 203
+ V NSPLSE A LG+E+G S+E P + +WEAQFGDF NGAQII D F+ E KW
Sbjct 631 FLEVSNSPLSEEAVLGFEYGMSIESPTLLPLWEAQFGDFFNGAQIIFDTFISGGEAKWLL 690
Query 204 QNGLVMLLPHGYDGQGPEHSSARIERFLQL 233
Q+GLV+LLPHGYDG GPEHSS RIERFLQ+
Sbjct 691 QSGLVILLPHGYDGAGPEHSSCRIERFLQM 720
> bbo:BBOV_I002070 19.m02351; 2-oxoglutarate dehydrogenase E1
component (EC:1.2.4.2); K00164 2-oxoglutarate dehydrogenase
E1 component [EC:1.2.4.2]
Length=891
Score = 187 bits (474), Expect = 4e-47, Method: Compositional matrix adjust.
Identities = 98/203 (48%), Positives = 132/203 (65%), Gaps = 10/203 (4%)
Query 33 VAAGVRLSALRELGRRIFTIPEGFVPHATIAKIMKQRLAAVEGPQHEKSLDFGAAENLAY 92
GV L ELG+ + +P+ + H I +I +R A+E + +D G AE LAY
Sbjct 483 TVTGVEPHRLVELGKALNGVPQDYQLHPAIRRIYNERSKAIEAGNN---IDTGLAEALAY 539
Query 93 ATLLSDGFHVRLAGQDAQRGTFSHRHAVLHDQAVEAQHCIFDGLKDLNLPH--CITVCNS 150
A+L DG+ VRL GQD++RGTFSHRH+ + Q IF+ N+P+ I V NS
Sbjct 540 ASLAEDGYRVRLVGQDSKRGTFSHRHSSVQCQKTFRFFNIFE-----NVPNGSNIEVYNS 594
Query 151 PLSEYAALGYEFGYSMEHPDTVAIWEAQFGDFSNGAQIIIDQFVVSAEVKWNRQNGLVML 210
LSE AA+ +E+GY +E + IWEAQFGDF+N AQ IID+FVVS E KW +++ + +
Sbjct 595 LLSETAAMAFEYGYGLEDSRVLNIWEAQFGDFANVAQPIIDEFVVSGEAKWAQKSAMCLF 654
Query 211 LPHGYDGQGPEHSSARIERFLQL 233
LPHG+DGQGP+HSSARIER+LQL
Sbjct 655 LPHGFDGQGPDHSSARIERYLQL 677
> xla:403360 dhtkd1, MGC68840; dehydrogenase E1 and transketolase
domain containing 1 (EC:1.2.4.2)
Length=927
Score = 181 bits (459), Expect = 2e-45, Method: Compositional matrix adjust.
Identities = 95/210 (45%), Positives = 128/210 (60%), Gaps = 3/210 (1%)
Query 24 ISATATAAVVAAGVRLSALRELGRRIFTIPEGFVPHATIAKIMKQRLAAVEGPQHEKSLD 83
I +A G+ L+ +G + +PE F H+ + K+ Q + V+ Q LD
Sbjct 521 IEPSARTTTWDTGLPADLLKFIGAKSVEVPEEFKMHSHLLKMHAQ--SRVQKLQEATKLD 578
Query 84 FGAAENLAYATLLSDGFHVRLAGQDAQRGTFSHRHAVLHDQAVEAQHCIFDGLKDLNLPH 143
+ AE LA+ +LL GF++R++GQD RGTFS RHA+L Q + + + +
Sbjct 579 WATAEALAFGSLLCQGFNIRISGQDVGRGTFSQRHAMLVCQETNDTYIPLNHMTP-DQKG 637
Query 144 CITVCNSPLSEYAALGYEFGYSMEHPDTVAIWEAQFGDFSNGAQIIIDQFVVSAEVKWNR 203
+ V NS LSE A LG+E+G S+E P + IWEAQFGDF NGAQII D F+ E KW
Sbjct 638 FLEVSNSALSEEAVLGFEYGMSIESPKLLPIWEAQFGDFFNGAQIIFDTFISGGEAKWLL 697
Query 204 QNGLVMLLPHGYDGQGPEHSSARIERFLQL 233
Q+G+V+LLPHGYDG GPEHSS RIERFLQ+
Sbjct 698 QSGIVILLPHGYDGAGPEHSSCRIERFLQM 727
> hsa:55526 DHTKD1, DKFZp762M115, KIAA1630, MGC3090; dehydrogenase
E1 and transketolase domain containing 1 (EC:1.2.4.2)
Length=919
Score = 181 bits (458), Expect = 3e-45, Method: Compositional matrix adjust.
