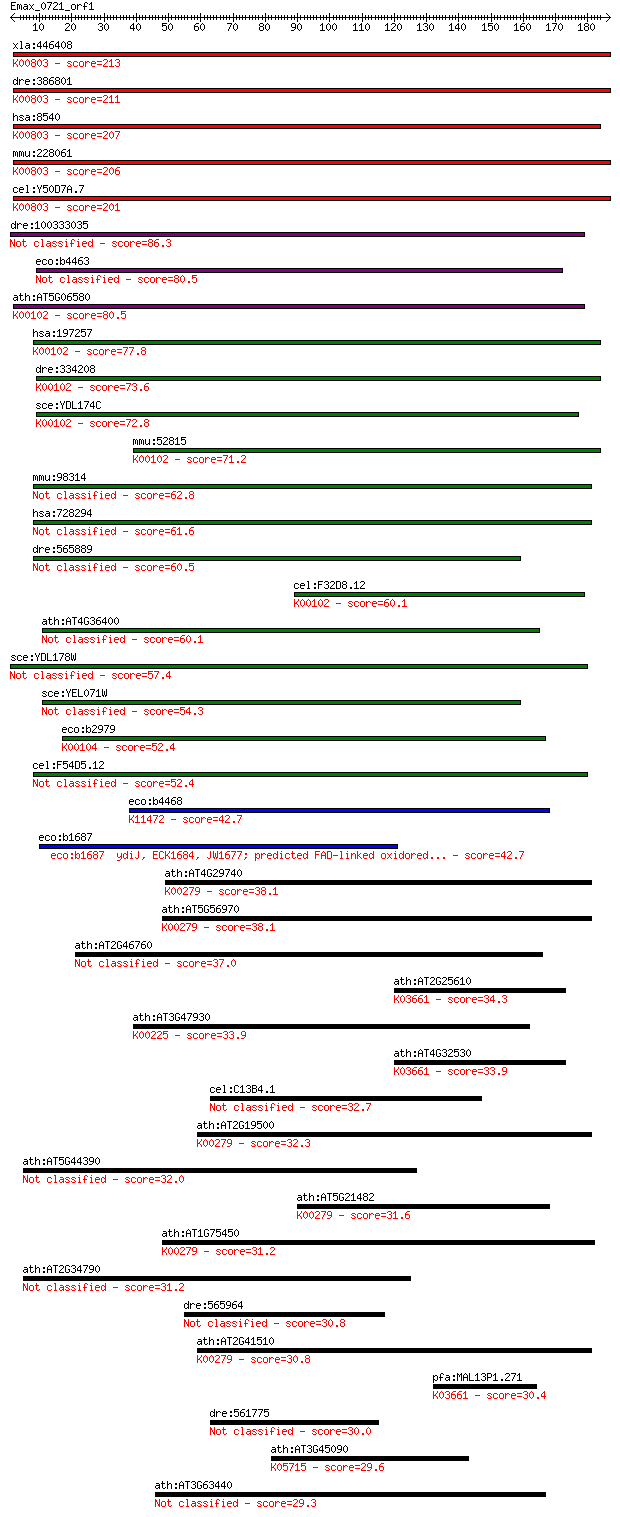
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0721_orf1
Length=186
Score E
Sequences producing significant alignments: (Bits) Value
xla:446408 agps, MGC83829; alkylglycerone phosphate synthase (... 213 2e-55
dre:386801 agps, fd16e06, wu:fd16e06, zgc:56718; alkylglyceron... 211 9e-55
hsa:8540 AGPS, ADAP-S, ADAS, ADHAPS, ADPS, ALDHPSY, DKFZp762O2... 207 2e-53
mmu:228061 Agps, 5832437L22, 9930035G10Rik, AW123847, Adaps, A... 206 5e-53
cel:Y50D7A.7 ads-1; Alkyl-Dihydroxyacetonephosphate Synthase f... 201 1e-51
dre:100333035 lactate dehydrogenase D-like 86.3 5e-17
eco:b4463 ygcU, ECK2767, JW5442, ygcT, ygcV; predicted FAD con... 80.5 3e-15
ath:AT5G06580 FAD linked oxidase family protein (EC:1.1.2.4); ... 80.5 3e-15
hsa:197257 LDHD, DLD, MGC57726; lactate dehydrogenase D (EC:1.... 77.8 2e-14
dre:334208 ldhd, wu:fi36b04, zgc:55447; lactate dehydrogenase ... 73.6 4e-13
sce:YDL174C DLD1; D-lactate dehydrogenase, oxidizes D-lactate ... 72.8 6e-13
mmu:52815 Ldhd, 4733401P21Rik, D8Bwg1320e; lactate dehydrogena... 71.2 2e-12
mmu:98314 D2hgdh, AA408776, AA408778, AI325464; D-2-hydroxyglu... 62.8 6e-10
hsa:728294 D2HGDH, D2HGD, FLJ42195, MGC25181; D-2-hydroxygluta... 61.6 1e-09
dre:565889 d2hgdh, zgc:158661; D-2-hydroxyglutarate dehydrogen... 60.5 3e-09
cel:F32D8.12 hypothetical protein; K00102 D-lactate dehydrogen... 60.1 4e-09
ath:AT4G36400 FAD linked oxidase family protein 60.1 4e-09
sce:YDL178W DLD2, AIP2; D-lactate dehydrogenase, located in th... 57.4 3e-08
sce:YEL071W DLD3; D-lactate dehydrogenase, part of the retrogr... 54.3 3e-07
eco:b2979 glcD, ECK2974, gox, JW2946, yghM; glycolate oxidase ... 52.4 1e-06
cel:F54D5.12 hypothetical protein 52.4 1e-06
eco:b4468 glcE, ECK2973, gox, JW5487, yghL; glycolate oxidase ... 42.7 6e-04
eco:b1687 ydiJ, ECK1684, JW1677; predicted FAD-linked oxidored... 42.7 7e-04
ath:AT4G29740 CKX4; CKX4 (CYTOKININ OXIDASE 4); amine oxidase/... 38.1 0.017
ath:AT5G56970 CKX3; CKX3 (CYTOKININ OXIDASE 3); amine oxidase/... 38.1 0.018
ath:AT2G46760 FAD-binding domain-containing protein 37.0 0.035
ath:AT2G25610 H+-transporting two-sector ATPase, C subunit fam... 34.3 0.28
ath:AT3G47930 ATGLDH; ATGLDH (L-GALACTONO-1,4-LACTONE DEHYDORO... 33.9 0.30
ath:AT4G32530 vacuolar ATP synthase, putative / V-ATPase, puta... 33.9 0.37
cel:C13B4.1 hypothetical protein 32.7 0.65
ath:AT2G19500 CKX2; CKX2 (CYTOKININ OXIDASE 2); amine oxidase/... 32.3 0.98
ath:AT5G44390 FAD-binding domain-containing protein 32.0 1.3
ath:AT5G21482 CKX7; CKX7 (CYTOKININ OXIDASE 7); cytokinin dehy... 31.6 1.7
ath:AT1G75450 CKX5; CKX5 (CYTOKININ OXIDASE 5); cytokinin dehy... 31.2 2.4
ath:AT2G34790 MEE23; MEE23 (MATERNAL EFFECT EMBRYO ARREST 23);... 31.2 2.4
dre:565964 MGC123280, sdr42e1, si:ch211-79l17.4; zgc:123280 (E... 30.8 2.5
ath:AT2G41510 CKX1; CKX1 (CYTOKININ OXIDASE/DEHYDROGENASE 1); ... 30.8 2.9
pfa:MAL13P1.271 V-type ATPase, putative; K03661 V-type H+-tran... 30.4 3.9
dre:561775 specc1la, cytsaa, im:7159316, specc1l; sperm antige... 30.0 5.0
ath:AT3G45090 2-phosphoglycerate kinase-related; K05715 2-phos... 29.6 5.4
ath:AT3G63440 CKX6; CKX6 (CYTOKININ OXIDASE/DEHYDROGENASE 6); ... 29.3 9.1
> xla:446408 agps, MGC83829; alkylglycerone phosphate synthase
(EC:2.5.1.26); K00803 alkyldihydroxyacetonephosphate synthase
[EC:2.5.1.26]
Length=627
Score = 213 bits (543), Expect = 2e-55, Method: Compositional matrix adjust.
