bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0734_orf1
Length=140
Score E
Sequences producing significant alignments: (Bits) Value
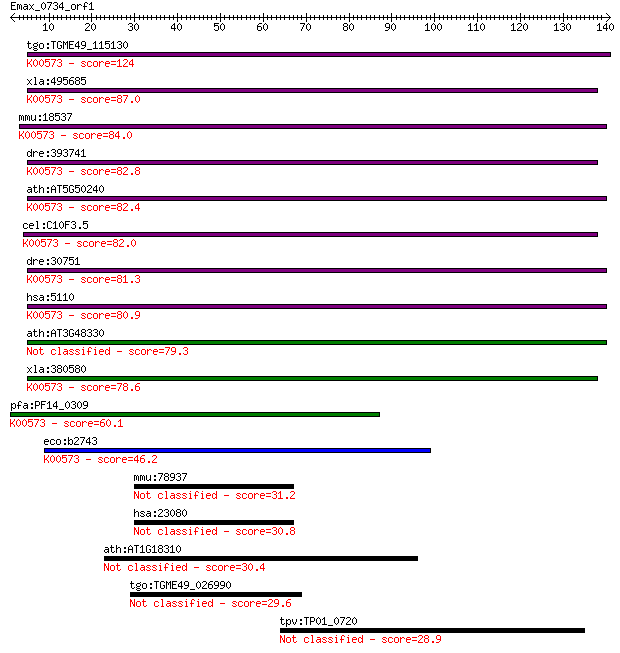
tgo:TGME49_115130 protein-L-isoaspartate O-methyltransferase, ... 124 8e-29
xla:495685 pcmt1; protein-L-isoaspartate (D-aspartate) O-methy... 87.0 1e-17
mmu:18537 Pcmt1, C79501, PIMT; protein-L-isoaspartate (D-aspar... 84.0 1e-16
dre:393741 pcmtl, MGC73268, zgc:73268; l-isoaspartyl protein c... 82.8 3e-16
ath:AT5G50240 PIMT2; PIMT2 (PROTEIN-L-ISOASPARTATE METHYLTRANS... 82.4 4e-16
cel:C10F3.5 pcm-1; Protein Carboxymethyltransferase family mem... 82.0 5e-16
dre:30751 pcmt, fj13d06, pimt, wu:fj13d06; l-isoaspartyl prote... 81.3 9e-16
hsa:5110 PCMT1; protein-L-isoaspartate (D-aspartate) O-methylt... 80.9 1e-15
ath:AT3G48330 PIMT1; PIMT1 (PROTEIN-L-ISOASPARTATE METHYLTRANS... 79.3 3e-15
xla:380580 pcmt1, MGC69121; protein-L-isoaspartate(D-aspartate... 78.6 6e-15
pfa:PF14_0309 protein-L-isoaspartate O-methyltransferase beta-... 60.1 2e-09
eco:b2743 pcm, ECK2738, JW2713; L-isoaspartate protein carboxy... 46.2 3e-05
mmu:78937 Avl9, 5830411G16Rik, D730049P16Rik, mKIAA0241; AVL9 ... 31.2 1.1
hsa:23080 AVL9, DKFZp686G0344, KIAA0241; AVL9 homolog (S. cere... 30.8 1.3
ath:AT1G18310 glycosyl hydrolase family 81 protein 30.4 1.8
tgo:TGME49_026990 hypothetical protein 29.6 2.8
tpv:TP01_0720 P-type ATPase 28.9 4.9
> tgo:TGME49_115130 protein-L-isoaspartate O-methyltransferase,
putative (EC:2.1.1.77); K00573 protein-L-isoaspartate(D-aspartate)
O-methyltransferase [EC:2.1.1.77]
Length=539
Score = 124 bits (311), Expect = 8e-29, Method: Composition-based stats.
