bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0724_orf2
Length=165
Score E
Sequences producing significant alignments: (Bits) Value
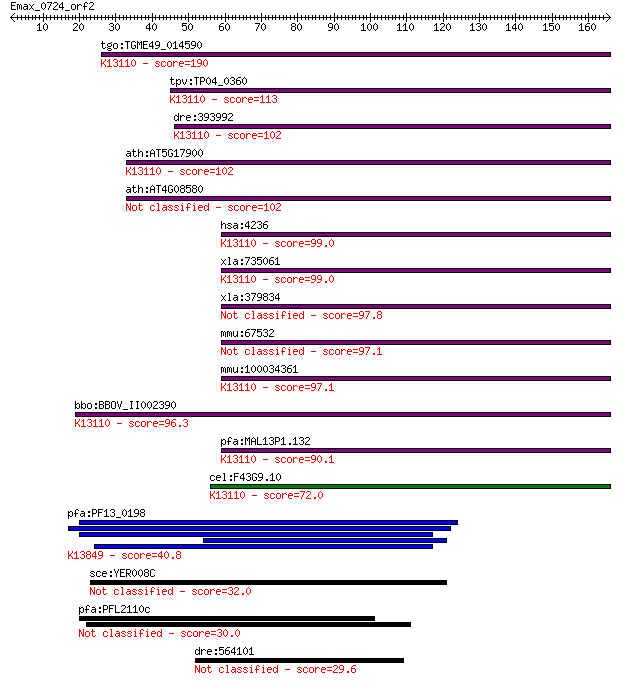
tgo:TGME49_014590 microfibrillar-associated protein 1, putativ... 190 2e-48
tpv:TP04_0360 hypothetical protein; K13110 microfibrillar-asso... 113 2e-25
dre:393992 mfap1, MGC56551, zgc:56551; microfibrillar-associat... 102 4e-22
ath:AT5G17900 hypothetical protein; K13110 microfibrillar-asso... 102 6e-22
ath:AT4G08580 hypothetical protein 102 7e-22
hsa:4236 MFAP1; microfibrillar-associated protein 1; K13110 mi... 99.0 6e-21
xla:735061 hypothetical protein MGC85054; K13110 microfibrilla... 99.0 7e-21
xla:379834 mfap1, MGC53463; microfibrillar-associated protein 1 97.8
mmu:67532 Mfap1a, 4432409M24Rik, Mfap1; microfibrillar-associa... 97.1 2e-20
mmu:100034361 Mfap1b; microfibrillar-associated protein 1B; K1... 97.1 2e-20
bbo:BBOV_II002390 18.m06195; micro-fibrillar-associated protei... 96.3 4e-20
pfa:MAL13P1.132 microfibril-associated protein homologue, puta... 90.1 3e-18
cel:F43G9.10 hypothetical protein; K13110 microfibrillar-assoc... 72.0 9e-13
pfa:PF13_0198 PfRh2a; reticulocyte binding protein 2 homolog A... 40.8 0.002
sce:YER008C SEC3, PSL1; Sec3p 32.0 0.99
pfa:PFL2110c hypothetical protein 30.0 3.4
dre:564101 si:dkey-230p4.1 29.6 4.8
> tgo:TGME49_014590 microfibrillar-associated protein 1, putative
; K13110 microfibrillar-associated protein 1
Length=438
Score = 190 bits (483), Expect = 2e-48, Method: Compositional matrix adjust.