Identities = 100/185 (54%), Positives = 131/185 (70%), Gaps = 0/185 (0%)
Query 2 IVDCAIKHNICLIPFGGGTSVTLGVCTPEEEERMVATVSLSFMQKILYLDRDALLMWVEA 61
IVD A KHN+CLIPFGGGTSV+ + PE+E+R +A++ S M +IL++D L +EA
Sbjct 190 IVDLACKHNVCLIPFGGGTSVSYALECPEDEKRTIASLDTSQMSRILWIDEKNLTAHIEA 249
Query 62 GAVGASIEERLRPYGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDLVVDIKVVT 121
G G +E +L G GHEPDSMEFST+GGWVATRASGMKKN YGNIEDLVV IK+VT
Sbjct 250 GITGQDLERQLGESGYCTGHEPDSMEFSTLGGWVATRASGMKKNIYGNIEDLVVHIKMVT 309
Query 122 TRGCMHACNSSPRISSGPSLQQLFLGSEGIYGIITQVLLKIKPIPEYKVYDCLAFPSFSL 181
+G + PR+S+GP + +GSEG G++T+V +KI+P+PEY+ Y + FP+F
Sbjct 310 PKGIIEKSCQGPRMSTGPDIHHFIMGSEGTLGVVTEVTIKIRPVPEYQKYGSVVFPNFER 369
Query 182 GVLFL 186
GV L
Sbjct 370 GVACL 374
> dre:386801 agps, fd16e06, wu:fd16e06, zgc:56718; alkylglycerone
phosphate synthase (EC:2.5.1.26); K00803 alkyldihydroxyacetonephosphate
synthase [EC:2.5.1.26]
Length=629
Score = 211 bits (538), Expect = 9e-55, Method: Compositional matrix adjust.
Identities = 100/185 (54%), Positives = 130/185 (70%), Gaps = 0/185 (0%)
Query 2 IVDCAIKHNICLIPFGGGTSVTLGVCTPEEEERMVATVSLSFMQKILYLDRDALLMWVEA 61
IVD A KHN+CLIP+GGGTSV+ + P+EE R + ++ S M +IL++D L VEA
Sbjct 192 IVDLACKHNVCLIPYGGGTSVSSALECPQEETRCIVSLDTSQMNRILWIDEKNLTAHVEA 251
Query 62 GAVGASIEERLRPYGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDLVVDIKVVT 121
G +G +E +L G GHEPDSMEFS++GGWVATRASGMKKN YGNIEDLVV IK+VT
Sbjct 252 GIIGQDLERQLNERGYCTGHEPDSMEFSSLGGWVATRASGMKKNIYGNIEDLVVHIKMVT 311
Query 122 TRGCMHACNSSPRISSGPSLQQLFLGSEGIYGIITQVLLKIKPIPEYKVYDCLAFPSFSL 181
RG + PR+S+GP + +GSEG G++T+V +KI+PIPEY+ Y + FP+F
Sbjct 312 PRGVIEKSCLGPRMSTGPDIHHFIMGSEGTLGVVTEVTMKIRPIPEYQKYGSVVFPNFQQ 371
Query 182 GVLFL 186
GV L
Sbjct 372 GVACL 376
> hsa:8540 AGPS, ADAP-S, ADAS, ADHAPS, ADPS, ALDHPSY, DKFZp762O2215,
FLJ99755; alkylglycerone phosphate synthase (EC:2.5.1.26);
K00803 alkyldihydroxyacetonephosphate synthase [EC:2.5.1.26]
Length=658
Score = 207 bits (526), Expect = 2e-53, Method: Compositional matrix adjust.
Identities = 96/182 (52%), Positives = 129/182 (70%), Gaps = 0/182 (0%)
Query 2 IVDCAIKHNICLIPFGGGTSVTLGVCTPEEEERMVATVSLSFMQKILYLDRDALLMWVEA 61
IV+ A K+N+C+IP GGGTSV+ G+ P +E R + ++ S M +IL++D + L VEA
Sbjct 221 IVNLACKYNLCIIPIGGGTSVSYGLMCPADETRTIISLDTSQMNRILWVDENNLTAHVEA 280
Query 62 GAVGASIEERLRPYGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDLVVDIKVVT 121
G G +E +L+ G GHEPDS+EFSTVGGWV+TRASGMKKN YGNIEDLVV IK+VT
Sbjct 281 GITGQELERQLKESGYCTGHEPDSLEFSTVGGWVSTRASGMKKNIYGNIEDLVVHIKMVT 340
Query 122 TRGCMHACNSSPRISSGPSLQQLFLGSEGIYGIITQVLLKIKPIPEYKVYDCLAFPSFSL 181
RG + PR+S+GP + +GSEG G+IT+ +KI+P+PEY+ Y +AFP+F
Sbjct 341 PRGIIEKSCQGPRMSTGPDIHHFIMGSEGTLGVITEATIKIRPVPEYQKYGSVAFPNFEQ 400
Query 182 GV 183
GV
Sbjct 401 GV 402
> mmu:228061 Agps, 5832437L22, 9930035G10Rik, AW123847, Adaps,
Adas, Adhaps, Adps, Aldhpsy, bs2; alkylglycerone phosphate
synthase (EC:2.5.1.26); K00803 alkyldihydroxyacetonephosphate
synthase [EC:2.5.1.26]
Length=671
Score = 206 bits (523), Expect = 5e-53, Method: Compositional matrix adjust.
Identities = 95/185 (51%), Positives = 130/185 (70%), Gaps = 0/185 (0%)
Query 2 IVDCAIKHNICLIPFGGGTSVTLGVCTPEEEERMVATVSLSFMQKILYLDRDALLMWVEA 61
IV+ A K+N+C+IP GGGTSV+ G+ P +E R + ++ S M +IL++D + L VEA
Sbjct 234 IVNLACKYNLCIIPIGGGTSVSYGLMCPADETRTIISLDTSQMNRILWVDENNLTAHVEA 293
Query 62 GAVGASIEERLRPYGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDLVVDIKVVT 121
G G +E +L+ G GHEPDS+EFSTVGGW++TRASGMKKN YGNIEDLVV +K+VT
Sbjct 294 GITGQDLERQLKESGYCTGHEPDSLEFSTVGGWISTRASGMKKNIYGNIEDLVVHMKMVT 353
Query 122 TRGCMHACNSSPRISSGPSLQQLFLGSEGIYGIITQVLLKIKPIPEYKVYDCLAFPSFSL 181
RG + + PR+S+GP + +GSEG G+IT+ +KI+P PEY+ Y +AFP+F
Sbjct 354 PRGVIEKSSQGPRMSTGPDIHHFIMGSEGTLGVITEATIKIRPTPEYQKYGSVAFPNFEQ 413
Query 182 GVLFL 186
GV L
Sbjct 414 GVACL 418
> cel:Y50D7A.7 ads-1; Alkyl-Dihydroxyacetonephosphate Synthase
family member (ads-1); K00803 alkyldihydroxyacetonephosphate
synthase [EC:2.5.1.26]
Length=597
Score = 201 bits (511), Expect = 1e-51, Method: Compositional matrix adjust.
