bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0700_orf1
Length=143
Score E
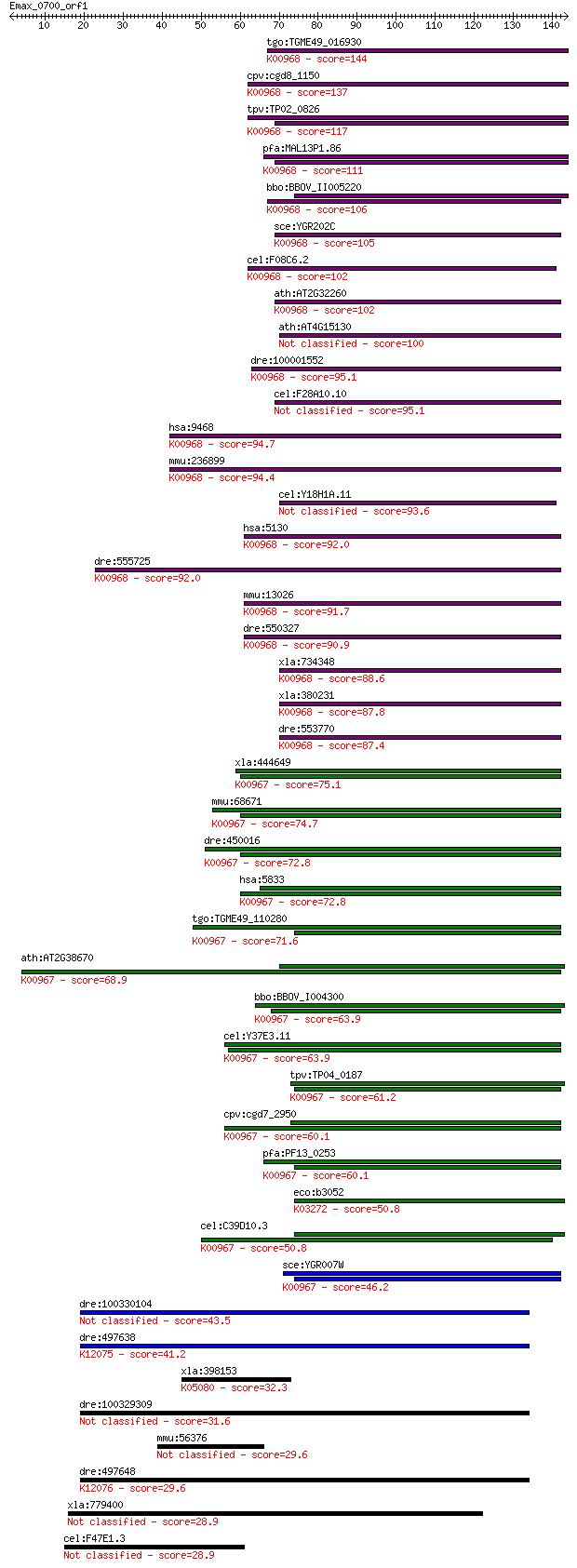
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_016930 cholinephosphate cytidylyltransferase, putat... 144 7e-35
cpv:cgd8_1150 choline-phosphate cytidylyltransferase ; K00968 ... 137 1e-32
tpv:TP02_0826 cholinephosphate cytidylyltransferase; K00968 ch... 117 1e-26
pfa:MAL13P1.86 ctp; cholinephosphate cytidylyltransferase (EC:... 111 7e-25
bbo:BBOV_II005220 18.m06431; choline-phosphate cytidylyltransf... 106 2e-23
sce:YGR202C PCT1, BSR2, CCT1; Cholinephosphate cytidylyltransf... 105 6e-23
cel:F08C6.2 hypothetical protein; K00968 choline-phosphate cyt... 102 4e-22
ath:AT2G32260 CCT1; cholinephosphate cytidylyltransferase, put... 102 5e-22
ath:AT4G15130 CCT2; catalytic/ choline-phosphate cytidylyltran... 100 1e-21
dre:100001552 pcyt1ba, pcyt1b; phosphate cytidylyltransferase ... 95.1 7e-20
cel:F28A10.10 hypothetical protein 95.1 7e-20
hsa:9468 PCYT1B, CCTB, CTB; phosphate cytidylyltransferase 1, ... 94.7 9e-20
mmu:236899 Pcyt1b, AW045697, CTTbeta; phosphate cytidylyltrans... 94.4 1e-19
cel:Y18H1A.11 hypothetical protein 93.6 2e-19
hsa:5130 PCYT1A, CCTA, CT, CTA, CTPCT, PCYT1; phosphate cytidy... 92.0 5e-19
dre:555725 pcyt1bb, MGC123291, zgc:123291; phosphate cytidylyl... 92.0 6e-19
mmu:13026 Pcyt1a, CTalpha, Cctalpha, Ctpct, Cttalpha; phosphat... 91.7 7e-19
dre:550327 pcyt1aa, zgc:110237; phosphate cytidylyltransferase... 90.9 1e-18
xla:734348 pcyt1a, MGC97881, ccta, cta, ctpct, pcyt1; phosphat... 88.6 6e-18
xla:380231 pcyt1b, MGC53725, pcyt1a; phosphate cytidylyltransf... 87.8 9e-18
dre:553770 pcyt1ab, MGC110012, zgc:110012; phosphate cytidylyl... 87.4 1e-17
xla:444649 pcyt2, MGC84177; phosphate cytidylyltransferase 2, ... 75.1 7e-14
mmu:68671 Pcyt2, 1110033E03Rik, ET; phosphate cytidylyltransfe... 74.7 1e-13
dre:450016 pcyt2, im:7158585, wu:fb39h11, zgc:103434; phosphat... 72.8 3e-13
hsa:5833 PCYT2, ET; phosphate cytidylyltransferase 2, ethanola... 72.8 3e-13
tgo:TGME49_110280 phosphoethanolamine cytidylyltransferase, pu... 71.6 8e-13
ath:AT2G38670 PECT1; PECT1 (PHOSPHORYLETHANOLAMINE CYTIDYLYLTR... 68.9 5e-12
bbo:BBOV_I004300 19.m02274; ethanolamine-phosphate cytidylyltr... 63.9 1e-10
cel:Y37E3.11 hypothetical protein; K00967 ethanolamine-phospha... 63.9 2e-10
tpv:TP04_0187 ethanolamine-phosphate cytidylyltransferase (EC:... 61.2 1e-09
cpv:cgd7_2950 phospholipid cytidyltransferase HIGH family ; K0... 60.1 2e-09
pfa:PF13_0253 ethanolamine-phosphate cytidylyltransferase, put... 60.1 2e-09
eco:b3052 rfaE, ECK3042, gmhC, hldE, JW3024, waaE, yqiF; fused... 50.8 1e-06
cel:C39D10.3 hypothetical protein; K00967 ethanolamine-phospha... 50.8 1e-06
sce:YGR007W MUQ1, ECT1; Choline phosphate cytidylyltransferase... 46.2 4e-05
dre:100330104 disks large homolog 2-like 43.5 2e-04
dre:497638 dlg2; discs, large (Drosophila) homolog 2; K12075 d... 41.2 0.001
xla:398153 ghr; growth hormone receptor; K05080 growth hormone... 32.3 0.47
dre:100329309 discs, large homolog 2 (Drosophila)-like 31.6 0.82
mmu:56376 Pdlim5, 1110001A05Rik, AI987914, C87059, Enh, Enh1, ... 29.6 3.3
dre:497648 dlg1l; discs, large (Drosophila) homolog 1, like; K... 29.6 3.7
xla:779400 nif3l1, MGC154449; NIF3 NGG1 interacting factor 3-l... 28.9 5.8
cel:F47E1.3 hypothetical protein 28.9 6.3
> tgo:TGME49_016930 cholinephosphate cytidylyltransferase, putative
(EC:2.7.7.15); K00968 choline-phosphate cytidylyltransferase
[EC:2.7.7.15]
Length=329
Score = 144 bits (364), Expect = 7e-35, Method: Compositional matrix adjust.