Identities = 95/145 (65%), Positives = 116/145 (80%), Gaps = 5/145 (3%)
Query 26 QQQQQQQQQNML-----EGEEEMPDDTDGLDPAQEYEDWKLRELERIRRDKEQLVAREKF 80
Q++ ++ + NML E + E+PDDTDGLD EYE WK REL RI+RD+E+ AR+ F
Sbjct 235 QKEDEEMRTNMLGEQLQEEDCELPDDTDGLDAEAEYEAWKARELLRIKRDQEERQARDNF 294
Query 81 LEAVERRRQMTEEERREGDKELDKMQPKREIKHKYQFMQKYYHRGAFFQDLARSGEEPIY 140
LEA+ERRRQMTEEERR DK+LD MQP+RE+K+KY FMQKYYHRGAFFQDLARSGEE +Y
Sbjct 295 LEALERRRQMTEEERRLDDKQLDAMQPRREVKYKYNFMQKYYHRGAFFQDLARSGEEALY 354
Query 141 LRDFNAPVGEDAVDKKALPKILQLR 165
LRD+NAPVGED DKK LP +++R
Sbjct 355 LRDYNAPVGEDKWDKKILPSAMRVR 379
> tpv:TP04_0360 hypothetical protein; K13110 microfibrillar-associated
protein 1
Length=408
Score = 113 bits (283), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 63/121 (52%), Positives = 83/121 (68%), Gaps = 4/121 (3%)
Query 45 DDTDGLDPAQEYEDWKLRELERIRRDKEQLVAREKFLEAVERRRQMTEEERREGDKELDK 104
DDTD D +EYE WK+REL+RI RDKE+ +K E V+ RR MT+EER ++++DK
Sbjct 244 DDTDTFD-EKEYELWKIRELKRILRDKEEREKFKKLEEEVKLRRSMTDEERELDNQKVDK 302
Query 105 MQPKREIKHKYQFMQKYYHRGAFFQDLARSGEEPIYLRDFNAPVGEDAVDKKALPKILQL 164
+ + K K +F+QKYYHRGAFF D + EP+Y RDFNAP ED VDK LPK +++
Sbjct 303 VVVE---KGKLRFLQKYYHRGAFFMDKLQDKSEPLYARDFNAPTAEDCVDKSLLPKPMRV 359
Query 165 R 165
R
Sbjct 360 R 360
> dre:393992 mfap1, MGC56551, zgc:56551; microfibrillar-associated
protein 1; K13110 microfibrillar-associated protein 1
Length=437
Score = 102 bits (255), Expect = 4e-22, Method: Compositional matrix adjust.
Identities = 58/120 (48%), Positives = 80/120 (66%), Gaps = 4/120 (3%)
Query 46 DTDGLDPAQEYEDWKLRELERIRRDKEQLVAREKFLEAVERRRQMTEEERREGDKELDKM 105
DTDG + +EYE WK+REL+RI+RD+E A EK +ER +TEEERR + K+
Sbjct 265 DTDGENEEEEYEAWKVRELKRIKRDRESREALEKEKAEIERFHNLTEEERRAELRTNGKV 324
Query 106 QPKREIKHKYQFMQKYYHRGAFFQDLARSGEEPIYLRDFNAPVGEDAVDKKALPKILQLR 165
+ K KY+F+QKYYHRGAFF D GE+ ++ RDF+AP ED +K LPK++Q++
Sbjct 325 ITNKASKGKYKFLQKYYHRGAFFMD----GEQDVFKRDFSAPTLEDHFNKTILPKVMQVK 380
> ath:AT5G17900 hypothetical protein; K13110 microfibrillar-associated
protein 1
Length=435
Score = 102 bits (254), Expect = 6e-22, Method: Compositional matrix adjust.
Identities = 63/146 (43%), Positives = 94/146 (64%), Gaps = 20/146 (13%)
Query 33 QQNMLEGEEEMPD--DTDGLDPAQEYEDWKLRELERIRRDKEQLVAREKFL---EAVERR 87
++N+L E + D D L+ A+EYE WK RE+ RI+R+++ ARE L E +E+
Sbjct 237 RKNILLEEANIGDVETDDELNEAEEYEVWKTREIGRIKRERD---AREAMLREREEIEKL 293
Query 88 RQMTEEERREGDKELDK---MQPKREIKHKYQFMQKYYHRGAFFQ-----DLARSGEEPI 139
R MTE+ERR+ +++ K QPK+ K+ FMQKYYH+GAFFQ + +G + I
Sbjct 294 RNMTEQERRDWERKNPKPLSAQPKK----KWNFMQKYYHKGAFFQADPDDEAGSAGTDGI 349
Query 140 YLRDFNAPVGEDAVDKKALPKILQLR 165
+ RDF+AP GED +DK LPK++Q++
Sbjct 350 FQRDFSAPTGEDRLDKSILPKVMQVK 375
> ath:AT4G08580 hypothetical protein
Length=435
Score = 102 bits (253), Expect = 7e-22, Method: Compositional matrix adjust.