Identities = 70/144 (48%), Positives = 91/144 (63%), Gaps = 9/144 (6%)
Query 5 GIAIGIDYIQGLVDLSKKNVTAGFPHLMQSRNFALLKGDGWAGGPSWGAPYDAIHVGAAA 64
G+A+GIDY+ LV S K V A +P L ++ F LL GDGW G P G PYDAIHVGAAA
Sbjct 391 GVAVGIDYLPDLVKYSVKKVKAAYPALSKNPRFKLLVGDGWRGHPELG-PYDAIHVGAAA 449
Query 65 AAVPAALLQQLAIGGKMVIPVE-CHEGLITLEG---LGDRQIDGD----CDGGQALVVIS 116
+++P LL QLA GGKMV+PVE +G + EG +RQ+ D Q V +S
Sbjct 450 SSIPRELLAQLAHGGKMVLPVETTSDGQVVFEGPEEETERQLSRGRGWWSDRNQVFVEVS 509
Query 117 KNKEGAVSVKYITSVVYVPLVRQT 140
K+ +G V VK + V+YVPLV+Q+
Sbjct 510 KDAQGKVRVKKLMGVMYVPLVKQS 533
> xla:495685 pcmt1; protein-L-isoaspartate (D-aspartate) O-methyltransferase;
K00573 protein-L-isoaspartate(D-aspartate) O-methyltransferase
[EC:2.1.1.77]
Length=228
Score = 87.0 bits (214), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 56/133 (42%), Positives = 71/133 (53%), Gaps = 19/133 (14%)
Query 5 GIAIGIDYIQGLVDLSKKNVTAGFPHLMQSRNFALLKGDGWAGGPSWGAPYDAIHVGAAA 64
G +GID+I+ LVD S NV P L+ S LL GDG G P APYDAIHVGAAA
Sbjct 104 GKVVGIDHIKELVDDSVNNVKKDDPTLLSSGRVKLLVGDGRMGYPEE-APYDAIHVGAAA 162
Query 65 AAVPAALLQQLAIGGKMVIPVECHEGLITLEGLGDRQIDGDCDGGQALVVISKNKEGAVS 124
VP AL+ QL GG++++PV G G Q L K ++G+V
Sbjct 163 PVVPQALIDQLKPGGRLILPV------------------GPAGGNQMLEQYDKLEDGSVK 204
Query 125 VKYITSVVYVPLV 137
+K + V+YVPL
Sbjct 205 MKPLMGVIYVPLT 217
> mmu:18537 Pcmt1, C79501, PIMT; protein-L-isoaspartate (D-aspartate)
O-methyltransferase 1 (EC:2.1.1.77); K00573 protein-L-isoaspartate(D-aspartate)
O-methyltransferase [EC:2.1.1.77]
Length=285
Score = 84.0 bits (206), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 56/137 (40%), Positives = 72/137 (52%), Gaps = 19/137 (13%)
Query 3 NGGIAIGIDYIQGLVDLSKKNVTAGFPHLMQSRNFALLKGDGWAGGPSWGAPYDAIHVGA 62
N G IGID+I+ LVD S NV P L+ S L+ GDG G APYDAIHVGA
Sbjct 160 NSGKVIGIDHIKELVDDSITNVKKDDPMLLSSGRVRLVVGDGRMGYAE-EAPYDAIHVGA 218
Query 63 AAAAVPAALLQQLAIGGKMVIPVECHEGLITLEGLGDRQIDGDCDGGQALVVISKNKEGA 122
AA VP AL+ QL GG++++PV G G Q L K ++G+
Sbjct 219 AAPVVPQALIDQLKPGGRLILPV------------------GPAGGNQMLEQYDKLQDGS 260
Query 123 VSVKYITSVVYVPLVRQ 139
V +K + V+YVPL +
Sbjct 261 VKMKPLMGVIYVPLTDK 277
> dre:393741 pcmtl, MGC73268, zgc:73268; l-isoaspartyl protein
carboxyl methyltransferase, like (EC:2.1.1.77); K00573 protein-L-isoaspartate(D-aspartate)
O-methyltransferase [EC:2.1.1.77]
Length=228
Score = 82.8 bits (203), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 55/133 (41%), Positives = 68/133 (51%), Gaps = 19/133 (14%)
Query 5 GIAIGIDYIQGLVDLSKKNVTAGFPHLMQSRNFALLKGDGWAGGPSWGAPYDAIHVGAAA 64
G +GID+I LV S KNV A P L+ + L+ GDG G P APYDAIHVGAAA
Sbjct 104 GKVVGIDHIDQLVQSSVKNVQADDPELLATGRIKLVVGDGRFGFPDE-APYDAIHVGAAA 162
Query 65 AAVPAALLQQLAIGGKMVIPVECHEGLITLEGLGDRQIDGDCDGGQALVVISKNKEGAVS 124
+P ALL+QL GG++V+PV G G Q L + +G
Sbjct 163 PTLPKALLEQLKPGGRLVLPV------------------GPEGGSQVLEQYDRQSDGTFL 204
Query 125 VKYITSVVYVPLV 137
K + VVYVPL
Sbjct 205 RKPLMGVVYVPLT 217
> ath:AT5G50240 PIMT2; PIMT2 (PROTEIN-L-ISOASPARTATE METHYLTRANSFERASE