Identities = 95/185 (51%), Positives = 129/185 (69%), Gaps = 0/185 (0%)
Query 2 IVDCAIKHNICLIPFGGGTSVTLGVCTPEEEERMVATVSLSFMQKILYLDRDALLMWVEA 61
I++ A+ HN +IP GGGTSVT + TPE E+R V ++ ++ + KIL++DR+ L +A
Sbjct 150 IIEGAMSHNCAIIPIGGGTSVTNALDTPETEKRAVISMDMALLDKILWIDRENLTCRAQA 209
Query 62 GAVGASIEERLRPYGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDLVVDIKVVT 121
G VG S+E +L G T GHEPDS+EFST+GGWV+TRASGMKKN+YGNIEDLVV + V
Sbjct 210 GIVGQSLERQLNKKGFTCGHEPDSIEFSTLGGWVSTRASGMKKNKYGNIEDLVVHLNFVC 269
Query 122 TRGCMHACNSSPRISSGPSLQQLFLGSEGIYGIITQVLLKIKPIPEYKVYDCLAFPSFSL 181
+G + PR+SSGP + Q+ LGSEG G++++V +KI PIPE K + FP+F
Sbjct 270 PKGIIQKQCQVPRMSSGPDIHQIILGSEGTLGVVSEVTIKIFPIPEVKRFGSFVFPNFES 329
Query 182 GVLFL 186
GV F
Sbjct 330 GVNFF 334
> dre:100333035 lactate dehydrogenase D-like
Length=484
Score = 86.3 bits (212), Expect = 5e-17, Method: Compositional matrix adjust.
Identities = 61/179 (34%), Positives = 92/179 (51%), Gaps = 7/179 (3%)
Query 1 VIVDCAIKHNICLIPFGGGTSVTLGVCTPEEEERMVATVSLSFMQKILYLDRDALLMWVE 60
+V +H + +IPFG G+S+ V P ++ S M +IL + + L + VE
Sbjct 80 AVVRACSEHGVPIIPFGTGSSLEGQVNAPSGG----ISIDFSRMNRILRVSPEDLDVTVE 135
Query 61 AGAVGASIEERLRPYGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDLVVDIKVV 120
G + LR G+ +P + +++GG ATRASG RYG ++D V+ + VV
Sbjct 136 PGVTREQLNVYLRDTGLFFPIDPGAN--ASIGGMTATRASGTNAVRYGTMKDNVLAMTVV 193
Query 121 TTRGCMHACNSSPRISS-GPSLQQLFLGSEGIYGIITQVLLKIKPIPEYKVYDCLAFPS 178
T G A S + SS G L +LF+GSEG G+ T + L+++ IPE AFPS
Sbjct 194 TADGEEIATGSRAKKSSAGYDLTRLFVGSEGTLGVFTSITLRLQGIPEKIGGGVCAFPS 252
> eco:b4463 ygcU, ECK2767, JW5442, ygcT, ygcV; predicted FAD containing
dehydrogenase
Length=484
Score = 80.5 bits (197), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 52/164 (31%), Positives = 83/164 (50%), Gaps = 4/164 (2%)
Query 9 HNICLIPFGGGTSVTLGVCTPEEEERMVATVSLSFMQKILYLDRDALLMWVEAGAVGASI 68
H I +P G ++ G+ T E + S M +I+ +D + + + G +
Sbjct 73 HKINGVPRTGASATEGGLETVVENS---VVLDGSAMNQIINIDIENMQATAQCGVPLEVL 129
Query 69 EERLRPYGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDLVVDIKVVTTRGCMHA 128
E LR G T GH P S + +GG VATR+ G YG IED+VV ++ V G +
Sbjct 130 ENALREKGYTTGHSPQSKPLAQMGGLVATRSIGQFSTLYGAIEDMVVGLEAVLADGTVTR 189
Query 129 CNSSPRISSGPSLQQLFLGSEGIYGIITQVLLKI-KPIPEYKVY 171
+ PR ++GP ++ + +G+EG IT+V +KI K PE ++
Sbjct 190 IKNVPRRAAGPDIRHIIIGNEGALCYITEVTVKIFKFTPENNLF 233
> ath:AT5G06580 FAD linked oxidase family protein (EC:1.1.2.4);
K00102 D-lactate dehydrogenase (cytochrome) [EC:1.1.2.4]
Length=567
Score = 80.5 bits (197), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 53/178 (29%), Positives = 92/178 (51%), Gaps = 7/178 (3%)
Query 2 IVDCAIKHNICLIPFGGGTSVTLGVCTPEEEERMVATVSLSFMQKILYLDRDALLMWVEA 61
I+ ++ + ++P+GG TS+ P + + +S M+++ L + + + VE
Sbjct 161 ILKSCNEYKVPIVPYGGATSIEGHTLAP----KGGVCIDMSLMKRVKALHVEDMDVIVEP 216
Query 62 GAVGASIEERLRPYGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDLVVDIKVVT 121
G + E L YG+ +P +++GG ATR SG RYG + D V+ +KVV
Sbjct 217 GIGWLELNEYLEEYGLFFPLDPGP--GASIGGMCATRCSGSLAVRYGTMRDNVISLKVVL 274
Query 122 TRGCMHACNSSPRISS-GPSLQQLFLGSEGIYGIITQVLLKIKPIPEYKVYDCLAFPS 178
G + S R S+ G L +L +GSEG G+IT++ L+++ IP++ V FP+
Sbjct 275 PNGDVVKTASRARKSAAGYDLTRLIIGSEGTLGVITEITLRLQKIPQHSVVAVCNFPT 332
> hsa:197257 LDHD, DLD, MGC57726; lactate dehydrogenase D (EC:1.1.2.4);
K00102 D-lactate dehydrogenase (cytochrome) [EC:1.1.2.4]
Length=507
Score = 77.8 bits (190), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 61/203 (30%), Positives = 94/203 (46%), Gaps = 33/203 (16%)
Query 8 KHNICLIPFGGGTSVTLGVCTPEEEERMVATVSLSFMQKILYLDRDALLMWVEAGAVGAS 67
+ + +IPFG GT + GVC + V+L+ M +IL L+++ + VE G +
Sbjct 87 RQGVPIIPFGTGTGLEGGVCAVQGG----VCVNLTHMDRILELNQEDFSVVVEPGVTRKA 142
Query 68 IEERLRPYGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDLVVDIKVVTTRGCM- 126
+ LR G+ +P + +++ G AT ASG RYG + D V++++VV G +
Sbjct 143 LNAHLRDSGLWFPVDPGAD--ASLCGMAATGASGTNAVRYGTMRDNVLNLEVVLPDGRLL 200
Query 127 -------------------HACNSSPRIS-------SGPSLQQLFLGSEGIYGIITQVLL 160
H SP +S +G +L LF+GSEG G+IT L
Sbjct 201 HTAGRGRHFRFGFWPEIPHHTAWYSPCVSLGRRKSAAGYNLTGLFVGSEGTLGLITATTL 260
Query 161 KIKPIPEYKVYDCLAFPSFSLGV 183
++ P PE V AFPS V
Sbjct 261 RLHPAPEATVAATCAFPSVQAAV 283
> dre:334208 ldhd, wu:fi36b04, zgc:55447; lactate dehydrogenase
D (EC:1.1.2.4); K00102 D-lactate dehydrogenase (cytochrome)
[EC:1.1.2.4]
Length=497
Score = 73.6 bits (179), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 55/179 (30%), Positives = 90/179 (50%), Gaps = 10/179 (5%)