Identities = 63/146 (43%), Positives = 94/146 (64%), Gaps = 20/146 (13%)
Query 33 QQNMLEGEEEMPD--DTDGLDPAQEYEDWKLRELERIRRDKEQLVAREKFL---EAVERR 87
++N+L E + D D L+ A+EYE WK RE+ RI+R+++ ARE L E +E+
Sbjct 237 RKNILLEEANIGDVETDDELNEAEEYEVWKTREIGRIKRERD---AREAMLREREEIEKL 293
Query 88 RQMTEEERREGDKELDK---MQPKREIKHKYQFMQKYYHRGAFFQ-----DLARSGEEPI 139
R MTE+ERR+ +++ K QPK+ K+ FMQKYYH+GAFFQ + +G + I
Sbjct 294 RNMTEQERRDWERKNPKPSSAQPKK----KWNFMQKYYHKGAFFQADPDDEAGSAGTDGI 349
Query 140 YLRDFNAPVGEDAVDKKALPKILQLR 165
+ RDF+AP GED +DK LPK++Q++
Sbjct 350 FQRDFSAPTGEDRLDKSILPKVMQVK 375
> hsa:4236 MFAP1; microfibrillar-associated protein 1; K13110
microfibrillar-associated protein 1
Length=439
Score = 99.0 bits (245), Expect = 6e-21, Method: Compositional matrix adjust.
Identities = 52/107 (48%), Positives = 72/107 (67%), Gaps = 4/107 (3%)
Query 59 WKLRELERIRRDKEQLVAREKFLEAVERRRQMTEEERREGDKELDKMQPKREIKHKYQFM 118
WK+REL+RI+RD+E A EK +ER R +TEEERR + K+ + +K KY+F+
Sbjct 279 WKVRELKRIKRDREDREALEKEKAEIERMRNLTEEERRAELRANGKVITNKAVKGKYKFL 338
Query 119 QKYYHRGAFFQDLARSGEEPIYLRDFNAPVGEDAVDKKALPKILQLR 165
QKYYHRGAFF D +E +Y RDF+AP ED +K LPK++Q++
Sbjct 339 QKYYHRGAFFMD----EDEEVYKRDFSAPTLEDHFNKTILPKVMQVK 381
> xla:735061 hypothetical protein MGC85054; K13110 microfibrillar-associated
protein 1
Length=442
Score = 99.0 bits (245), Expect = 7e-21, Method: Compositional matrix adjust.
Identities = 52/107 (48%), Positives = 73/107 (68%), Gaps = 4/107 (3%)
Query 59 WKLRELERIRRDKEQLVAREKFLEAVERRRQMTEEERREGDKELDKMQPKREIKHKYQFM 118
WK+REL+RI+RD+E+ A EK V+R R MT+EERR + K+ + +K KY+F+
Sbjct 282 WKVRELKRIKRDREEREALEKEKAEVDRLRNMTDEERRAELRANGKIITNKALKGKYKFL 341
Query 119 QKYYHRGAFFQDLARSGEEPIYLRDFNAPVGEDAVDKKALPKILQLR 165
QKYYHRGAFF D +E +Y RDF+AP ED +K LPK++Q++
Sbjct 342 QKYYHRGAFFMD----EDENVYKRDFSAPTLEDHFNKTILPKVMQVK 384
> xla:379834 mfap1, MGC53463; microfibrillar-associated protein
1
Length=441
Score = 97.8 bits (242), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 52/107 (48%), Positives = 73/107 (68%), Gaps = 4/107 (3%)
Query 59 WKLRELERIRRDKEQLVAREKFLEAVERRRQMTEEERREGDKELDKMQPKREIKHKYQFM 118
WK+REL+RI+RD+E+ A EK V+R R MT+EERR + K+ + +K KY+F+
Sbjct 281 WKVRELKRIKRDREEREAMEKEKAEVDRLRNMTDEERRAELRANGKIITNKAMKGKYKFL 340
Query 119 QKYYHRGAFFQDLARSGEEPIYLRDFNAPVGEDAVDKKALPKILQLR 165
QKYYHRGAFF D +E +Y RDF+AP ED +K LPK++Q++
Sbjct 341 QKYYHRGAFFLD----EDENVYKRDFSAPTLEDHFNKTILPKVMQVK 383
> mmu:67532 Mfap1a, 4432409M24Rik, Mfap1; microfibrillar-associated
protein 1A
Length=439
Score = 97.1 bits (240), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 51/107 (47%), Positives = 72/107 (67%), Gaps = 4/107 (3%)