Identities = 66/80 (82%), Positives = 75/80 (93%), Gaps = 3/80 (3%)
Query 67 DPN---RPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQS 123
DPN RP+R+Y DGVYDLLHLGHMRQLEQAKK+FK+V+L+AGVASDE+THRLKGQTVQ+
Sbjct 43 DPNEWGRPIRVYADGVYDLLHLGHMRQLEQAKKLFKNVHLIAGVASDEDTHRLKGQTVQT 102
Query 124 LEERAETLRHIKWVDEVIAP 143
+ ERAETLRHIKWVDEVIAP
Sbjct 103 MTERAETLRHIKWVDEVIAP 122
> cpv:cgd8_1150 choline-phosphate cytidylyltransferase ; K00968
choline-phosphate cytidylyltransferase [EC:2.7.7.15]
Length=341
Score = 137 bits (346), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 62/88 (70%), Positives = 75/88 (85%), Gaps = 6/88 (6%)
Query 62 LAEGE------DPNRPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHR 115
LA+G+ D NR +RIY DGVYDLLHLGHMRQLEQAKKM+ + +L+ GVASDEETHR
Sbjct 52 LADGQENNKECDTNRKIRIYADGVYDLLHLGHMRQLEQAKKMYPNTHLIVGVASDEETHR 111
Query 116 LKGQTVQSLEERAETLRHIKWVDEVIAP 143
LKG+TVQ+L+ER ETLRH+KWVDE+I+P
Sbjct 112 LKGRTVQTLQERTETLRHVKWVDEIISP 139
> tpv:TP02_0826 cholinephosphate cytidylyltransferase; K00968
choline-phosphate cytidylyltransferase [EC:2.7.7.15]
Length=523
Score = 117 bits (293), Expect = 1e-26, Method: Composition-based stats.
Identities = 55/82 (67%), Positives = 65/82 (79%), Gaps = 1/82 (1%)
Query 62 LAEGEDPNRPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTV 121
+AE +D R RIY+DGV+D+LHLG MR LEQAKKMFK+VYL+AGV D+ET R K V
Sbjct 1 MAENDD-GRVYRIYSDGVFDMLHLGQMRHLEQAKKMFKNVYLIAGVTEDDETVRYKSHIV 59
Query 122 QSLEERAETLRHIKWVDEVIAP 143
++ ERAE LRHIKWVDEVIAP
Sbjct 60 NTMVERAEMLRHIKWVDEVIAP 81
Score = 89.4 bits (220), Expect = 3e-18, Method: Composition-based stats.
Identities = 41/75 (54%), Positives = 51/75 (68%), Gaps = 0/75 (0%)
Query 69 NRPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQSLEERA 128
+ V +YT GV+DLLH GH R E KKMF V L+ GV SDE+T KG+ +Q L RA
Sbjct 280 DEEVVVYTYGVFDLLHYGHARHFEYVKKMFARVKLIVGVLSDEDTVTCKGRLIQPLHIRA 339
Query 129 ETLRHIKWVDEVIAP 143
TL HIKWVDE+++P
Sbjct 340 ATLEHIKWVDEIVSP 354
> pfa:MAL13P1.86 ctp; cholinephosphate cytidylyltransferase (EC:2.7.7.15);
K00968 choline-phosphate cytidylyltransferase [EC:2.7.7.15]
Length=896
Score = 111 bits (278), Expect = 7e-25, Method: Composition-based stats.
Identities = 49/78 (62%), Positives = 63/78 (80%), Gaps = 0/78 (0%)
Query 66 EDPNRPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQSLE 125
++ + +RIY DGVYDLLHLGHM+QLEQAK + K+V L+ GV D ET + KGQ VQ+LE
Sbjct 27 DNEEKSIRIYADGVYDLLHLGHMKQLEQAKHVDKNVTLIVGVTGDNETRKFKGQIVQTLE 86
Query 126 ERAETLRHIKWVDEVIAP 143
ER ETL+HI+WVDE+I+P
Sbjct 87 ERTETLKHIRWVDEIISP 104
Score = 110 bits (276), Expect = 1e-24, Method: Composition-based stats.
Identities = 49/75 (65%), Positives = 62/75 (82%), Gaps = 0/75 (0%)
Query 69 NRPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQSLEERA 128
++ V IY DGVYD+LHLGHM+QLEQAKK+F++ L+ GV SD ET KGQ VQ+LEER
Sbjct 615 SKNVVIYADGVYDMLHLGHMKQLEQAKKLFENTTLIVGVTSDNETKLFKGQVVQTLEERT 674
Query 129 ETLRHIKWVDEVIAP 143
ETL+HI+WVDE+I+P
Sbjct 675 ETLKHIRWVDEIISP 689
> bbo:BBOV_II005220 18.m06431; choline-phosphate cytidylyltransferase
(EC:2.7.7.15); K00968 choline-phosphate cytidylyltransferase
[EC:2.7.7.15]
Length=546
Score = 106 bits (265), Expect = 2e-23, Method: Composition-based stats.
Identities = 48/70 (68%), Positives = 57/70 (81%), Gaps = 0/70 (0%)
Query 74 IYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQSLEERAETLRH 133
+Y+DGV+D+ HLGHMRQLEQAKKMF L GV DEET +LKGQTV ++ ERAE LRH
Sbjct 9 VYSDGVFDMPHLGHMRQLEQAKKMFPKCILKVGVTDDEETLQLKGQTVNTMAERAEFLRH 68
Query 134 IKWVDEVIAP 143
++WVDEVIAP
Sbjct 69 VRWVDEVIAP 78
Score = 79.3 bits (194), Expect = 3e-15, Method: Composition-based stats.