2); protein-L-isoaspartate (D-aspartate) O-methyltransferase;
K00573 protein-L-isoaspartate(D-aspartate) O-methyltransferase
[EC:2.1.1.77]
Length=306
Score = 82.4 bits (202), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 53/136 (38%), Positives = 73/136 (53%), Gaps = 22/136 (16%)
Query 5 GIAIGIDYIQGLVDLSKKNVTAGFP-HLMQSRNFALLKGDGWAGGPSWGAPYDAIHVGAA 63
G +G+D+I LVD+S KN+ ++ + +L GDG G + APYDAIHVGAA
Sbjct 185 GRVVGVDHIPELVDMSIKNIEKSVAASFLKKGSLSLHVGDGRKGWQEF-APYDAIHVGAA 243
Query 64 AAAVPAALLQQLAIGGKMVIPVECHEGLITLEGLGDRQIDGDCDGGQALVVISKNKEGAV 123
A+ +P LL QL GG+MVIP+ + Q L VI KN++G++
Sbjct 244 ASEIPQPLLDQLKPGGRMVIPLGTY--------------------FQELKVIDKNEDGSI 283
Query 124 SVKYITSVVYVPLVRQ 139
V TSV YVPL +
Sbjct 284 KVHTETSVRYVPLTSR 299
> cel:C10F3.5 pcm-1; Protein Carboxymethyltransferase family member
(pcm-1); K00573 protein-L-isoaspartate(D-aspartate) O-methyltransferase
[EC:2.1.1.77]
Length=219
Score = 82.0 bits (201), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 50/134 (37%), Positives = 70/134 (52%), Gaps = 20/134 (14%)
Query 4 GGIAIGIDYIQGLVDLSKKNVTAGFPHLMQSRNFALLKGDGWAGGPSWGAPYDAIHVGAA 63
G +GI+++ LV+LS+KN+ ++ N +++GDG G APY+AIHVGAA
Sbjct 97 NGTVVGIEHMPQLVELSEKNIRKHHSEQLERGNVIIIEGDGRQGFAE-KAPYNAIHVGAA 155
Query 64 AAAVPAALLQQLAIGGKMVIPVECHEGLITLEGLGDRQIDGDCDGGQALVVISKNKEGAV 123
+ VP AL QLA GG+M+IPVE DG Q + I K G +
Sbjct 156 SKGVPKALTDQLAEGGRMMIPVE------------------QVDGNQVFMQIDK-INGKI 196
Query 124 SVKYITSVVYVPLV 137
K + V+YVPL
Sbjct 197 EQKIVEHVIYVPLT 210
> dre:30751 pcmt, fj13d06, pimt, wu:fj13d06; l-isoaspartyl protein
carboxyl methyltransferase (EC:2.1.1.77); K00573 protein-L-isoaspartate(D-aspartate)
O-methyltransferase [EC:2.1.1.77]
Length=228
Score = 81.3 bits (199), Expect = 9e-16, Method: Compositional matrix adjust.
Identities = 54/135 (40%), Positives = 71/135 (52%), Gaps = 19/135 (14%)
Query 5 GIAIGIDYIQGLVDLSKKNVTAGFPHLMQSRNFALLKGDGWAGGPSWGAPYDAIHVGAAA 64
G IGID+I+ LV+ S NV P L+ S L+ GDG G + APYDAIHVGAAA
Sbjct 104 GKVIGIDHIKELVEDSIANVKKDDPSLITSGRIKLIVGDGRMGF-TEEAPYDAIHVGAAA 162
Query 65 AAVPAALLQQLAIGGKMVIPVECHEGLITLEGLGDRQIDGDCDGGQALVVISKNKEGAVS 124
VP ALL QL GG++++PV G G Q L K ++G+
Sbjct 163 PTVPQALLDQLKPGGRLILPV------------------GPAGGNQMLEQYDKLEDGSTK 204
Query 125 VKYITSVVYVPLVRQ 139
+K + V+YVPL +
Sbjct 205 MKPLMGVIYVPLTDK 219
> hsa:5110 PCMT1; protein-L-isoaspartate (D-aspartate) O-methyltransferase
(EC:2.1.1.77); K00573 protein-L-isoaspartate(D-aspartate)
O-methyltransferase [EC:2.1.1.77]
Length=285
Score = 80.9 bits (198), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 54/135 (40%), Positives = 71/135 (52%), Gaps = 19/135 (14%)
Query 5 GIAIGIDYIQGLVDLSKKNVTAGFPHLMQSRNFALLKGDGWAGGPSWGAPYDAIHVGAAA 64
G IGID+I+ LVD S NV P L+ S L+ GDG G APYDAIHVGAAA
Sbjct 162 GKVIGIDHIKELVDDSVNNVRKDDPTLLSSGRVQLVVGDGRMGYAE-EAPYDAIHVGAAA 220
Query 65 AAVPAALLQQLAIGGKMVIPVECHEGLITLEGLGDRQIDGDCDGGQALVVISKNKEGAVS 124
VP AL+ QL GG++++PV G G Q L K ++G++
Sbjct 221 PVVPQALIDQLKPGGRLILPV------------------GPAGGNQMLEQYDKLQDGSIK 262
Query 125 VKYITSVVYVPLVRQ 139