Query 59 WKLRELERIRRDKEQLVAREKFLEAVERRRQMTEEERREGDKELDKMQPKREIKHKYQFM 118
WK+REL+RI+R++E A EK +ER R +TEEERR + K+ + +K KY+F+
Sbjct 279 WKVRELKRIKREREDREALEKEKAEIERMRNLTEEERRAELRANGKVITNKAVKGKYKFL 338
Query 119 QKYYHRGAFFQDLARSGEEPIYLRDFNAPVGEDAVDKKALPKILQLR 165
QKYYHRGAFF D +E +Y RDF+AP ED +K LPK++Q++
Sbjct 339 QKYYHRGAFFMD----EDEEVYKRDFSAPTLEDHFNKTILPKVMQVK 381
> mmu:100034361 Mfap1b; microfibrillar-associated protein 1B;
K13110 microfibrillar-associated protein 1
Length=439
Score = 97.1 bits (240), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 51/107 (47%), Positives = 72/107 (67%), Gaps = 4/107 (3%)
Query 59 WKLRELERIRRDKEQLVAREKFLEAVERRRQMTEEERREGDKELDKMQPKREIKHKYQFM 118
WK+REL+RI+R++E A EK +ER R +TEEERR + K+ + +K KY+F+
Sbjct 279 WKVRELKRIKREREDREALEKEKAEIERMRNLTEEERRAELRANGKVITNKAVKGKYKFL 338
Query 119 QKYYHRGAFFQDLARSGEEPIYLRDFNAPVGEDAVDKKALPKILQLR 165
QKYYHRGAFF D +E +Y RDF+AP ED +K LPK++Q++
Sbjct 339 QKYYHRGAFFMD----EDEEVYKRDFSAPTLEDHFNKTILPKVMQVK 381
> bbo:BBOV_II002390 18.m06195; micro-fibrillar-associated protein
1 C-terminus containing protein; K13110 microfibrillar-associated
protein 1
Length=437
Score = 96.3 bits (238), Expect = 4e-20, Method: Compositional matrix adjust.
Identities = 72/160 (45%), Positives = 100/160 (62%), Gaps = 14/160 (8%)
Query 19 IEAQQQQQQQQQQQQQ----------NM---LEGEEEMPDDTDGLDPAQEYEDWKLRELE 65
IEA+Q++ +++++Q + NM +E E DD D L +EYE WK+REL+
Sbjct 225 IEAEQKRLEERRKQSKELVIQTLVAENMHQEIENEVNCVDDKDELT-EEEYELWKIRELK 283
Query 66 RIRRDKEQLVAREKFLEAVERRRQMTEEERREGDKELDKMQPKREIKHKYQFMQKYYHRG 125
RI RD+ + A E+ VERRR+MTEEER E D+ + + + K K +F+QKYYH+G
Sbjct 284 RIIRDRNERNAHERLAAEVERRREMTEEERLEDDERIRQEKGPIAPKTKIKFLQKYYHKG 343
Query 126 AFFQDLARSGEEPIYLRDFNAPVGEDAVDKKALPKILQLR 165
AFF D G EPIY RDFNAP +D VDK +PK +Q+R
Sbjct 344 AFFMDKLEDGSEPIYKRDFNAPTADDCVDKSLMPKSMQVR 383
> pfa:MAL13P1.132 microfibril-associated protein homologue, putative;
K13110 microfibrillar-associated protein 1
Length=492
Score = 90.1 bits (222), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 48/107 (44%), Positives = 72/107 (67%), Gaps = 2/107 (1%)
Query 59 WKLRELERIRRDKEQLVAREKFLEAVERRRQMTEEERREGDKELDKMQPKREIKHKYQFM 118
WK+R + R++RD+ E +++RR+MT++E + +K L + K++ K FM
Sbjct 335 WKIRHINRLKRDELDRKKHEILELEIKKRRKMTDKEIIQDNKTLPNKEKKKK--RKMLFM 392
Query 119 QKYYHRGAFFQDLARSGEEPIYLRDFNAPVGEDAVDKKALPKILQLR 165
QKYYH+G F+QDL G+E IYLRD+N PV ED VD++ LPK+LQ+R
Sbjct 393 QKYYHKGGFYQDLFEEGKEEIYLRDYNEPVYEDKVDRQNLPKVLQVR 439
> cel:F43G9.10 hypothetical protein; K13110 microfibrillar-associated
protein 1
Length=466
Score = 72.0 bits (175), Expect = 9e-13, Method: Compositional matrix adjust.