Identities = 35/75 (46%), Positives = 49/75 (65%), Gaps = 0/75 (0%)
Query 67 DPNRPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQSLEE 126
D + +YT GV+DLLH GH R EQ KK F V+L+ GV SD+ +LKG+ +Q +
Sbjct 315 DTREDIFVYTAGVFDLLHYGHARHFEQIKKCFNKVHLIVGVVSDDVALKLKGRVMQPCRD 374
Query 127 RAETLRHIKWVDEVI 141
RA L HI+WVD+++
Sbjct 375 RAAALYHIRWVDDIL 389
> sce:YGR202C PCT1, BSR2, CCT1; Cholinephosphate cytidylyltransferase,
also known as CTP:phosphocholine cytidylyltransferase,
rate-determining enzyme of the CDP-choline pathway for phosphatidylcholine
synthesis, inhibited by Sec14p, activated
upon lipid-binding (EC:2.7.7.15); K00968 choline-phosphate
cytidylyltransferase [EC:2.7.7.15]
Length=424
Score = 105 bits (261), Expect = 6e-23, Method: Compositional matrix adjust.
Identities = 45/73 (61%), Positives = 58/73 (79%), Gaps = 0/73 (0%)
Query 69 NRPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQSLEERA 128
+RP+RIY DGV+DL HLGHM+QLEQ KK F +V L+ GV SD+ TH+LKG TV + ++R
Sbjct 101 DRPIRIYADGVFDLFHLGHMKQLEQCKKAFPNVTLIVGVPSDKITHKLKGLTVLTDKQRC 160
Query 129 ETLRHIKWVDEVI 141
ETL H +WVDEV+
Sbjct 161 ETLTHCRWVDEVV 173
> cel:F08C6.2 hypothetical protein; K00968 choline-phosphate cytidylyltransferase
[EC:2.7.7.15]
Length=362
Score = 102 bits (254), Expect = 4e-22, Method: Compositional matrix adjust.
Identities = 45/79 (56%), Positives = 57/79 (72%), Gaps = 0/79 (0%)
Query 62 LAEGEDPNRPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTV 121
+AE + RPVRIY DG+YDL H GH QL Q KKMF +VYL+ GV D +TH+ KG+TV
Sbjct 71 MAEANEAGRPVRIYADGIYDLFHHGHANQLRQVKKMFPNVYLIVGVCGDRDTHKYKGRTV 130
Query 122 QSLEERAETLRHIKWVDEV 140
S EER + +RH ++VDEV
Sbjct 131 TSEEERYDGVRHCRYVDEV 149
> ath:AT2G32260 CCT1; cholinephosphate cytidylyltransferase, putative
/ phosphorylcholine transferase, putative / CTP:phosphocholine
cytidylyltransferase, putative (EC:2.7.7.15); K00968
choline-phosphate cytidylyltransferase [EC:2.7.7.15]
Length=332
Score = 102 bits (253), Expect = 5e-22, Method: Compositional matrix adjust.
Identities = 45/74 (60%), Positives = 55/74 (74%), Gaps = 1/74 (1%)
Query 69 NRPVRIYTDGVYDLLHLGHMRQLEQAKKMF-KHVYLMAGVASDEETHRLKGQTVQSLEER 127
+RPVR+Y DG+YDL H GH R LEQAK F + YL+ G +DE TH+ KG+TV + EER
Sbjct 32 DRPVRVYADGIYDLFHFGHARSLEQAKLAFPNNTYLLVGCCNDETTHKYKGRTVMTAEER 91
Query 128 AETLRHIKWVDEVI 141
E+LRH KWVDEVI
Sbjct 92 YESLRHCKWVDEVI 105
> ath:AT4G15130 CCT2; catalytic/ choline-phosphate cytidylyltransferase/
nucleotidyltransferase (EC:2.7.7.15)
Length=304
Score = 100 bits (250), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 42/72 (58%), Positives = 54/72 (75%), Gaps = 0/72 (0%)
Query 70 RPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQSLEERAE 129
RPVR+Y DG++DL H GH R +EQAKK F + YL+ G +DE T++ KG+TV + ER E
Sbjct 19 RPVRVYADGIFDLFHFGHARAIEQAKKSFPNTYLLVGCCNDEITNKFKGKTVMTESERYE 78
Query 130 TLRHIKWVDEVI 141
+LRH KWVDEVI
Sbjct 79 SLRHCKWVDEVI 90
> dre:100001552 pcyt1ba, pcyt1b; phosphate cytidylyltransferase
1, choline, beta a (EC:2.7.7.15); K00968 choline-phosphate
cytidylyltransferase [EC:2.7.7.15]
Length=343
Score = 95.1 bits (235), Expect = 7e-20, Method: Compositional matrix adjust.
Identities = 42/79 (53%), Positives = 56/79 (70%), Gaps = 0/79 (0%)
Query 63 AEGEDPNRPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQ 122
+ G +RPVR+Y DG++DL H GH R L QAK +F + YL+ GV SDE TH+ KG TV
Sbjct 42 SHGTPVDRPVRVYADGIFDLFHSGHARALMQAKNLFPNTYLIVGVCSDELTHKYKGFTVM 101
Query 123 SLEERAETLRHIKWVDEVI 141
+ +ER E LRH ++VDEV+
Sbjct 102 TEDERYEALRHCRYVDEVL 120
> cel:F28A10.10 hypothetical protein
Length=183
Score = 95.1 bits (235), Expect = 7e-20, Method: Compositional matrix adjust.
Identities = 40/73 (54%), Positives = 54/73 (73%), Gaps = 0/73 (0%)
Query 69 NRPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQSLEERA 128
+RP+R+Y DG+YD+ H GH +QL Q K+MF VYL+ GV SDE T + KG TVQS ER
Sbjct 39 SRPIRVYADGIYDMFHYGHAKQLLQIKQMFPMVYLIVGVCSDENTLKFKGPTVQSENERY 98
Query 129 ETLRHIKWVDEVI 141
E++R ++VDEV+
Sbjct 99 ESVRQCRYVDEVL 111
> hsa:9468 PCYT1B, CCTB, CTB; phosphate cytidylyltransferase 1,
choline, beta (EC:2.7.7.15); K00968 choline-phosphate cytidylyltransferase
[EC:2.7.7.15]
Length=351
Score = 94.7 bits (234), Expect = 9e-20, Method: Compositional matrix adjust.
