bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0691_orf1
Length=119
Score E
Sequences producing significant alignments: (Bits) Value
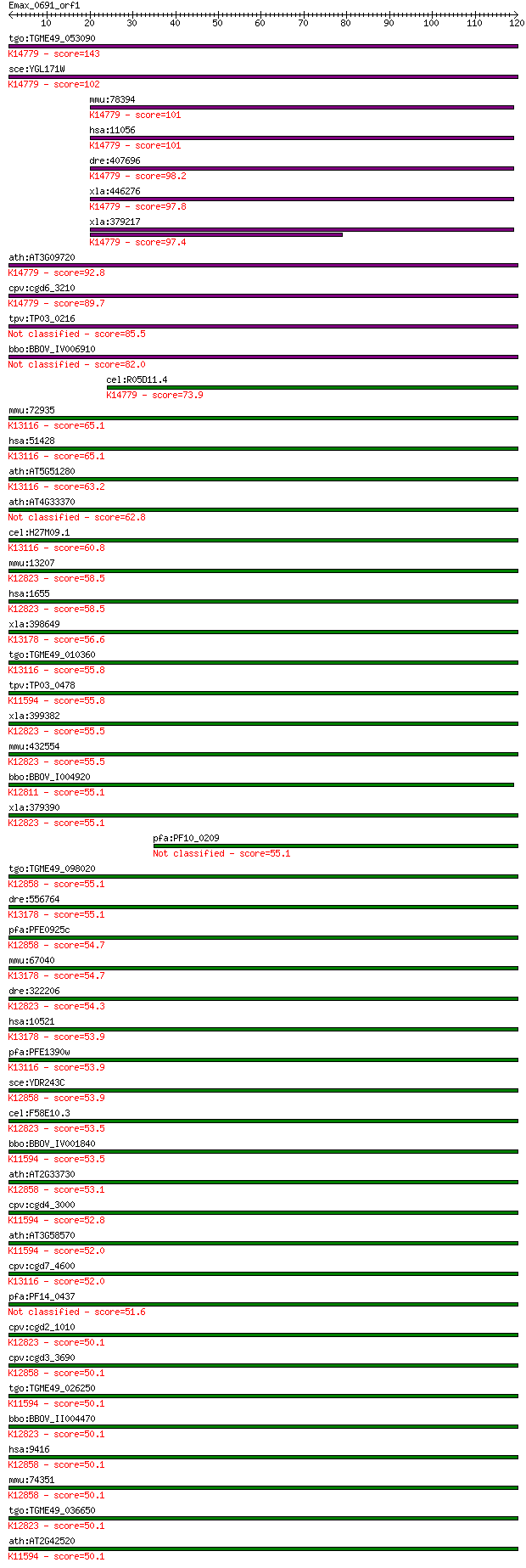
tgo:TGME49_053090 ATP-dependent RNA helicase, putative ; K1477... 143 1e-34
sce:YGL171W ROK1; ATP-dependent RNA helicase of the DEAD box f... 102 3e-22
mmu:78394 Ddx52, 2700029C06Rik, ROK1; DEAD (Asp-Glu-Ala-Asp) b... 101 4e-22
hsa:11056 DDX52, HUSSY19, ROK1; DEAD (Asp-Glu-Ala-Asp) box pol... 101 6e-22
dre:407696 ddx52, MGC136495; DEAD (Asp-Glu-Ala-Asp) box polype... 98.2 6e-21
xla:446276 ddx52; DEAD (Asp-Glu-Ala-Asp) box polypeptide 52; K... 97.8 7e-21
xla:379217 MGC53409; similar to ATP-dependent, RNA helicase; K... 97.4 9e-21
ath:AT3G09720 DEAD/DEAH box helicase, putative; K14779 ATP-dep... 92.8 2e-19
cpv:cgd6_3210 Rok1p, eIF4A-1-family RNA SFII helicase ; K14779... 89.7 2e-18
tpv:TP03_0216 RNA helicase 85.5 4e-17
bbo:BBOV_IV006910 23.m06397; DEAD box RNA helicase (EC:3.6.1.-) 82.0 4e-16
cel:R05D11.4 hypothetical protein; K14779 ATP-dependent RNA he... 73.9 1e-13
mmu:72935 Ddx41, 2900024F02Rik, AA958953, ABS, AI324246; DEAD ... 65.1 6e-11
hsa:51428 DDX41, ABS, MGC8828; DEAD (Asp-Glu-Ala-Asp) box poly... 65.1 6e-11
ath:AT5G51280 DEAD-box protein abstrakt, putative; K13116 ATP-... 63.2 2e-10
ath:AT4G33370 DEAD-box protein abstrakt, putative 62.8 2e-10
cel:H27M09.1 hypothetical protein; K13116 ATP-dependent RNA he... 60.8 9e-10
mmu:13207 Ddx5, 2600009A06Rik, G17P1, HUMP68, Hlr1, MGC118083,... 58.5 5e-09
hsa:1655 DDX5, DKFZp434E109, DKFZp686J01190, G17P1, HLR1, HUMP... 58.5 5e-09
xla:398649 ddx17, MGC80019; DEAD (Asp-Glu-Ala-Asp) box polypep... 56.6 2e-08
tgo:TGME49_010360 DEAD/DEAH box helicase, putative (EC:5.99.1.... 55.8 3e-08
tpv:TP03_0478 RNA helicase; K11594 ATP-dependent RNA helicase ... 55.8 3e-08
xla:399382 ddx5, MGC81559; DEAD (Asp-Glu-Ala-Asp) box polypept... 55.5 4e-08
mmu:432554 Gm12183, OTTMUSG00000005521; predicted gene 12183; ... 55.5 4e-08