Identities = 43/112 (38%), Positives = 71/112 (63%), Gaps = 8/112 (7%)
Query 56 YEDWKLRELERIRRDKEQL--VAREKFLEAVERRRQMTEEERREGDKELDKMQPKREIKH 113
YE WKLRE++R++R++++ AREK +++ M+EEER + + K+ ++ K
Sbjct 304 YEAWKLREMKRLKRNRDEREEAAREK--AELDKIHAMSEEERLKYLRLNPKVITNKQDKG 361
Query 114 KYQFMQKYYHRGAFFQDLARSGEEPIYLRDFNAPVGEDAVDKKALPKILQLR 165
KY+F+QKY+HRGAFF D E+ + R+F +D DK LPK++Q++
Sbjct 362 KYKFLQKYFHRGAFFLD----EEDEVLKRNFAEATNDDQFDKTILPKVMQVK 409
> pfa:PF13_0198 PfRh2a; reticulocyte binding protein 2 homolog
A; K13849 reticulocyte-binding protein
Length=3130
Score = 40.8 bits (94), Expect = 0.002, Method: Composition-based stats.
Identities = 28/111 (25%), Positives = 61/111 (54%), Gaps = 9/111 (8%)
Query 20 EAQQQQQQQQQQQQQNMLEGEEEMPDDTDGLDPAQEYEDWKLRELERIRRDKEQLVAREK 79
+ Q Q++++ ++Q+Q L+ EE + + Q+ E+ K +E ER+ R+K++ + +E+
Sbjct 2766 QEQLQKEEELKRQEQERLQKEEALKRQEQ--ERLQKEEELKRQEQERLEREKQEQLQKEE 2823
Query 80 FLEAVERRRQMTEE-------ERREGDKELDKMQPKREIKHKYQFMQKYYH 123
L+ E+ R EE ER + ++EL + + +R + K + ++ H
Sbjct 2824 ELKRQEQERLQKEEALKRQEQERLQKEEELKRQEQERLERKKIELAEREQH 2874
Score = 36.6 bits (83), Expect = 0.034, Method: Composition-based stats.
Identities = 30/107 (28%), Positives = 59/107 (55%), Gaps = 5/107 (4%)
Query 17 LSIEAQQQQQQQQQ--QQQQNMLEGEEEMPDDTDGLDPAQEYEDWKLRELERIRRDKEQL 74
L E Q+Q Q++++ +Q+Q L+ EE + + Q+ E+ K +E ER+ R K +L
Sbjct 2811 LEREKQEQLQKEEELKRQEQERLQKEEALKRQEQ--ERLQKEEELKRQEQERLERKKIEL 2868
Query 75 VAREKFLEAVERRRQMTEEERREGDKELDKMQPKREIKHKYQFMQKY 121
RE+ +++ + M + + E KE D++ ++IK ++ QK+
Sbjct 2869 AEREQHIKS-KLESDMVKIIKDELTKEKDEIIKNKDIKLRHSLEQKW 2914
Score = 35.0 bits (79), Expect = 0.12, Method: Composition-based stats.
Identities = 24/97 (24%), Positives = 56/97 (57%), Gaps = 4/97 (4%)
Query 20 EAQQQQQQQQQQQQQNMLEGEEEMPDDTDGLDPAQEYEDWKLRELERIRRDKEQLVAREK 79
E ++Q+Q++ ++++Q L+ EEE+ + Q+ E K +E ER+++++E ++
Sbjct 2752 ELKRQEQERLEREKQEQLQKEEELKRQEQ--ERLQKEEALKRQEQERLQKEEELKRQEQE 2809
Query 80 FLEAVERRRQMTEEERREGDKELDKMQPKREIKHKYQ 116
LE ++ + EEE + ++E ++Q + +K + Q
Sbjct 2810 RLEREKQEQLQKEEELKRQEQE--RLQKEEALKRQEQ 2844
Score = 33.5 bits (75), Expect = 0.36, Method: Composition-based stats.
Identities = 22/74 (29%), Positives = 43/74 (58%), Gaps = 7/74 (9%)
Query 54 QEYEDWKLRELERIRRDKEQLVAREKFLEAVERRRQMTEE-------ERREGDKELDKMQ 106
Q+ E+ K +E ER+ R+K++ + +E+ L+ E+ R EE ER + ++EL + +
Sbjct 2748 QKEEELKRQEQERLEREKQEQLQKEEELKRQEQERLQKEEALKRQEQERLQKEEELKRQE 2807
Query 107 PKREIKHKYQFMQK 120
+R + K + +QK
Sbjct 2808 QERLEREKQEQLQK 2821
Score = 33.1 bits (74), Expect = 0.47, Method: Composition-based stats.