Identities = 52/109 (47%), Positives = 67/109 (61%), Gaps = 9/109 (8%)
Query 42 RKATEDPSPQPD---SQGTAPVE---LAE---GEDPNRPVRIYTDGVYDLLHLGHMRQLE 92
RK P+P D Q AP E +A+ G +RPVR+Y DG++DL H GH R L
Sbjct 20 RKTLTAPAPFADETNCQCQAPHEKLTIAQARLGTPADRPVRVYADGIFDLFHSGHARALM 79
Query 93 QAKKMFKHVYLMAGVASDEETHRLKGQTVQSLEERAETLRHIKWVDEVI 141
QAK +F + YL+ GV SD+ TH+ KG TV + ER E LRH ++VDEVI
Sbjct 80 QAKTLFPNSYLLVGVCSDDLTHKFKGFTVMNEAERYEALRHCRYVDEVI 128
> mmu:236899 Pcyt1b, AW045697, CTTbeta; phosphate cytidylyltransferase
1, choline, beta isoform (EC:2.7.7.15); K00968 choline-phosphate
cytidylyltransferase [EC:2.7.7.15]
Length=339
Score = 94.4 bits (233), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 52/109 (47%), Positives = 67/109 (61%), Gaps = 9/109 (8%)
Query 42 RKATEDPSPQPD---SQGTAPVE---LAE---GEDPNRPVRIYTDGVYDLLHLGHMRQLE 92
RK P+P D Q AP E +A+ G +RPVR+Y DG++DL H GH R L
Sbjct 8 RKTLTAPAPFADESSCQCQAPHEKLTVAQARLGTPVDRPVRVYADGIFDLFHSGHARALM 67
Query 93 QAKKMFKHVYLMAGVASDEETHRLKGQTVQSLEERAETLRHIKWVDEVI 141
QAK +F + YL+ GV SD+ TH+ KG TV + ER E LRH ++VDEVI
Sbjct 68 QAKTLFPNSYLLVGVCSDDLTHKFKGFTVMNEAERYEALRHCRYVDEVI 116
> cel:Y18H1A.11 hypothetical protein
Length=272
Score = 93.6 bits (231), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 41/71 (57%), Positives = 51/71 (71%), Gaps = 0/71 (0%)
Query 70 RPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQSLEERAE 129
RPVR+Y DGVYD+ H GH Q Q K+ +VYL+ GV SDEET + KG+TVQ EER E
Sbjct 45 RPVRVYADGVYDMFHYGHANQFLQIKQTLPNVYLIVGVCSDEETMKNKGRTVQGEEERYE 104
Query 130 TLRHIKWVDEV 140
+RH ++VDEV
Sbjct 105 AIRHCRYVDEV 115
> hsa:5130 PCYT1A, CCTA, CT, CTA, CTPCT, PCYT1; phosphate cytidylyltransferase
1, choline, alpha (EC:2.7.7.15); K00968 choline-phosphate
cytidylyltransferase [EC:2.7.7.15]
Length=367
Score = 92.0 bits (227), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 40/81 (49%), Positives = 54/81 (66%), Gaps = 0/81 (0%)
Query 61 ELAEGEDPNRPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQT 120
E + G RPVR+Y DG++DL H GH R L QAK +F + YL+ GV SDE TH KG T
Sbjct 66 EASRGTPCERPVRVYADGIFDLFHSGHARALMQAKNLFPNTYLIVGVCSDELTHNFKGFT 125
Query 121 VQSLEERAETLRHIKWVDEVI 141
V + ER + ++H ++VDEV+
Sbjct 126 VMNENERYDAVQHCRYVDEVV 146
> dre:555725 pcyt1bb, MGC123291, zgc:123291; phosphate cytidylyltransferase
1, choline, beta b; K00968 choline-phosphate cytidylyltransferase
[EC:2.7.7.15]
Length=299
Score = 92.0 bits (227), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 53/133 (39%), Positives = 74/133 (55%), Gaps = 14/133 (10%)
Query 23 SRATGNSARPALLKTSMKKRK-----ATEDPSPQPDSQGTAP------VELAE---GEDP 68
SR G +AR + ++RK A +P+ + G+A V LA+ G
Sbjct 9 SRGNGRTARGGVQAQQQQQRKRPPIKALREPAIFANRSGSASDTPQEKVTLAQARRGTPA 68
Query 69 NRPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQSLEERA 128
RPVR+Y DG++DL H GH R L QAK +F + L+ GV SD TH+ KG TV + +ER
Sbjct 69 QRPVRVYADGIFDLFHSGHARALMQAKNLFPNTQLIVGVCSDALTHKYKGYTVMTEDERY 128
Query 129 ETLRHIKWVDEVI 141
E L H ++VDEV+
Sbjct 129 EALIHCRYVDEVV 141
> mmu:13026 Pcyt1a, CTalpha, Cctalpha, Ctpct, Cttalpha; phosphate
cytidylyltransferase 1, choline, alpha isoform (EC:2.7.7.15);
K00968 choline-phosphate cytidylyltransferase [EC:2.7.7.15]
Length=367
Score = 91.7 bits (226), Expect = 7e-19, Method: Compositional matrix adjust.
Identities = 40/81 (49%), Positives = 53/81 (65%), Gaps = 0/81 (0%)
Query 61 ELAEGEDPNRPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQT 120
E G RPVR+Y DG++DL H GH R L QAK +F + YL+ GV SDE TH KG T
Sbjct 66 EACRGTPCERPVRVYADGIFDLFHSGHARALMQAKNLFPNTYLIVGVCSDELTHNFKGFT 125
Query 121 VQSLEERAETLRHIKWVDEVI 141
V + ER + ++H ++VDEV+
Sbjct 126 VMNENERYDAVQHCRYVDEVV 146
> dre:550327 pcyt1aa, zgc:110237; phosphate cytidylyltransferase
1, choline, alpha a (EC:2.7.7.15); K00968 choline-phosphate
cytidylyltransferase [EC:2.7.7.15]
Length=374
Score = 90.9 bits (224), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 39/81 (48%), Positives = 57/81 (70%), Gaps = 0/81 (0%)
Query 61 ELAEGEDPNRPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQT 120
E G P+RPVR+Y DG++D+ H GH R L QAK +F + +L+ GV SD+ TH+LKG T
Sbjct 63 EARRGTPPDRPVRVYADGIFDMFHSGHARALMQAKCLFPNTHLIVGVCSDDLTHKLKGFT 122
Query 121 VQSLEERAETLRHIKWVDEVI 141
V + +ER + + H ++VDEV+
Sbjct 123 VMNEDERYDAVSHCRYVDEVV 143
> xla:734348 pcyt1a, MGC97881, ccta, cta, ctpct, pcyt1; phosphate
cytidylyltransferase 1, choline, alpha (EC:2.7.7.15); K00968
choline-phosphate cytidylyltransferase [EC:2.7.7.15]
Length=367
Score = 88.6 bits (218), Expect = 6e-18, Method: Compositional matrix adjust.
Identities = 38/72 (52%), Positives = 52/72 (72%), Gaps = 0/72 (0%)
Query 70 RPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQSLEERAE 129
RPVR+Y DG++DL H GH R L QAK +F + +L+ GV SDE TH LKG TV + ER +
Sbjct 75 RPVRVYADGIFDLFHSGHARALMQAKNLFPNTHLIVGVCSDELTHNLKGFTVMNEAERYD 134
Query 130 TLRHIKWVDEVI 141
++H ++VDEV+
Sbjct 135 AVQHCRYVDEVV 146
> xla:380231 pcyt1b, MGC53725, pcyt1a; phosphate cytidylyltransferase
1, choline, beta (EC:2.7.7.15); K00968 choline-phosphate
cytidylyltransferase [EC:2.7.7.15]
Length=366
Score = 87.8 bits (216), Expect = 9e-18, Method: Compositional matrix adjust.