Query 9 HNICLIPFGGGTSVTLGVCTPEEEERMVATVSLSFMQKILYLDRDALLMWVEAGAVGASI 68
+ + +IPFG GT + GV + SL M++++ L ++ + VE G S+
Sbjct 92 YRLPIIPFGTGTGLEGGVSALQGG----VCFSLRKMEQVVDLHQEDFDVTVEPGVTRKSL 147
Query 69 EERLRPYGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDLVVDIKVVTTRGC-MH 127
LR G+ +P + +++ G AT ASG RYG + + V++++VV G +H
Sbjct 148 NSYLRDTGLWFPVDPGAD--ASLCGMAATSASGTNAVRYGTMRENVLNLEVVLADGTILH 205
Query 128 ACNSSPR---ISSGPSLQQLFLGSEGIYGIITQVLLKIKPIPEYKVYDCLAFPSFSLGV 183
R ++G +L LF+GSEG GIIT+ L++ +PE V +FPS V
Sbjct 206 TAGKGRRPRKTAAGYNLTNLFVGSEGTLGIITKATLRLYGVPESMVSAVCSFPSVQSAV 264
> sce:YDL174C DLD1; D-lactate dehydrogenase, oxidizes D-lactate
to pyruvate, transcription is heme-dependent, repressed by
glucose, and derepressed in ethanol or lactate; located in
the mitochondrial inner membrane (EC:1.1.2.4); K00102 D-lactate
dehydrogenase (cytochrome) [EC:1.1.2.4]
Length=587
Score = 72.8 bits (177), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 52/170 (30%), Positives = 89/170 (52%), Gaps = 6/170 (3%)
Query 9 HNICLIPFGGGTSVTLGVCTPEEEERMVATVSLS-FMQKILYLDRDALLMWVEAGAVGAS 67
+N+ ++PF GGTS+ G P + TV LS FM ++ D+ L + V+AG
Sbjct 173 NNMPVVPFSGGTSLE-GHFLPTRIGDTI-TVDLSKFMNNVVKFDKLDLDITVQAGLPWED 230
Query 68 IEERLRPYGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDLVVDIKVVTTRGCMH 127
+ + L +G+ G +P + +GG +A SG RYG +++ ++++ +V G +
Sbjct 231 LNDYLSDHGLMFGCDPGPG--AQIGGCIANSCSGTNAYRYGTMKENIINMTIVLPDGTIV 288
Query 128 ACNSSPRISS-GPSLQQLFLGSEGIYGIITQVLLKIKPIPEYKVYDCLAF 176
PR SS G +L LF+GSEG GI+T+ +K P+ + ++F
Sbjct 289 KTKKRPRKSSAGYNLNGLFVGSEGTLGIVTEATVKCHVKPKAETVAVVSF 338
> mmu:52815 Ldhd, 4733401P21Rik, D8Bwg1320e; lactate dehydrogenase
D (EC:1.1.2.4); K00102 D-lactate dehydrogenase (cytochrome)
[EC:1.1.2.4]
Length=484
Score = 71.2 bits (173), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 48/149 (32%), Positives = 76/149 (51%), Gaps = 6/149 (4%)
Query 39 VSLSFMQKILYLDRDALLMWVEAGAVGASIEERLRPYGVTLGHEPDSMEFSTVGGWVATR 98
++L+ M +I L+ + + VE G ++ LR G+ +P + +++ G AT
Sbjct 114 INLTHMDQITELNTEDFSVVVEPGVTRKALNTHLRDSGLWFPVDPGA--DASLCGMAATG 171
Query 99 ASGMKKNRYGNIEDLVVDIKVVTTRG-CMHACNSSP---RISSGPSLQQLFLGSEGIYGI 154
ASG RYG + D V++++VV G +H + ++G +L LF+GSEG GI
Sbjct 172 ASGTNAVRYGTMRDNVINLEVVLPDGRLLHTAGRGRHYRKSAAGYNLTGLFVGSEGTLGI 231
Query 155 ITQVLLKIKPIPEYKVYDCLAFPSFSLGV 183
IT L++ P PE V AFPS V
Sbjct 232 ITSTTLRLHPAPEATVAATCAFPSVQAAV 260
> mmu:98314 D2hgdh, AA408776, AA408778, AI325464; D-2-hydroxyglutarate
dehydrogenase (EC:1.1.99.-)
Length=535
Score = 62.8 bits (151), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 48/174 (27%), Positives = 89/174 (51%), Gaps = 5/174 (2%)
Query 8 KHNICLIPFGGGTSVTLGVCTPEEEERMVATVSLSFMQKILYLDRDALLMWVEAGAVGAS 67
K N+ + P GG T + +G P +E +++T + M +++ + ++ +AG V
Sbjct 135 KRNLAVNPQGGNTGM-VGGSVPVFDEVILST---ALMNQVISFHDVSGILVCQAGCVLEE 190
Query 68 IEERLRPYGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDLVVDIKVVTTRGCMH 127
+ ++ + + + +GG VAT A G++ RYG++ V+ ++VV G +
Sbjct 191 LSRYVQERDFIMPLDLGAKGSCHIGGNVATNAGGLRFLRYGSLRGTVLGLEVVLADGTIL 250
Query 128 ACNSSPRI-SSGPSLQQLFLGSEGIYGIITQVLLKIKPIPEYKVYDCLAFPSFS 180
C +S R ++G L+Q+F+GSEG G+IT V + P P+ L P F+
Sbjct 251 NCLTSLRKDNTGYDLKQMFIGSEGTLGVITAVSIVCPPRPKAVNVAFLGCPGFA 304
> hsa:728294 D2HGDH, D2HGD, FLJ42195, MGC25181; D-2-hydroxyglutarate
dehydrogenase (EC:1.1.99.-)
Length=521
Score = 61.6 bits (148), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 50/174 (28%), Positives = 86/174 (49%), Gaps = 5/174 (2%)
Query 8 KHNICLIPFGGGTSVTLGVCTPEEEERMVATVSLSFMQKILYLDRDALLMWVEAGAVGAS 67
+ N+ + P GG T + +G P +E +++T M ++L + ++ +AG V
Sbjct 121 ERNLAVNPQGGNTGM-VGGSVPVFDEIILSTAR---MNRVLSFHSVSGILVCQAGCVLEE 176
Query 68 IEERLRPYGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDLVVDIKVVTTRGCMH 127
+ + + + + +GG VAT A G++ RYG++ V+ ++VV G +
Sbjct 177 LSRYVEERDFIMPLDLGAKGSCHIGGNVATNAGGLRFLRYGSLHGTVLGLEVVLADGTVL 236
Query 128 ACNSSPRI-SSGPSLQQLFLGSEGIYGIITQVLLKIKPIPEYKVYDCLAFPSFS 180
C +S R ++G L+QLF+GSEG GIIT V + P P L P F+
Sbjct 237 DCLTSLRKDNTGYDLKQLFIGSEGTLGIITTVSILCPPKPRAVNVAFLGCPGFA 290
> dre:565889 d2hgdh, zgc:158661; D-2-hydroxyglutarate dehydrogenase
(EC:1.1.99.-)
Length=533
Score = 60.5 bits (145), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 44/152 (28%), Positives = 81/152 (53%), Gaps = 5/152 (3%)
Query 8 KHNICLIPFGGGTSVTLGVCTPEEEERMVATVSLSFMQKILYLDRDALLMWVEAGAVGAS 67
+ N+ + P GG T + +G P +E +++T S M ++ D + ++ +AG V +