Identities = 95/200 (47%), Positives = 126/200 (63%), Gaps = 3/200 (1%)
Query 34 AAGVRLSALRELGRRIFTIPEGFVPHATIAKIMKQRLAAVEGPQHEKSLDFGAAENLAYA 93
+ GV L LR +G + +P H+ + K Q + +E LD+ AE LA
Sbjct 523 STGVPLDLLRFVGMKSVEVPRELQMHSHLLKTHVQ--SRMEKMMDGIKLDWATAEALALG 580
Query 94 TLLSDGFHVRLAGQDAQRGTFSHRHAVLHDQAVEAQHCIFDGLKDLNLPHCITVCNSPLS 153
+LL+ GF+VRL+GQD RGTFS RHA++ Q + + + + D N + V NSPLS
Sbjct 581 SLLAQGFNVRLSGQDVGRGTFSQRHAIVVCQETDDTYIPLNHM-DPNQKGFLEVSNSPLS 639
Query 154 EYAALGYEFGYSMEHPDTVAIWEAQFGDFSNGAQIIIDQFVVSAEVKWNRQNGLVMLLPH 213
E A LG+E+G S+E P + +WEAQFGDF NGAQII D F+ E KW Q+G+V+LLPH
Sbjct 640 EEAVLGFEYGMSIESPKLLPLWEAQFGDFFNGAQIIFDTFISGGEAKWLLQSGIVILLPH 699
Query 214 GYDGQGPEHSSARIERFLQL 233
GYDG GP+HSS RIERFLQ+
Sbjct 700 GYDGAGPDHSSCRIERFLQM 719
> dre:494076 dhtkd1, zgc:101818; dehydrogenase E1 and transketolase
domain containing 1 (EC:1.2.4.2)
Length=925
Score = 179 bits (455), Expect = 5e-45, Method: Compositional matrix adjust.
Identities = 96/199 (48%), Positives = 123/199 (61%), Gaps = 3/199 (1%)
Query 35 AGVRLSALRELGRRIFTIPEGFVPHATIAKIMKQRLAAVEGPQHEKSLDFGAAENLAYAT 94
GV L+ +G + IPE + H+ + K Q A ++ + LD+ AE LA+ T
Sbjct 530 TGVAQPLLQFVGAKSVDIPEEIILHSHLRKTHVQ--ARLQKLEEGTKLDWSTAEALAFGT 587
Query 95 LLSDGFHVRLAGQDAQRGTFSHRHAVLHDQAVEAQHCIFDGLKDLNLPHCITVCNSPLSE 154
LL GF++R++GQD RGTFS RHA++ Q + + H + VCNS LSE
Sbjct 588 LLCQGFNIRISGQDVGRGTFSQRHAMVVCQETNDMFIPLNHISSEQKGH-LEVCNSALSE 646
Query 155 YAALGYEFGYSMEHPDTVAIWEAQFGDFSNGAQIIIDQFVVSAEVKWNRQNGLVMLLPHG 214
A LG+E+G S+ P + IWEAQFGDF NGAQII D F+ E KW Q+GLV+LLPHG
Sbjct 647 EAVLGFEYGMSIAQPRLLPIWEAQFGDFFNGAQIIFDTFLSGGEAKWLLQSGLVILLPHG 706
Query 215 YDGQGPEHSSARIERFLQL 233
YDG GPEHSS RIERFLQL
Sbjct 707 YDGAGPEHSSCRIERFLQL 725
> cel:ZK836.2 hypothetical protein
Length=911
Score = 177 bits (449), Expect = 3e-44, Method: Compositional matrix adjust.