Identities = 38/72 (52%), Positives = 52/72 (72%), Gaps = 0/72 (0%)
Query 70 RPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQSLEERAE 129
RPVR+Y DG++DL H GH R L QAK +F + +L+ GV SDE TH LKG TV + ER +
Sbjct 75 RPVRVYADGIFDLFHSGHARALMQAKTLFPNTHLIVGVCSDELTHNLKGFTVMNEAERYD 134
Query 130 TLRHIKWVDEVI 141
++H ++VDEV+
Sbjct 135 AVQHCRYVDEVV 146
> dre:553770 pcyt1ab, MGC110012, zgc:110012; phosphate cytidylyltransferase
1, choline, alpha b; K00968 choline-phosphate
cytidylyltransferase [EC:2.7.7.15]
Length=359
Score = 87.4 bits (215), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 36/72 (50%), Positives = 52/72 (72%), Gaps = 0/72 (0%)
Query 70 RPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQSLEERAE 129
RPVR+Y DG++D+ H GH R L QAK +F + YL+ GV SD+ TH+ KG TV + +ER +
Sbjct 72 RPVRVYADGIFDMFHSGHARALMQAKCLFPNTYLIVGVCSDDLTHKFKGFTVMNEDERYD 131
Query 130 TLRHIKWVDEVI 141
+ H ++VDEV+
Sbjct 132 AVCHCRYVDEVV 143
> xla:444649 pcyt2, MGC84177; phosphate cytidylyltransferase 2,
ethanolamine (EC:2.7.7.14); K00967 ethanolamine-phosphate
cytidylyltransferase [EC:2.7.7.14]
Length=383
Score = 75.1 bits (183), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 36/83 (43%), Positives = 53/83 (63%), Gaps = 2/83 (2%)
Query 59 PVELAEGEDPNRPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKG 118
P + + E RPVR++ DG YD++H GH QL QA+ M YL+ GV +DEE + KG
Sbjct 7 PEQALDKESVRRPVRVWCDGCYDMVHYGHSNQLRQARAMGD--YLIVGVHTDEEISQHKG 64
Query 119 QTVQSLEERAETLRHIKWVDEVI 141
V + +ER + ++ IKWVDE++
Sbjct 65 PPVFTQDERYKMVKAIKWVDEIV 87
Score = 46.2 bits (108), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 23/85 (27%), Positives = 48/85 (56%), Gaps = 3/85 (3%)
Query 60 VELAEGEDPN-RPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKG 118
++ A G++P+ IY G +DL H+GH+ LE+ + + Y++ G+ D+E + K
Sbjct 197 MQFASGKEPSPEDTIIYVAGAFDLFHIGHIDFLEKVYSLVEKPYVIVGLHFDQEVNHYKR 256
Query 119 QT--VQSLEERAETLRHIKWVDEVI 141
+ + ++ ER ++ ++V EV+
Sbjct 257 KNYPIMNIHERTLSVLACRYVAEVV 281
> mmu:68671 Pcyt2, 1110033E03Rik, ET; phosphate cytidylyltransferase
2, ethanolamine (EC:2.7.7.14); K00967 ethanolamine-phosphate
cytidylyltransferase [EC:2.7.7.14]
Length=404
Score = 74.7 bits (182), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 38/89 (42%), Positives = 53/89 (59%), Gaps = 2/89 (2%)
Query 53 DSQGTAPVELAEGEDPNRPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEE 112
+ G A +G R VR++ DG YD++H GH QL QA+ M YL+ GV +DEE
Sbjct 4 NGHGAASAAGLKGPGDQRIVRVWCDGCYDMVHYGHSNQLRQARAMGD--YLIVGVHTDEE 61
Query 113 THRLKGQTVQSLEERAETLRHIKWVDEVI 141
+ KG V + EER + ++ IKWVDEV+
Sbjct 62 IAKHKGPPVFTQEERYKMVQAIKWVDEVV 90
Score = 57.8 bits (138), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 28/85 (32%), Positives = 51/85 (60%), Gaps = 3/85 (3%)
Query 60 VELAEGEDPN-RPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKG 118
++ A G++P IY G +DL H+GH+ L++ K+ K Y++AG+ D+E +R KG
Sbjct 219 IQFASGKEPQPGETVIYVAGAFDLFHIGHVDFLQEVHKLAKRPYVIAGLHFDQEVNRYKG 278
Query 119 QT--VQSLEERAETLRHIKWVDEVI 141
+ + +L ER ++ ++V EV+
Sbjct 279 KNYPIMNLHERTLSVLACRYVSEVV 303
> dre:450016 pcyt2, im:7158585, wu:fb39h11, zgc:103434; phosphate
cytidylyltransferase 2, ethanolamine (EC:2.7.7.14); K00967
ethanolamine-phosphate cytidylyltransferase [EC:2.7.7.14]
Length=397
Score = 72.8 bits (177), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 39/91 (42%), Positives = 53/91 (58%), Gaps = 8/91 (8%)
Query 51 QPDSQGTAPVELAEGEDPNRPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASD 110
QP S +P E R +R++ DG YD++H GH QL QAK M YL+ GV +D
Sbjct 16 QPSSSTHSP------EKRKRVIRVWCDGCYDMVHYGHSNQLRQAKAMGD--YLVVGVHTD 67
Query 111 EETHRLKGQTVQSLEERAETLRHIKWVDEVI 141
EE + KG V + ER + +R IKWVDE++
Sbjct 68 EEIAKHKGPPVFTQAERYKMIRAIKWVDEIV 98
Score = 49.7 bits (117), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 25/85 (29%), Positives = 47/85 (55%), Gaps = 3/85 (3%)
Query 60 VELAEGEDPN-RPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKG 118
++ A G++P IY G +DL H+GH+ LE + Y++ G+ D+E +R KG
Sbjct 208 IQFASGKEPQPGDTIIYVAGAFDLFHIGHVDFLETVHGQAEKPYVIVGLHFDQEVNRYKG 267
Query 119 QT--VQSLEERAETLRHIKWVDEVI 141
+ + ++ ER ++ ++V EV+
Sbjct 268 KNYPIMNIHERILSVLACRYVSEVV 292
> hsa:5833 PCYT2, ET; phosphate cytidylyltransferase 2, ethanolamine
(EC:2.7.7.14); K00967 ethanolamine-phosphate cytidylyltransferase
[EC:2.7.7.14]
Length=407
Score = 72.8 bits (177), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 36/77 (46%), Positives = 49/77 (63%), Gaps = 2/77 (2%)
Query 65 GEDPNRPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQSL 124
G R VR++ DG YD++H GH QL QA+ M YL+ GV +DEE + KG V +
Sbjct 16 GPGGRRAVRVWCDGCYDMVHYGHSNQLRQARAMGD--YLIVGVHTDEEIAKHKGPPVFTQ 73
Query 125 EERAETLRHIKWVDEVI 141
EER + ++ IKWVDEV+
Sbjct 74 EERYKMVQAIKWVDEVV 90
Score = 53.9 bits (128), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 26/85 (30%), Positives = 50/85 (58%), Gaps = 3/85 (3%)
Query 60 VELAEGEDPN-RPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKG 118
++ A G++P IY G +DL H+GH+ LE+ ++ + Y++AG+ D+E + KG
Sbjct 219 IQFASGKEPQPGETVIYVAGAFDLFHIGHVDFLEKVHRLAERPYIIAGLHFDQEVNHYKG 278
Query 119 QT--VQSLEERAETLRHIKWVDEVI 141