+K + V+YVPL +
Sbjct 263 MKPLMGVIYVPLTDK 277
> ath:AT3G48330 PIMT1; PIMT1 (PROTEIN-L-ISOASPARTATE METHYLTRANSFERASE
1); protein-L-isoaspartate (D-aspartate) O-methyltransferase
Length=230
Score = 79.3 bits (194), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 59/137 (43%), Positives = 77/137 (56%), Gaps = 24/137 (17%)
Query 5 GIAIGIDYIQGLVDLSKKNV--TAGFPHLMQSRNFALLKGDGWAGGPSWGAPYDAIHVGA 62
G AIG+++I LV S KN+ +A P L + + A+ GDG G + APYDAIHVGA
Sbjct 109 GRAIGVEHIPELVASSVKNIEASAASPFLKEG-SLAVHVGDGRQGWAEF-APYDAIHVGA 166
Query 63 AAAAVPAALLQQLAIGGKMVIPVECHEGLITLEGLGDRQIDGDCDGGQALVVISKNKEGA 122
AA +P AL+ QL GG++VIPV G I Q L V+ KN +G+
Sbjct 167 AAPEIPEALIDQLKPGGRLVIPV----GNIF----------------QDLQVVDKNSDGS 206
Query 123 VSVKYITSVVYVPLVRQ 139
VS+K TSV YVPL +
Sbjct 207 VSIKDETSVRYVPLTSR 223
> xla:380580 pcmt1, MGC69121; protein-L-isoaspartate(D-aspartate)O-methyltransferase
(EC:2.1.1.77); K00573 protein-L-isoaspartate(D-aspartate)
O-methyltransferase [EC:2.1.1.77]
Length=228
Score = 78.6 bits (192), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 51/133 (38%), Positives = 68/133 (51%), Gaps = 19/133 (14%)
Query 5 GIAIGIDYIQGLVDLSKKNVTAGFPHLMQSRNFALLKGDGWAGGPSWGAPYDAIHVGAAA 64
G +GI++I LV + +NV P L+ S + GDG G P G PYDAIHVGAAA
Sbjct 104 GKVVGIEHINHLVHDAIQNVKQDDPTLLSSGRIKFVVGDGRLGYPDEG-PYDAIHVGAAA 162
Query 65 AAVPAALLQQLAIGGKMVIPVECHEGLITLEGLGDRQIDGDCDGGQALVVISKNKEGAVS 124
A VP LL+QL GG++++PV G G Q L K+ EG ++
Sbjct 163 AIVPQELLKQLKPGGRLILPV------------------GPEGGSQVLEQYDKDNEGKIT 204
Query 125 VKYITSVVYVPLV 137
+ V+YVPL
Sbjct 205 RARLMGVMYVPLT 217
> pfa:PF14_0309 protein-L-isoaspartate O-methyltransferase beta-aspartate
methyltransferase, putative; K00573 protein-L-isoaspartate(D-aspartate)
O-methyltransferase [EC:2.1.1.77]
Length=240
Score = 60.1 bits (144), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 28/89 (31%), Positives = 49/89 (55%), Gaps = 3/89 (3%)
Query 1 EENGGIAIGIDYIQGLVDLSKKNVTAGFPHLMQSRNFALLKGDGWAGGPSWGAP---YDA 57
E IG++ ++ LV+ S +N+ P L++ NF ++ + + +DA
Sbjct 119 ENKNSYVIGLERVKDLVNFSLENIKRDKPELLKIDNFKIIHKNIYQVNEEEKKELGLFDA 178
Query 58 IHVGAAAAAVPAALLQQLAIGGKMVIPVE 86
IHVGA+A+ +P L+ LA GK++IP+E
Sbjct 179 IHVGASASELPEILVDLLAENGKLIIPIE 207
> eco:b2743 pcm, ECK2738, JW2713; L-isoaspartate protein carboxylmethyltransferase
type II (EC:2.1.1.77); K00573 protein-L-isoaspartate(D-aspartate)
O-methyltransferase [EC:2.1.1.77]
Length=208
Score = 46.2 bits (108), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 32/91 (35%), Positives = 50/91 (54%), Gaps = 7/91 (7%)
Query 9 GIDYIQGLVDLSKKNVTAGFPHLMQSRNFALLKGDGWAGGPSWGAPYDAIHVGAAAAAVP 68
++ I+GL +++ + H + +R+ GDGW G + AP+DAI V AA +P
Sbjct 102 SVERIKGLQWQARRRLKNLDLHNVSTRH-----GDGWQGWQA-RAPFDAIIVTAAPPEIP 155
Query 69 AALLQQLAIGGKMVIPV-ECHEGLITLEGLG 98
AL+ QL GG +V+PV E H+ L + G
Sbjct 156 TALMTQLDEGGILVLPVGEEHQYLKRVRRRG 186
> mmu:78937 Avl9, 5830411G16Rik, D730049P16Rik, mKIAA0241; AVL9
homolog (S. cerevisiase)
Length=649
Score = 31.2 bits (69), Expect = 1.1, Method: Composition-based stats.