Sbjct 132 ERNLAVCPQGGNTGL-VGGSVPVFDEIILST---SLMNQVFAFDNISGILTCQAGCVLEN 187
Query 68 IEERLRPYGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDLVVDIKVVTTRGCMH 127
+ L + + + +GG V+T A G++ RYG++ V+ ++VV G +
Sbjct 188 LSHYLEERDFIMPLDLGAKGSCHIGGNVSTNAGGLRLLRYGSLRGTVLGLEVVLADGHVL 247
Query 128 ACNSSPRI-SSGPSLQQLFLGSEGIYGIITQV 158
C ++ R ++G L+QLF+GSEG G+IT V
Sbjct 248 NCLATLRKDNTGYDLKQLFIGSEGTLGVITAV 279
> cel:F32D8.12 hypothetical protein; K00102 D-lactate dehydrogenase
(cytochrome) [EC:1.1.2.4]
Length=460
Score = 60.1 bits (144), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 36/94 (38%), Positives = 55/94 (58%), Gaps = 4/94 (4%)
Query 89 STVGGWVATRASGMKKNRYGNIEDLVVDIKVVTTRGCM---HACNSSPRISS-GPSLQQL 144
++V G VAT ASG RYG +++ VV+++VV G + PR SS G + +L
Sbjct 134 ASVCGMVATSASGTNAIRYGTMKENVVNLEVVLADGTIIDTKGKGRCPRKSSAGFNFTEL 193
Query 145 FLGSEGIYGIITQVLLKIKPIPEYKVYDCLAFPS 178
F+GSEG GIIT+ +K+ P P++ +FP+
Sbjct 194 FVGSEGTLGIITEATVKVHPRPQFLSAAVCSFPT 227
> ath:AT4G36400 FAD linked oxidase family protein
Length=559
Score = 60.1 bits (144), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 44/155 (28%), Positives = 81/155 (52%), Gaps = 5/155 (3%)
Query 11 ICLIPFGGGTSVTLGVCTPEEEERMVATVSLSFMQKILYLDRDALLMWVEAGAVGASIEE 70
+ ++P GG T + +G P +E V++ M KIL D + ++ EAG + ++
Sbjct 158 LAVVPQGGNTGL-VGGSVPVFDE---VIVNVGLMNKILSFDEVSGVLVCEAGCILENLAT 213
Query 71 RLRPYGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDLVVDIKVVTTRG-CMHAC 129
L G + + + +GG V+T A G++ RYG++ V+ ++ VT G +
Sbjct 214 FLDTKGFIMPLDLGAKGSCHIGGNVSTNAGGLRLIRYGSLHGTVLGLEAVTANGNVLDML 273
Query 130 NSSPRISSGPSLQQLFLGSEGIYGIITQVLLKIKP 164
+ + ++G L+ LF+GSEG GI+T+V + +P
Sbjct 274 GTLRKDNTGYDLKHLFIGSEGSLGIVTKVSILTQP 308
> sce:YDL178W DLD2, AIP2; D-lactate dehydrogenase, located in
the mitochondrial matrix (EC:1.1.2.4)
Length=530
Score = 57.4 bits (137), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 51/180 (28%), Positives = 91/180 (50%), Gaps = 5/180 (2%)
Query 1 VIVDCAIKHNICLIPFGGGTSVTLGVCTPEEEERMVATVSLSFMQKILYLDRDALLMWVE 60
+I++ I ++P GG T + +G P +E ++ SL+ + KI D + ++ +
Sbjct 116 LILNYCNDEKIAVVPQGGNTGL-VGGSVPIFDELIL---SLANLNKIRDFDPVSGILKCD 171
Query 61 AGAVGASIEERLRPYGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDLVVDIKVV 120
AG + + + + + VGG VAT A G++ RYG++ V+ ++VV
Sbjct 172 AGVILENANNYVMEQNYMFPLDLGAKGSCHVGGVVATNAGGLRLLRYGSLHGSVLGLEVV 231
Query 121 TTRG-CMHACNSSPRISSGPSLQQLFLGSEGIYGIITQVLLKIKPIPEYKVYDCLAFPSF 179
G +++ +S + ++G L+QLF+GSEG GIIT V + P P+ L+ SF
Sbjct 232 MPNGQIVNSMHSMRKDNTGYDLKQLFIGSEGTIGIITGVSILTVPKPKAFNVSYLSVESF 291
> sce:YEL071W DLD3; D-lactate dehydrogenase, part of the retrograde
regulon which consists of genes whose expression is stimulated
by damage to mitochondria and reduced in cells grown
with glutamate as the sole nitrogen source, located in the
cytoplasm (EC:1.1.2.4)
Length=496
Score = 54.3 bits (129), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 43/149 (28%), Positives = 73/149 (48%), Gaps = 5/149 (3%)
Query 11 ICLIPFGGGTSVTLGVCTPEEEERMVATVSLSFMQKILYLDRDALLMWVEAGAVGASIEE 70
+ ++P GG T + +G P +E +SL M K+ D + +AG V +
Sbjct 92 LAVVPQGGNTDL-VGASVPVFDE---IVLSLRNMNKVRDFDPVSGTFKCDAGVVMRDAHQ 147
Query 71 RLRPYGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDLVVDIKVVTTRG-CMHAC 129
L + + S VGG V+T A G+ RYG++ V+ ++VV G +
Sbjct 148 FLHDHDHIFPLDLPSRNNCQVGGVVSTNAGGLNFLRYGSLHGNVLGLEVVLPNGEIISNI 207
Query 130 NSSPRISSGPSLQQLFLGSEGIYGIITQV 158
N+ + ++G L+QLF+G+EG G++T V
Sbjct 208 NALRKDNTGYDLKQLFIGAEGTIGVVTGV 236
> eco:b2979 glcD, ECK2974, gox, JW2946, yghM; glycolate oxidase
subunit, FAD-linked; K00104 glycolate oxidase [EC:1.1.3.15]
Length=499
Score = 52.4 bits (124), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 42/150 (28%), Positives = 71/150 (47%), Gaps = 4/150 (2%)
Query 17 GGGTSVTLGVCTPEEEERMVATVSLSFMQKILYLDRDALLMWVEAGAVGASIEERLRPYG 76
G GT ++ G E+ +V ++ ++IL ++ V+ G +I + + P+
Sbjct 86 GAGTGLSGGALPLEKGVLLV----MARFKEILDINPVGRRARVQPGVRNLAISQAVAPHN 141
Query 77 VTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDLVVDIKVVTTRGCMHACNSSPRIS 136
+ +P S ++GG VA A G+ +YG ++ I+V T G S S
Sbjct 142 LYYAPDPSSQIACSIGGNVAENAGGVHCLKYGLTVHNLLKIEVQTLDGEALTLGSDALDS 201
Query 137 SGPSLQQLFLGSEGIYGIITQVLLKIKPIP 166
G L LF GSEG+ G+ T+V +K+ P P
Sbjct 202 PGFDLLALFTGSEGMLGVTTEVTVKLLPKP 231
> cel:F54D5.12 hypothetical protein
Length=487
Score = 52.4 bits (124), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 43/175 (24%), Positives = 82/175 (46%), Gaps = 7/175 (4%)