+ + +L ER ++ ++V EV+
Sbjct 279 KNYPIMNLHERTLSVLACRYVSEVV 303
> tgo:TGME49_110280 phosphoethanolamine cytidylyltransferase,
putative (EC:4.1.1.70 2.7.7.39 2.7.7.14); K00967 ethanolamine-phosphate
cytidylyltransferase [EC:2.7.7.14]
Length=1128
Score = 71.6 bits (174), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 42/95 (44%), Positives = 52/95 (54%), Gaps = 8/95 (8%)
Query 48 PSPQPDSQGTAPVELAEGEDPNRPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGV 107
PSPQ +P P PVRIY DGV+DLLH GH L QA+++ L+ GV
Sbjct 535 PSPQ-----LSPFPARSACAPGAPVRIYVDGVFDLLHSGHFNALRQARQLGGK--LVVGV 587
Query 108 ASDEETHRLKG-QTVQSLEERAETLRHIKWVDEVI 141
SD T K + + + ERAE +R KWVDEVI
Sbjct 588 CSDAATFAAKKVRPIYTETERAEIVRGCKWVDEVI 622
Score = 60.5 bits (145), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 30/70 (42%), Positives = 45/70 (64%), Gaps = 4/70 (5%)
Query 74 IYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQT--VQSLEERAETL 131
+Y DG +D+ H+GH+R LE+AK++ YL+ G+ DE R+KG V +L ERA +
Sbjct 918 VYVDGSFDVFHVGHLRILEKAKQLGD--YLIVGIHDDETVSRIKGPGFPVLNLHERALNV 975
Query 132 RHIKWVDEVI 141
++ VDEVI
Sbjct 976 LAMRVVDEVI 985
> ath:AT2G38670 PECT1; PECT1 (PHOSPHORYLETHANOLAMINE CYTIDYLYLTRANSFERASE
1); ethanolamine-phosphate cytidylyltransferase
(EC:2.7.7.14); K00967 ethanolamine-phosphate cytidylyltransferase
[EC:2.7.7.14]
Length=421
Score = 68.9 bits (167), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 33/73 (45%), Positives = 45/73 (61%), Gaps = 2/73 (2%)
Query 70 RPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQSLEERAE 129
+PVR+Y DG +D++H GH L QA+ + L+ GV SDEE KG V L ER
Sbjct 53 KPVRVYMDGCFDMMHYGHCNALRQARALGDQ--LVVGVVSDEEIIANKGPPVTPLHERMT 110
Query 130 TLRHIKWVDEVIA 142
++ +KWVDEVI+
Sbjct 111 MVKAVKWVDEVIS 123
Score = 52.8 bits (125), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 37/140 (26%), Positives = 67/140 (47%), Gaps = 22/140 (15%)
Query 4 SELRRAQTHCAAPPYFELTSRATGNSARPALLKTSMKKRKATEDPSPQPDSQGTAPVELA 63
S L+R +H + P FE + + G L TS + + + P PD++
Sbjct 205 SSLQRQFSHGHSSPKFEDGASSAGTRV-SHFLPTSRRIVQFSNGKGPGPDAR-------- 255
Query 64 EGEDPNRPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKG--QTV 121
IY DG +DL H GH+ L +A+++ +L+ G+ +D+ +G + +
Sbjct 256 ---------IIYIDGAFDLFHAGHVEILRRARELGD--FLLVGIHNDQTVSAKRGAHRPI 304
Query 122 QSLEERAETLRHIKWVDEVI 141
+L ER+ ++ ++VDEVI
Sbjct 305 MNLHERSLSVLACRYVDEVI 324
> bbo:BBOV_I004300 19.m02274; ethanolamine-phosphate cytidylyltransferase
(EC:2.7.7.14); K00967 ethanolamine-phosphate cytidylyltransferase
[EC:2.7.7.14]
Length=386
Score = 63.9 bits (154), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 33/80 (41%), Positives = 48/80 (60%), Gaps = 3/80 (3%)
Query 64 EGEDPNRPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQT-VQ 122
E +D RIY DGV+DL+H GH+ L QA ++ + + GV SD+ET KG +
Sbjct 2 EDQDMTSHRRIYVDGVFDLVHWGHLNALRQAHQLGGKI--VVGVVSDKETQDTKGIAPIY 59
Query 123 SLEERAETLRHIKWVDEVIA 142
+ +ERAE + +WVD+VI
Sbjct 60 NSQERAELISGCRWVDDVIV 79
Score = 57.0 bits (136), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 29/76 (38%), Positives = 47/76 (61%), Gaps = 4/76 (5%)
Query 68 PNRPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQ--TVQSLE 125
P+ IY DG +D+ H+GH+R L++AK++ YL+ G+ D+ +KG V L
Sbjct 225 PDGAKVIYVDGTFDVFHVGHLRFLQRAKELGD--YLIVGLYDDQTVRTIKGNPFPVNHLM 282
Query 126 ERAETLRHIKWVDEVI 141
+RA T+ +K+VD+VI
Sbjct 283 DRALTVLAMKYVDDVI 298
> cel:Y37E3.11 hypothetical protein; K00967 ethanolamine-phosphate
cytidylyltransferase [EC:2.7.7.14]
Length=377
Score = 63.9 bits (154), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 36/86 (41%), Positives = 49/86 (56%), Gaps = 8/86 (9%)
Query 56 GTAPVELAEGEDPNRPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHR 115
G AP L +G R++ DG YD++H GH QL QAK+ + L+ GV +DEE
Sbjct 3 GLAPDGLPKGN------RVWADGCYDMVHFGHANQLRQAKQFGQK--LIVGVHNDEEIRL 54
Query 116 LKGQTVQSLEERAETLRHIKWVDEVI 141
KG V + +ER + IKWVDEV+
Sbjct 55 HKGPPVFNEQERYRMVAGIKWVDEVV 80
Score = 53.1 bits (126), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 32/89 (35%), Positives = 51/89 (57%), Gaps = 7/89 (7%)
Query 57 TAPVELAEGEDPNRPVR--IYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETH 114
T +E AEG P +P +Y G +DL H+GH+ LE+AK+ YL+ G+ SD+ +
Sbjct 195 TTILEFAEGRPP-KPTDKVVYVTGSFDLFHIGHLAFLEKAKEFGD--YLIVGILSDQTVN 251
Query 115 RLKGQT--VQSLEERAETLRHIKWVDEVI 141
+ KG + S+ ER ++ K V+EV+
Sbjct 252 QYKGSNHPIMSIHERVLSVLAYKPVNEVV 280
> tpv:TP04_0187 ethanolamine-phosphate cytidylyltransferase (EC:2.7.7.14);
K00967 ethanolamine-phosphate cytidylyltransferase
[EC:2.7.7.14]
Length=385
Score = 61.2 bits (147), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 30/71 (42%), Positives = 47/71 (66%), Gaps = 3/71 (4%)
Query 73 RIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQ-TVQSLEERAETL 131
RIY DGV+DL+H GH+ L Q+ ++ L+ GV SD++T R KG + + +ERAE +
Sbjct 15 RIYVDGVFDLIHWGHLNALRQSYELGGQ--LVIGVISDDDTQRAKGIPPIYTDQERAEIV 72
Query 132 RHIKWVDEVIA 142
+ KWV++V+
Sbjct 73 QACKWVNDVMV 83
Score = 45.8 bits (107), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 22/70 (31%), Positives = 40/70 (57%), Gaps = 4/70 (5%)