Identities = 13/37 (35%), Positives = 20/37 (54%), Gaps = 0/37 (0%)
Query 30 HLMQSRNFALLKGDGWAGGPSWGAPYDAIHVGAAAAA 66
H+ ++R+ L G GW GG W A+++ A AA
Sbjct 484 HVTENRDDVFLDGTGWEGGDEWIRAQFAVYIHALLAA 520
> hsa:23080 AVL9, DKFZp686G0344, KIAA0241; AVL9 homolog (S. cerevisiase)
Length=648
Score = 30.8 bits (68), Expect = 1.3, Method: Composition-based stats.
Identities = 13/37 (35%), Positives = 20/37 (54%), Gaps = 0/37 (0%)
Query 30 HLMQSRNFALLKGDGWAGGPSWGAPYDAIHVGAAAAA 66
H+ ++R+ L G GW GG W A+++ A AA
Sbjct 483 HVTENRDDVFLDGTGWEGGDEWIRAQFAVYIHALLAA 519
> ath:AT1G18310 glycosyl hydrolase family 81 protein
Length=649
Score = 30.4 bits (67), Expect = 1.8, Method: Composition-based stats.
Identities = 28/75 (37%), Positives = 34/75 (45%), Gaps = 8/75 (10%)
Query 23 NVTAGFPHLMQSRNFALLKGDGWAGGPS--WGAPYDAIHVGAAAAAVPAALLQQLAIGGK 80
N + +P L RNF L K WAGG + W A A AALL LA G K
Sbjct 459 NSNSSYPRL---RNFDLFKLHSWAGGLTEFWDGRNQESTSEAVNAYYSAALL-GLAYGDK 514
Query 81 MVIPVECHEGLITLE 95
+ VE ++TLE
Sbjct 515 HL--VETASTIMTLE 527
> tgo:TGME49_026990 hypothetical protein
Length=591
Score = 29.6 bits (65), Expect = 2.8, Method: Composition-based stats.
Identities = 17/40 (42%), Positives = 22/40 (55%), Gaps = 1/40 (2%)
Query 29 PHLMQSRNFALLKGDGWAGGPSWGAPYDAIHVGAAAAAVP 68
P L +SR F L KG GG + G+P ++ V AA A P
Sbjct 488 PALKESRTFGLSKGSPLEGGKAPGSP-RSLDVPAAGRATP 526
> tpv:TP01_0720 P-type ATPase
Length=1361
Score = 28.9 bits (63), Expect = 4.9, Method: Composition-based stats.
Identities = 19/71 (26%), Positives = 33/71 (46%), Gaps = 0/71 (0%)
Query 64 AAAVPAALLQQLAIGGKMVIPVECHEGLITLEGLGDRQIDGDCDGGQALVVISKNKEGAV 123
A +P + + LA+ ++I + G+ITL+ L DR D +V + E +
Sbjct 1095 AGNLPHTIFEALAVIASILIGLYLCTGVITLKDLNDRCRYVKVDDSTKMVYFCSSHEYLI 1154
Query 124 SVKYITSVVYV 134
+ KY+ V V
Sbjct 1155 TPKYVGWVTNV 1165
Lambda K H
0.318 0.139 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2552834388
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40