Identities = 24/93 (25%), Positives = 56/93 (60%), Gaps = 2/93 (2%)
Query 24 QQQQQQQQQQQNMLEGEEEMPDDTDGLDPAQEYEDWKLRELERIRRDKEQLVAREKFLEA 83
++Q+Q++ Q++ L+ +E+ + + + Q+ E+ K +E ER++++ E L +E+
Sbjct 2740 KRQEQERLQKEEELKRQEQERLEREKQEQLQKEEELKRQEQERLQKE-EALKRQEQERLQ 2798
Query 84 VERRRQMTEEERREGDKELDKMQPKREIKHKYQ 116
E + E+ER E +K+ +++Q + E+K + Q
Sbjct 2799 KEEELKRQEQERLEREKQ-EQLQKEEELKRQEQ 2830
> sce:YER008C SEC3, PSL1; Sec3p
Length=1336
Score = 32.0 bits (71), Expect = 0.99, Method: Composition-based stats.
Identities = 24/98 (24%), Positives = 53/98 (54%), Gaps = 5/98 (5%)
Query 23 QQQQQQQQQQQQNMLEGEEEMPDDTDGLDPAQEYEDWKLRELERIRRDKEQLVAREKFLE 82
Q++ Q Q++ + LE E + + E++ R+LE R ++ + +K +E
Sbjct 348 QKRLQLQKENEMKRLEEERRIKQEERKRQMELEHQ----RQLEEEERKRQMELEAKKQME 403
Query 83 AVERRRQMTEEERREGDKELDKMQPKREIKHKYQFMQK 120
++R+RQ EE+R + ++EL ++Q K+ + + ++K
Sbjct 404 -LKRQRQFEEEQRLKKERELLEIQRKQREQETAERLKK 440
> pfa:PFL2110c hypothetical protein
Length=1846
Score = 30.0 bits (66), Expect = 3.4, Method: Composition-based stats.
Identities = 23/81 (28%), Positives = 49/81 (60%), Gaps = 5/81 (6%)
Query 20 EAQQQQQQQQQQQQQNMLEGEEEMPDDTDGLDPAQEYEDWKLRELERIRRDKEQLVAREK 79
E Q+ +++Q+ +++Q M E E++M ++ + + E+ K+RE +++R +EQ V E+
Sbjct 1532 EEQKMREEQKMREEQKMRE-EQKMREEQKVREEQKMREEQKMREEQKMR--EEQKVREEQ 1588
Query 80 FLEAVERRRQMTEEERREGDK 100
L ++ R+ E++ RE K
Sbjct 1589 KLREEQKMRE--EQKMREEQK 1607
Score = 28.5 bits (62), Expect = 9.3, Method: Composition-based stats.
Identities = 24/97 (24%), Positives = 53/97 (54%), Gaps = 8/97 (8%)
Query 22 QQQQQQQQQQQQQNMLEGEEEMPDDTDGLDPAQEY-EDWKLRELERIRRDK----EQLVA 76
++Q+ +++Q+ + GEE+ + L Q+ E+ K+RE +++R ++ EQ V
Sbjct 1502 EEQKMREEQKVGEEQKVGEEQKVGEEQKLREEQKMREEQKMREEQKMREEQKMREEQKVR 1561
Query 77 REKFLEAVERRRQ---MTEEERREGDKELDKMQPKRE 110
E+ + ++ R+ M EE++ +++L + Q RE
Sbjct 1562 EEQKMREEQKMREEQKMREEQKVREEQKLREEQKMRE 1598
> dre:564101 si:dkey-230p4.1
Length=2775
Score = 29.6 bits (65), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 25/57 (43%), Positives = 33/57 (57%), Gaps = 3/57 (5%)
Query 52 PAQEYEDWKLRELERIRRDKEQLVAREKFLEAVERRRQMTEEERREGDKELDKMQPK 108
E E WK RE +++R+KE+L +KFLE VER Q E +RE K D M+ K
Sbjct 958 ACDEVERWKERE-NKVQREKEEL--NQKFLERVERESQNLEITQREKAKMSDLMKKK 1011
Lambda K H
0.314 0.131 0.361
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3962792044
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40