Identities = 94/200 (47%), Positives = 129/200 (64%), Gaps = 5/200 (2%)
Query 35 AGVRLSALRELGRRIFTIPEGFVPHATIAKI-MKQRLAAVEGPQHEKSLDFGAAENLAYA 93
GV LR +G +PE F H + K+ + R+ + Q + +D+ AE +A+
Sbjct 514 TGVATDLLRFIGAGSVKVPEDFDTHKHLYKMHIDSRMQKM---QTGEGIDWATAEAMAFG 570
Query 94 TLLSDGFHVRLAGQDAQRGTFSHRHAVLHDQAVEAQHCIFDGLKDLNLPHCITVCNSPLS 153
++L +G VR++GQD RGTF HRHA++ DQ+ + H + L + + + V N+ LS
Sbjct 571 SILLEGNDVRISGQDVGRGTFCHRHAMMVDQSTDHIHIPLNELVE-EQKNQLEVANNLLS 629
Query 154 EYAALGYEFGYSMEHPDTVAIWEAQFGDFSNGAQIIIDQFVVSAEVKWNRQNGLVMLLPH 213
E A LG+E+G+S E+P + IWEAQFGDF NGAQIIID F+ SAE KW +GL MLLPH
Sbjct 630 EEAILGFEWGFSSENPRRLCIWEAQFGDFFNGAQIIIDTFLASAESKWLTSSGLTMLLPH 689
Query 214 GYDGQGPEHSSARIERFLQL 233
G+DG GPEHSS R+ERFLQL
Sbjct 690 GFDGAGPEHSSCRMERFLQL 709
> dre:100334699 probable 2-oxoglutarate dehydrogenase E1 component
DHKTD1, mitochondrial-like
Length=657
Score = 61.6 bits (148), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 34/87 (39%), Positives = 51/87 (58%), Gaps = 2/87 (2%)
Query 35 AGVRLSALRELGRRIFTIPEGFVPHATIAKIMKQRLAAVEGPQHEKSLDFGAAENLAYAT 94
GV L+ +G + IPE + H+ + K Q A ++ + LD+ AE LA+ T
Sbjct 530 TGVAQPLLQFVGAKSVDIPEEIILHSHLRKTHVQ--ARLQKLEEGTKLDWSTAEALAFGT 587
Query 95 LLSDGFHVRLAGQDAQRGTFSHRHAVL 121
LL GF++R++GQD RGTFS RHA++
Sbjct 588 LLCQGFNIRISGQDVGRGTFSQRHAMV 614
> dre:100331749 oxoglutarate (alpha-ketoglutarate) dehydrogenase
(lipoamide)-like
Length=687
Score = 54.3 bits (129), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 27/82 (32%), Positives = 50/82 (60%), Gaps = 5/82 (6%)
Query 34 AAGVRLSALRELGRRIFTIP-EGFVPHATIAKIMKQRLAAVEGPQHEKSLDFGAAENLAY 92
+ G+ L ++G+ ++P E F H +++I+K R ++ +++D+ E +A+
Sbjct 606 STGLPEEELAQIGQVASSVPVEDFTIHGGLSRILKGRGDMIKN----RTVDWALGEYMAF 661
Query 93 ATLLSDGFHVRLAGQDAQRGTF 114
+LL +G HVRL+GQD +RGTF
Sbjct 662 GSLLKEGIHVRLSGQDVERGTF 683
> eco:b0807 rlmF, ECK0796, JW5107, ybiN; 23S rRNA mA1618 methyltransferase,
SAM-dependent; K06970 ribosomal RNA large subunit
methyltransferase F [EC:2.1.1.181]
Length=308
Score = 30.0 bits (66), Expect = 8.1, Method: Compositional matrix adjust.
Identities = 31/125 (24%), Positives = 49/125 (39%), Gaps = 17/125 (13%)
Query 50 FTIPEGF----VP-HATIAKIMKQRLAAVEG--PQHEKSLDFGAAENLAYATLLSDGFHV 102
+ IP+GF VP A + LA G P + LD G N Y + +
Sbjct 69 WDIPDGFLCPPVPGRADYIHHLADLLAEASGTIPANASILDIGVGANCIYPLIGVHEYGW 128
Query 103 RLAGQDAQRGTFSHRHAVLH-----DQAVEAQH-----CIFDGLKDLNLPHCITVCNSPL 152
R G + S A++ ++A+ + IF+G+ N + T+CN P
Sbjct 129 RFTGSETSSQALSSAQAIISSNPGLNRAIRLRRQKESGAIFNGIIHKNEQYDATLCNPPF 188
Query 153 SEYAA 157
+ AA
Sbjct 189 HDSAA 193
> mmu:12317 Calr, CRT, Calregulin; calreticulin; K08057 calreticulin
Length=416
Score = 30.0 bits (66), Expect = 8.3, Method: Compositional matrix adjust.
Identities = 16/40 (40%), Positives = 20/40 (50%), Gaps = 0/40 (0%)
Query 177 AQFGDFSNGAQIIIDQFVVSAEVKWNRQNGLVMLLPHGYD 216
A+F FSN Q ++ QF V E + G V L P G D
Sbjct 79 AKFEPFSNKGQTLVVQFTVKHEQNIDCGGGYVKLFPSGLD 118
> bbo:BBOV_IV001720 21.m02935; hypothetical protein
Length=1567
Score = 29.6 bits (65), Expect = 8.5, Method: Composition-based stats.
Identities = 21/75 (28%), Positives = 33/75 (44%), Gaps = 4/75 (5%)
Query 126 VEAQHCIFDGLKDLNLPHCITVCNSPLSEYAALGYEFGYSMEHPDTVAIWEAQFGDFSNG 185
+E H IF + N+ C+ V + + Y + H +TV + QF F
Sbjct 1239 IEQMHRIF---HNFNVHQCLRVLAEVIPTLVKFSWP-SYLISHLETVVQFLEQFRTFPLD 1294
Query 186 AQIIIDQFVVSAEVK 200
A I I +FV SA+ +
Sbjct 1295 ADIYIGRFVASAKYR 1309
Lambda K H
0.321 0.135 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 7953524176
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40