Query 74 IYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQ--TVQSLEERAETL 131
+Y DG +DL H GH+R L++A+ + YL+ G+ D+ +KG ++ +R +
Sbjct 227 VYVDGSFDLFHNGHVRFLKKARALGD--YLIVGIYDDQTVRTIKGSPFPFTNMLDRCLVV 284
Query 132 RHIKWVDEVI 141
+K+ D+VI
Sbjct 285 SAMKYTDDVI 294
> cpv:cgd7_2950 phospholipid cytidyltransferase HIGH family ;
K00967 ethanolamine-phosphate cytidylyltransferase [EC:2.7.7.14]
Length=405
Score = 60.1 bits (144), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 27/70 (38%), Positives = 42/70 (60%), Gaps = 3/70 (4%)
Query 73 RIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQT-VQSLEERAETL 131
RI+ DGV+DL+H GH L +AK+ L+ G+ SD + LKG + + +ER E +
Sbjct 15 RIFVDGVFDLMHAGHFNALRKAKQFGNE--LVVGINSDLDCFNLKGCYPIYNQDERGELM 72
Query 132 RHIKWVDEVI 141
+ KW DE++
Sbjct 73 KGCKWADEIV 82
Score = 56.2 bits (134), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 31/88 (35%), Positives = 47/88 (53%), Gaps = 6/88 (6%)
Query 56 GTAPVELAEGEDPNRPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHR 115
G A + +PN Y DG +D+ H+GH+R LE+ KK+F V L+ G+ D
Sbjct 204 GIASAKQVYNSNPNVT---YIDGSFDIFHIGHLRFLERVKKIFGGV-LIIGIYDDSTAQL 259
Query 116 LKGQT--VQSLEERAETLRHIKWVDEVI 141
+ G + + ERA TL ++ VD+VI
Sbjct 260 IYGDGFPILKMMERALTLLSMRVVDDVI 287
> pfa:PF13_0253 ethanolamine-phosphate cytidylyltransferase, putative
(EC:2.7.7.14); K00967 ethanolamine-phosphate cytidylyltransferase
[EC:2.7.7.14]
Length=573
Score = 60.1 bits (144), Expect = 2e-09, Method: Composition-based stats.
Identities = 31/77 (40%), Positives = 45/77 (58%), Gaps = 3/77 (3%)
Query 66 EDPNRPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKG-QTVQSL 124
++ + RIY DG++DL H GH + QAKK+ V + G+ SDE+ KG + + +
Sbjct 125 QEKTKETRIYVDGIFDLSHSGHFNAMRQAKKLGDIV--VVGINSDEDALNSKGVKPIYTQ 182
Query 125 EERAETLRHIKWVDEVI 141
EER + KWVDEVI
Sbjct 183 EERGALIAGCKWVDEVI 199
Score = 58.2 bits (139), Expect = 9e-09, Method: Composition-based stats.
Identities = 30/70 (42%), Positives = 45/70 (64%), Gaps = 4/70 (5%)
Query 74 IYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQ--TVQSLEERAETL 131
+Y DG +D+ H+GH+R LE AKK+ YL+ G+ SDE ++KG+ V SL ER +
Sbjct 409 VYVDGSFDIFHIGHLRILENAKKLGD--YLLVGMHSDEVVQKMKGKYFPVVSLLERTLNV 466
Query 132 RHIKWVDEVI 141
+K VD+V+
Sbjct 467 LAMKVVDDVV 476
> eco:b3052 rfaE, ECK3042, gmhC, hldE, JW3024, waaE, yqiF; fused
heptose 7-phosphate kinase/heptose 1-phosphate adenyltransferase
(EC:2.7.1.- 2.7.7.-); K03272 D-beta-D-heptose 7-phosphate
kinase / D-beta-D-heptose 1-phosphate adenosyltransferase
[EC:2.7.1.- 2.7.7.-]
Length=477
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 28/71 (39%), Positives = 42/71 (59%), Gaps = 4/71 (5%)
Query 74 IYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQT--VQSLEERAETL 131
+ T+GV+D+LH GH+ L A+K+ L+ V SD T RLKG + V LE+R L
Sbjct 343 VMTNGVFDILHAGHVSYLANARKLGDR--LIVAVNSDASTKRLKGDSRPVNPLEQRMIVL 400
Query 132 RHIKWVDEVIA 142
++ VD V++
Sbjct 401 GALEAVDWVVS 411
> cel:C39D10.3 hypothetical protein; K00967 ethanolamine-phosphate
cytidylyltransferase [EC:2.7.7.14]
Length=361
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 26/71 (36%), Positives = 38/71 (53%), Gaps = 4/71 (5%)
Query 74 IYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQT--VQSLEERAETL 131
+Y G +DL H GH+ LE AK + YL+ G+ D++ + KG V +L ER +
Sbjct 210 VYVSGAFDLFHAGHLSFLEAAKDLGD--YLIVGIVGDDDVNEEKGTIFPVMNLLERTLNI 267
Query 132 RHIKWVDEVIA 142
+K VDEV
Sbjct 268 SSLKIVDEVFV 278
Score = 50.4 bits (119), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 31/90 (34%), Positives = 46/90 (51%), Gaps = 8/90 (8%)
Query 50 PQPDSQGTAPVELAEGEDPNRPVRIYTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVAS 109
P+ QG P +GE + R+YTDG +D +H + R L AK+ K L+ G+ S
Sbjct 2 PKAIVQGLTP----DGE--KKKARVYTDGCFDFVHFANARLLWPAKQYGKK--LIVGIHS 53
Query 110 DEETHRLKGQTVQSLEERAETLRHIKWVDE 139
D+E + + EER + I+WVDE
Sbjct 54 DDELDNNGILPIFTDEERYRLISAIRWVDE 83
> sce:YGR007W MUQ1, ECT1; Choline phosphate cytidylyltransferase,
catalyzes the second step of phosphatidylethanolamine biosynthesis;
involved in the maintenance of plasma membrane;
similar to mammalian CTP: phosphocholine cytidylyl-transferases
(EC:2.7.7.14); K00967 ethanolamine-phosphate cytidylyltransferase
[EC:2.7.7.14]
Length=323
Score = 46.2 bits (108), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 24/73 (32%), Positives = 38/73 (52%), Gaps = 2/73 (2%)
Query 71 PVRIYTDGVYDLLHLGHMRQLEQAKKMF--KHVYLMAGVASDEETHRLKGQTVQSLEERA 128
P +++ DG +D H GH + QA++ ++ L GV +DE+ KG V + ER
Sbjct 7 PDKVWIDGCFDFTHHGHAGAILQARRTVSKENGKLFCGVHTDEDIQHNKGTPVMNSSERY 66
Query 129 ETLRHIKWVDEVI 141
E R +W EV+
Sbjct 67 EHTRSNRWCSEVV 79
Score = 37.7 bits (86), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 20/69 (28%), Positives = 38/69 (55%), Gaps = 6/69 (8%)
Query 74 IYTDGVYDLLHLGHMRQLEQAK-KMFKHVYLMAGVASDEETHRLKGQTVQSLEERAETLR 132
+Y DG +DL H+G + QL + K + L+ G+ + + + T+ +++ER ++
Sbjct 200 VYVDGDFDLFHMGDIDQLRKLKMDLHPDKKLIVGITTSDYS-----STIMTMKERVLSVL 254
Query 133 HIKWVDEVI 141
K+VD VI
Sbjct 255 SCKYVDAVI 263
> dre:100330104 disks large homolog 2-like
Length=584
Score = 43.5 bits (101), Expect = 2e-04, Method: Composition-based stats.