Query 8 KHNICLIPFGGGTSVTLGVCTPEEEERMVATVSLSFMQKILYLDRDALLMWVEAGAVGAS 67
K+ + ++P GG T + +G P +E +S++ + K D ++ ++G +
Sbjct 88 KNKLAVVPQGGNTGL-VGGSIPVHDE---VVISMNKINKQFSFDDTMGILKCDSGFILED 143
Query 68 IEERLRPYGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDLVVDIKVVTTR---G 124
++ +L G + + + +GG +AT A G++ RYG++ ++ + VV
Sbjct 144 LDNKLAKLGYMMPFDLGAKGSCQIGGNIATCAGGIRLIRYGSLHAHLLGLTVVLPDEHGT 203
Query 125 CMHACNSSPRISSGPSLQQLFLGSEGIYGIITQVLLKIKPIPEYKVYDCLAFPSF 179
+H +S + ++ LFLGSEG G+IT V + P P+ L SF
Sbjct 204 VLHLGSSIRKDNTTLHTPHLFLGSEGQLGVITSVTMTAVPKPKSVQSAMLGIESF 258
> eco:b4468 glcE, ECK2973, gox, JW5487, yghL; glycolate oxidase
FAD binding subunit; K11472 glycolate oxidase FAD binding
subunit
Length=350
Score = 42.7 bits (99), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 35/132 (26%), Positives = 63/132 (47%), Gaps = 2/132 (1%)
Query 38 TVSLSFMQKILYLDRDALLMWVEAGAVGASIEERLRPYGVTLGHEPDSM-EFSTVGGWVA 96
T+ + + I+ D L++ G +IE L G L EP E +T GG VA
Sbjct 44 TLDVRCHRGIVNYDPTELVITARVGTPLVTIEAALESAGQMLPCEPPHYGEEATWGGMVA 103
Query 97 TRASGMKKNRYGNIEDLVVDIKVVTTRG-CMHACNSSPRISSGPSLQQLFLGSEGIYGII 155
+G ++ G++ D V+ +++T G + + +G L +L +GS G G++
Sbjct 104 CGLAGPRRPWSGSVRDFVLGTRIITGAGKHLRFGGEVMKNVAGYDLSRLMVGSYGCLGVL 163
Query 156 TQVLLKIKPIPE 167
T++ +K+ P P
Sbjct 164 TEISMKVLPRPR 175
> eco:b1687 ydiJ, ECK1684, JW1677; predicted FAD-linked oxidoreductase;
K06911
Length=1018
Score = 42.7 bits (99), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 29/111 (26%), Positives = 53/111 (47%), Gaps = 5/111 (4%)
Query 10 NICLIPFGGGTSVTLGVCTPEEEERMVATVSLSFMQKILYLDRDALLMWVEAGAVGASIE 69
++ P GGGT G + ++ +S M +I+ ++ + + VEAG + +
Sbjct 78 SLIFTPRGGGT----GTNGQALNQGIIVDMS-RHMNRIIEINPEEGWVRVEAGVIKDQLN 132
Query 70 ERLRPYGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDLVVDIKVV 120
+ L+P+G E + +T+GG + T ASG YG D V+ ++ V
Sbjct 133 QYLKPFGYFFAPELSTSNRATLGGMINTDASGQGSLVYGKTSDHVLGVRAV 183
> ath:AT4G29740 CKX4; CKX4 (CYTOKININ OXIDASE 4); amine oxidase/
cytokinin dehydrogenase (EC:1.5.99.12); K00279 cytokinin
dehydrogenase [EC:1.5.99.12]
Length=524
Score = 38.1 bits (87), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 37/132 (28%), Positives = 62/132 (46%), Gaps = 12/132 (9%)
Query 49 YLDRDALLMWVEAGAVGASIEERLRPYGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYG 108
Y D A MWV+ + A+++ + P T D + S VGG ++ G + R+G
Sbjct 145 YADVAAGTMWVDV--LKAAVDRGVSPVTWT-----DYLYLS-VGGTLSNAGIGGQTFRHG 196
Query 109 NIEDLVVDIKVVTTRGCMHACNSSPRISSGPSLQQLFLGSEGIYGIITQVLLKIKPIPEY 168
V ++ V+T +G M C SP+++ P L LG G +GIIT+ + + P
Sbjct 197 PQISNVHELDVITGKGEMMTC--SPKLN--PELFYGVLGGLGQFGIITRARIALDHAPTR 252
Query 169 KVYDCLAFPSFS 180
+ + + FS
Sbjct 253 VKWSRILYSDFS 264
> ath:AT5G56970 CKX3; CKX3 (CYTOKININ OXIDASE 3); amine oxidase/
cytokinin dehydrogenase (EC:1.5.99.12); K00279 cytokinin
dehydrogenase [EC:1.5.99.12]
Length=523
Score = 38.1 bits (87), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 35/133 (26%), Positives = 60/133 (45%), Gaps = 12/133 (9%)
Query 48 LYLDRDALLMWVEAGAVGASIEERLRPYGVTLGHEPDSMEFSTVGGWVATRASGMKKNRY 107
LY+D DA +W+E + ++E L P T + TVGG ++ + RY
Sbjct 138 LYVDVDAAWLWIEV--LNKTLELGLTPVSWT------DYLYLTVGGTLSNGGISGQTFRY 189
Query 108 GNIEDLVVDIKVVTTRGCMHACNSSPRISSGPSLQQLFLGSEGIYGIITQVLLKIKPIPE 167
G V+++ V+T +G + C+ L LG G +GIIT+ +K++ P+
Sbjct 190 GPQITNVLEMDVITGKGEIATCSK----DMNSDLFFAVLGGLGQFGIITRARIKLEVAPK 245
Query 168 YKVYDCLAFPSFS 180
+ + FS
Sbjct 246 RAKWLRFLYIDFS 258
> ath:AT2G46760 FAD-binding domain-containing protein
Length=603
Score = 37.0 bits (84), Expect = 0.035, Method: Compositional matrix adjust.
Identities = 37/149 (24%), Positives = 71/149 (47%), Gaps = 9/149 (6%)
Query 21 SVTLGVCTPEEEERMVATVSLSFMQKILYLDRDALLMWVEAGAVGASIEERLRPYGVTLG 80
S+T CT + +++T F+ + D A+ + VE+G + G+ L
Sbjct 103 SITKLACTDGTDGLLIST---KFLNHTVRTDATAMTLTVESGVTLRQLIAEAAKVGLALP 159
Query 81 HEPDSMEFSTVGGWVATRASGMKKNRYGN-IEDLVVDIKVVTTRGCMHACNSSPRISSGP 139
+ P TVGG + T A G G+ + D V +I++V+ G ++ + R+
Sbjct 160 YAPYWWGL-TVGGMMGTGAHGSSLWGKGSAVHDYVTEIRIVSP-GSVNDGFAKVRVLRET 217
Query 140 SLQQLFLGSE---GIYGIITQVLLKIKPI 165
+ + F ++ G+ G+I+QV LK++P+
Sbjct 218 TTPKEFNAAKVSLGVLGVISQVTLKLQPM 246
> ath:AT2G25610 H+-transporting two-sector ATPase, C subunit family
protein; K03661 V-type H+-transporting ATPase 21kDa proteolipid
subunit [EC:3.6.3.14]
Length=178
Score = 34.3 bits (77), Expect = 0.28, Method: Compositional matrix adjust.