Identities = 36/117 (30%), Positives = 54/117 (46%), Gaps = 8/117 (6%)
Query 19 FELTSRATGNSARPALLKTSMKKRKATEDPSPQPDSQGTA--PVELAEGEDPNRPVRIYT 76
+E R N ARP ++ MK R + S PD G+ P ++ ED RP R Y
Sbjct 375 YEPVIRQEINYARPVIILGPMKDRINDDLISEFPDKFGSCVPPANSSDQEDTTRPKRDYE 434
Query 77 DGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQSLEERAETLRH 133
D + Q+E K + +H ++ AG +D L G +VQS++ AE +H
Sbjct 435 VDGRDYHFMASREQME--KDIQEHKFIEAGQYNDN----LYGTSVQSVKYVAERGKH 485
> dre:497638 dlg2; discs, large (Drosophila) homolog 2; K12075
discs large protein
Length=881
Score = 41.2 bits (95), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 36/117 (30%), Positives = 54/117 (46%), Gaps = 8/117 (6%)
Query 19 FELTSRATGNSARPALLKTSMKKRKATEDPSPQPDSQGTA--PVELAEGEDPNRPVRIYT 76
+E R N ARP ++ MK R + S PD G+ P ++ ED RP R Y
Sbjct 672 YEPVIRQEINYARPVIILGPMKDRINDDLISEFPDKFGSCVPPANSSDQEDTTRPKRDYE 731
Query 77 DGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQSLEERAETLRH 133
D + Q+E K + +H ++ AG +D L G +VQS++ AE +H
Sbjct 732 VDGRDYHFMASREQME--KDIQEHKFIEAGQYNDN----LYGTSVQSVKYVAERGKH 782
> xla:398153 ghr; growth hormone receptor; K05080 growth hormone
receptor
Length=603
Score = 32.3 bits (72), Expect = 0.47, Method: Composition-based stats.
Identities = 13/28 (46%), Positives = 20/28 (71%), Gaps = 0/28 (0%)
Query 45 TEDPSPQPDSQGTAPVELAEGEDPNRPV 72
TED SP+P+++ T PV ++E E + PV
Sbjct 415 TEDGSPKPEAEKTCPVAVSENEPTSLPV 442
> dre:100329309 discs, large homolog 2 (Drosophila)-like
Length=334
Score = 31.6 bits (70), Expect = 0.82, Method: Compositional matrix adjust.
Identities = 33/115 (28%), Positives = 49/115 (42%), Gaps = 12/115 (10%)
Query 19 FELTSRATGNSARPALLKTSMKKRKATEDPSPQPDSQGTAPVELAEGEDPNRPVRIYTDG 78
+E R N ARP ++ MK R + S PD G+ RP R Y
Sbjct 133 YEPVIRQEINYARPVIILGPMKDRINDDLISEFPDKFGSCVPH------TTRPKRDYEVD 186
Query 79 VYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQSLEERAETLRH 133
D + Q+E K + +H ++ AG +D L G +VQS++ AE +H
Sbjct 187 GRDYHFMASREQME--KDIQEHKFIEAGQYNDN----LYGTSVQSVKYVAERGKH 235
> mmu:56376 Pdlim5, 1110001A05Rik, AI987914, C87059, Enh, Enh1,
Enh2, Enh3, LIM; PDZ and LIM domain 5
Length=614
Score = 29.6 bits (65), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 12/27 (44%), Positives = 19/27 (70%), Gaps = 0/27 (0%)
Query 39 MKKRKATEDPSPQPDSQGTAPVELAEG 65
+KK +T++PS QP S G +P+ +EG
Sbjct 326 VKKSSSTQEPSQQPASSGASPLSASEG 352
> dre:497648 dlg1l; discs, large (Drosophila) homolog 1, like;
K12076 disks large protein 1
Length=827
Score = 29.6 bits (65), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 31/115 (26%), Positives = 49/115 (42%), Gaps = 12/115 (10%)
Query 19 FELTSRATGNSARPALLKTSMKKRKATEDPSPQPDSQGTAPVELAEGEDPNRPVRIYTDG 78
+E S+ N +RP ++ MK R + S PD G+ RP R Y
Sbjct 626 YEPVSQQEVNYSRPVIILGPMKDRVNDDLISEFPDKFGSCVPHTT------RPKRDYEVD 679
Query 79 VYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTVQSLEERAETLRH 133
D + Q+E + + +H ++ AG + L G +VQS+ E AE +H
Sbjct 680 GRDYHFVVSREQME--RDIQEHKFIEAG----QYNSHLYGTSVQSVREVAEKGKH 728
> xla:779400 nif3l1, MGC154449; NIF3 NGG1 interacting factor 3-like
1
Length=344
Score = 28.9 bits (63), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 28/114 (24%), Positives = 48/114 (42%), Gaps = 12/114 (10%)
Query 16 PPYFELTSRATGNSARPALLKTSMKKRKATEDPSPQPDSQGTAPVE-LAEGEDPNRPVRI 74
PP F+ R T + + L+ +++KR A P D+ + LA P++ V +
Sbjct 67 PPVFKALKRVTQKNWKERLVVKALEKRLAIYSPHTSCDALANGVNDWLARALGPSKSVPL 126
Query 75 -------YTDGVYDLLHLGHMRQLEQAKKMFKHVYLMAGVASDEETHRLKGQTV 121
Y G LL +L+ A+ +F + + GV+ T +GQ V
Sbjct 127 HASTSLTYPGGFGHLLEF----RLDTAENIFSRLKSIQGVSVCTSTASPEGQNV 176
> cel:F47E1.3 hypothetical protein
Length=594
Score = 28.9 bits (63), Expect = 6.3, Method: Composition-based stats.
Identities = 17/46 (36%), Positives = 25/46 (54%), Gaps = 1/46 (2%)
Query 15 APPYFELTSRATGNSARPALLKTSMKKRKATEDPSPQPDSQGTAPV 60
+PP F +TS AT N++ KTS A +P P S+ T+P+
Sbjct 90 SPPPF-MTSPATSNNSVSVARKTSAPPGFAQYEPDTAPPSRSTSPL 134
Lambda K H
0.314 0.129 0.372
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2749206264
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40