Identities = 17/55 (30%), Positives = 31/55 (56%), Gaps = 2/55 (3%)
Query 120 VTTRGCMHACNSSPRISSGPSLQQLFLGSEGIYGIITQVLL--KIKPIPEYKVYD 172
+T + A +PRI+S + +F + IYG+I ++L K++ +P K+YD
Sbjct 45 ITGSSLIGAAIEAPRITSKNLISVIFCEAVAIYGVIVAIILQTKLESVPSSKMYD 99
> ath:AT3G47930 ATGLDH; ATGLDH (L-GALACTONO-1,4-LACTONE DEHYDOROGENASE);
L-gulono-1,4-lactone dehydrogenase/ galactonolactone
dehydrogenase; K00225 L-galactono-1,4-lactone dehydrogenase
[EC:1.3.2.3]
Length=512
Score = 33.9 bits (76), Expect = 0.30, Method: Compositional matrix adjust.
Identities = 32/124 (25%), Positives = 60/124 (48%), Gaps = 8/124 (6%)
Query 39 VSLSFMQKILYLDRDALLMWVEAGAVGASIEERLRPYGVTLGHEPDSMEFSTVGGWVATR 98
V+L+ M K+L +D++ + V+AG + + ++ YG+TL + S+ +GG +
Sbjct 169 VNLALMDKVLEVDKEKKRVTVQAGIRVQQLVDAIKDYGLTLQNFA-SIREQQIGGIIQVG 227
Query 99 ASGMKKNRYGNIEDLVVDIKVVT-TRGCMHACNSSPRISSGPSLQQLFLGSEGIYGIITQ 157
A G R I++ V+ +K+VT +G + P L L G G++ +
Sbjct 228 AHGTGA-RLPPIDEQVISMKLVTPAKGTIELSR-----EKDPELFHLARCGLGGLGVVAE 281
Query 158 VLLK 161
V L+
Sbjct 282 VTLQ 285
> ath:AT4G32530 vacuolar ATP synthase, putative / V-ATPase, putative;
K03661 V-type H+-transporting ATPase 21kDa proteolipid
subunit [EC:3.6.3.14]
Length=210
Score = 33.9 bits (76), Expect = 0.37, Method: Compositional matrix adjust.
Identities = 17/55 (30%), Positives = 31/55 (56%), Gaps = 2/55 (3%)
Query 120 VTTRGCMHACNSSPRISSGPSLQQLFLGSEGIYGIITQVLL--KIKPIPEYKVYD 172
+T + A +PRI+S + +F + IYG+I ++L K++ +P K+YD
Sbjct 47 ITGSSLIGAAIEAPRITSKNLISVIFCEAVAIYGVIVAIILQTKLESVPSSKMYD 101
> cel:C13B4.1 hypothetical protein
Length=1010
Score = 32.7 bits (73), Expect = 0.65, Method: Compositional matrix adjust.
Identities = 28/91 (30%), Positives = 40/91 (43%), Gaps = 7/91 (7%)
Query 63 AVGASIEERLRPYGV----TLGHEPDSMEFSTVGGWVATRASG---MKKNRYGNIEDLVV 115
A+G S + ++ P V +LG +P SM+FST G+ R SG ++ N E V
Sbjct 618 ALGFSNDGKMNPANVIECSSLGSQPLSMKFSTNSGYSNDRISGEEAIRSQYITNTETSYV 677
Query 116 DIKVVTTRGCMHACNSSPRISSGPSLQQLFL 146
D K+ NS+ I QQ L
Sbjct 678 DGKIYCKGTVRSDGNSNAAIFKYTPKQQYHL 708
> ath:AT2G19500 CKX2; CKX2 (CYTOKININ OXIDASE 2); amine oxidase/
cytokinin dehydrogenase (EC:1.5.99.12); K00279 cytokinin
dehydrogenase [EC:1.5.99.12]
Length=501
Score = 32.3 bits (72), Expect = 0.98, Method: Compositional matrix adjust.
Identities = 30/122 (24%), Positives = 55/122 (45%), Gaps = 5/122 (4%)
Query 59 VEAGAVGASIEERLRPYGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDLVVDIK 118
V AG + + ++ GV+ D + TVGG ++ G + R G + V+++
Sbjct 125 VAAGTLWVDVLKKTAEKGVSPVSWTDYLHI-TVGGTLSNGGIGGQVFRNGPLVSNVLELD 183
Query 119 VVTTRGCMHACNSSPRISSGPSLQQLFLGSEGIYGIITQVLLKIKPIPEYKVYDCLAFPS 178
V+T +G M C+ P L LG G +GIIT+ + + P+ + + +
Sbjct 184 VITGKGEMLTCSR----QLNPELFYGVLGGLGQFGIITRARIVLDHAPKRAKWFRMLYSD 239
Query 179 FS 180
F+
Sbjct 240 FT 241
> ath:AT5G44390 FAD-binding domain-containing protein
Length=542
Score = 32.0 bits (71), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 37/126 (29%), Positives = 54/126 (42%), Gaps = 10/126 (7%)
Query 5 CAIKHNICLIPFGGGTSVTLGVCTPEEEERMVATVSLSFMQKILYLDRDALLMWVEAGA- 63
C+ K I GG GV + E+ + LS +++I +D WVEAGA
Sbjct 104 CSKKLEIHFRVRSGGHDYE-GVSYVSQIEKPFVLIDLSKLRQI-NVDIKDTSAWVEAGAT 161
Query 64 VGA---SIEERLRPYGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDLVVDIKVV 120
VG I E+ + +G G P +GG + A G +YG D V+D K+V
Sbjct 162 VGELYYRIAEKSKFHGFPAGVYPSL----GIGGHITGGAYGSLMRKYGLAADNVLDAKIV 217
Query 121 TTRGCM 126
G +
Sbjct 218 DANGKL 223
> ath:AT5G21482 CKX7; CKX7 (CYTOKININ OXIDASE 7); cytokinin dehydrogenase/
oxidoreductase (EC:1.5.99.12); K00279 cytokinin
dehydrogenase [EC:1.5.99.12]
Length=524
Score = 31.6 bits (70), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 24/81 (29%), Positives = 38/81 (46%), Gaps = 10/81 (12%)
Query 90 TVGGWVATRASGMKKNRYGNIEDLVVDIKVVTTRGCMHACNSSPRISSGPSLQQLF---L 146
TVGG ++ + RYG V ++ VVT G + C+ +LF L
Sbjct 167 TVGGTLSNAGVSGQAFRYGPQTSNVTELDVVTGNGDVVTCSEIEN-------SELFFSVL 219
Query 147 GSEGIYGIITQVLLKIKPIPE 167
G G +GIIT+ + ++P P+
Sbjct 220 GGLGQFGIITRARVLLQPAPD 240
> ath:AT1G75450 CKX5; CKX5 (CYTOKININ OXIDASE 5); cytokinin dehydrogenase
(EC:1.5.99.12); K00279 cytokinin dehydrogenase [EC:1.5.99.12]
Length=540
Score = 31.2 bits (69), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 31/134 (23%), Positives = 59/134 (44%), Gaps = 12/134 (8%)
Query 48 LYLDRDALLMWVEAGAVGASIEERLRPYGVTLGHEPDSMEFSTVGGWVATRASGMKKNRY 107
+Y+D +WV+ + ++E L P T + TVGG ++ + +
Sbjct 136 MYVDVWGGELWVDV--LKKTLEHGLAPKSWT------DYLYLTVGGTLSNAGISGQAFHH 187
Query 108 GNIEDLVVDIKVVTTRGCMHACNSSPRISSGPSLQQLFLGSEGIYGIITQVLLKIKPIPE 167
G V+++ VVT +G + C+ L LG G +GIIT+ + ++P P+
Sbjct 188 GPQISNVLELDVVTGKGEVMRCSEE----ENTRLFHGVLGGLGQFGIITRARISLEPAPQ 243
Query 168 YKVYDCLAFPSFSL 181
+ + + SF +
Sbjct 244 RVRWIRVLYSSFKV 257
> ath:AT2G34790 MEE23; MEE23 (MATERNAL EFFECT EMBRYO ARREST 23);
FAD binding / catalytic/ electron carrier/ oxidoreductase
Length=532
Score = 31.2 bits (69), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 31/120 (25%), Positives = 50/120 (41%), Gaps = 2/120 (1%)
Query 5 CAIKHNICLIPFGGGTSVTLGVCTPEEEERMVATVSLSFMQKILYLDRDALLMWVEAGAV 64
CA K + L GG G+ E+E V LS ++++ +D D+ W AGA
Sbjct 100 CAKKLQLHLRLRSGGHDYE-GLSFVAEDETPFVIVDLSKLRQV-DVDLDSNSAWAHAGAT 157
Query 65 GASIEERLRPYGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDLVVDIKVVTTRG 124
+ R++ T G +GG + A G ++G D V+D ++V G
Sbjct 158 IGEVYYRIQEKSQTHGFPAGLCSSLGIGGHLVGGAYGSMMRKFGLGADNVLDARIVDANG 217
> dre:565964 MGC123280, sdr42e1, si:ch211-79l17.4; zgc:123280
(EC:1.1.1.-)
Length=402
Score = 30.8 bits (68), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 18/62 (29%), Positives = 27/62 (43%), Gaps = 1/62 (1%)
Query 55 LLMWVEAGAVGASIEERLRPYGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDLV 114
LL E G + +R LG+EP + V W R G K++R +I L+
Sbjct 322 LLTRTEVYKTGVTHYFSMRKAQEELGYEPKLYDLEDVVQWFQARGHGKKRSR-SSIRKLI 380
Query 115 VD 116
+D
Sbjct 381 LD 382
> ath:AT2G41510 CKX1; CKX1 (CYTOKININ OXIDASE/DEHYDROGENASE 1);
cytokinin dehydrogenase (EC:1.5.99.12); K00279 cytokinin dehydrogenase
[EC:1.5.99.12]
Length=575
Score = 30.8 bits (68), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 32/125 (25%), Positives = 57/125 (45%), Gaps = 11/125 (8%)
Query 59 VEAGAVGASIEERLRPYGVTLGHEPDSMEFSTVGGWVATRASGMKKNRYGNIEDLVVDIK 118
V G + +I YG++ D + TVGG ++ + ++G + V ++
Sbjct 161 VSGGEIWINILRETLKYGLSPKSWTDYLHL-TVGGTLSNAGISGQAFKHGPQINNVYQLE 219
Query 119 VVTTRGCMHACNSSPRISSGPSLQQLF---LGSEGIYGIITQVLLKIKPIPEYKVYDCLA 175
+VT +G + C S R S +LF LG G +GIIT+ + ++P P + +
Sbjct 220 IVTGKGEVVTC-SEKRNS------ELFFSVLGGLGQFGIITRARISLEPAPHMVKWIRVL 272
Query 176 FPSFS 180
+ FS
Sbjct 273 YSDFS 277
> pfa:MAL13P1.271 V-type ATPase, putative; K03661 V-type H+-transporting
ATPase 21kDa proteolipid subunit [EC:3.6.3.14]
Length=181
Score = 30.4 bits (67), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 15/32 (46%), Positives = 21/32 (65%), Gaps = 0/32 (0%)
Query 132 SPRISSGPSLQQLFLGSEGIYGIITQVLLKIK 163
SPRI S + +F + G+YG+IT V L+IK
Sbjct 51 SPRIISKNLISIIFCEALGMYGVITAVFLQIK 82
> dre:561775 specc1la, cytsaa, im:7159316, specc1l; sperm antigen
with calponin homology and coiled-coil domains 1-like a
Length=1132
Score = 30.0 bits (66), Expect = 5.0, Method: Composition-based stats.
Identities = 21/56 (37%), Positives = 31/56 (55%), Gaps = 4/56 (7%)
Query 63 AVGASIEERLRPYGVTLGHEPDSMEFSTVG----GWVATRASGMKKNRYGNIEDLV 114
A+G S+E+RL T G S E S+ G G AT AS ++ + G++EDL+
Sbjct 261 ALGFSLEQRLDGADKTFGFPSTSPELSSGGAGAHGDCATVASSVEGSAPGSMEDLL 316
> ath:AT3G45090 2-phosphoglycerate kinase-related; K05715 2-phosphoglycerate
kinase [EC:2.7.2.-]
Length=717
Score = 29.6 bits (65), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 21/63 (33%), Positives = 32/63 (50%), Gaps = 7/63 (11%)
Query 82 EPDSMEFSTVG--GWVATRASGMKKNRYGNIEDLVVDIKVVTTRGCMHACNSSPRISSGP 139
EP +++F G W + A+ G+++DL DI +R +C SSPR S GP
Sbjct 492 EPVNLQFGHFGISAWPSDGATS----HAGSVDDLRADIIETGSRH-YSSCCSSPRTSDGP 546
Query 140 SLQ 142
S +
Sbjct 547 SKE 549
> ath:AT3G63440 CKX6; CKX6 (CYTOKININ OXIDASE/DEHYDROGENASE 6);
cytokinin dehydrogenase (EC:1.5.99.12)
Length=533
Score = 29.3 bits (64), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 29/121 (23%), Positives = 51/121 (42%), Gaps = 5/121 (4%)
Query 46 KILYLDRDALLMWVEAGAVGASIEERLRPYGVTLGHEPDSMEFSTVGGWVATRASGMKKN 105
++ +D A + V G + +I YG+ D + TVGG ++ +
Sbjct 134 QVYSVDSPAPYVDVSGGELWINILHETLKYGLAPKSWTDYLHL-TVGGTLSNAGISGQAF 192
Query 106 RYGNIEDLVVDIKVVTTRGCMHACNSSPRISSGPSLQQLFLGSEGIYGIITQVLLKIKPI 165
R+G V +++VT +G + C L LG G +GIIT+ + ++P
Sbjct 193 RHGPQISNVHQLEIVTGKGEILNCTKR----QNSDLFNGVLGGLGQFGIITRARIALEPA 248
Query 166 P 166
P
Sbjct 249 P 249
Lambda K H
0.322 0.139 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5170784960
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40