bbo:BBOV_I004920 19.m02284; DEAD/DEAH box helicase and helicas... 55.1 5e-08
xla:379390 MGC53795; similar to DEAD/H (Asp-Glu-Ala-Asp/His) b... 55.1 5e-08
pfa:PF10_0209 RNA helicase, putative 55.1 5e-08
tgo:TGME49_098020 DEAD-box ATP-dependent RNA helicase, putativ... 55.1 6e-08
dre:556764 similar to Probable RNA-dependent helicase p72 (DEA... 55.1 6e-08
pfa:PFE0925c snrnp protein, putative; K12858 ATP-dependent RNA... 54.7 7e-08
mmu:67040 Ddx17, 2610007K22Rik, A430025E01Rik, AI047725, C8092... 54.7 7e-08
dre:322206 ddx5, wu:fa56a07, wu:fb11e01, wu:fb16c10, wu:fb53b0... 54.3 1e-07
hsa:10521 DDX17, DKFZp761H2016, P72, RH70; DEAD (Asp-Glu-Ala-A... 53.9 1e-07
pfa:PFE1390w RNA helicase-1; K13116 ATP-dependent RNA helicase... 53.9 1e-07
sce:YDR243C PRP28; Prp28p (EC:3.6.1.-); K12858 ATP-dependent R... 53.9 1e-07
cel:F58E10.3 hypothetical protein; K12823 ATP-dependent RNA he... 53.5 2e-07
bbo:BBOV_IV001840 21.m02846; DEAD/DEAH box helicase (EC:3.6.1.... 53.5 2e-07
ath:AT2G33730 DEAD box RNA helicase, putative; K12858 ATP-depe... 53.1 2e-07
cpv:cgd4_3000 Dbp1p, eIF4a-1 family RNA SFII helicase (DEXDC+H... 52.8 3e-07
ath:AT3G58570 DEAD box RNA helicase, putative; K11594 ATP-depe... 52.0 4e-07
cpv:cgd7_4600 abstrakt protein SF II helicase + Znknuckle C2HC... 52.0 5e-07
pfa:PF14_0437 helicase, putative 51.6 6e-07
cpv:cgd2_1010 hypothetical protein ; K12823 ATP-dependent RNA ... 50.1 2e-06
cpv:cgd3_3690 U5 snRNP 100 kD protein ; K12858 ATP-dependent R... 50.1 2e-06
tgo:TGME49_026250 ATP-dependent RNA helicase, putative ; K1159... 50.1 2e-06
bbo:BBOV_II004470 18.m06373; p68-like protein; K12823 ATP-depe... 50.1 2e-06
hsa:9416 DDX23, MGC8416, PRPF28, U5-100K, U5-100KD, prp28; DEA... 50.1 2e-06
mmu:74351 Ddx23, 3110082M05Rik, 4921506D17Rik; DEAD (Asp-Glu-A... 50.1 2e-06
tgo:TGME49_036650 DEAD/DEAH box helicase, putative (EC:5.99.1.... 50.1 2e-06
ath:AT2G42520 DEAD box RNA helicase, putative; K11594 ATP-depe... 50.1 2e-06
> tgo:TGME49_053090 ATP-dependent RNA helicase, putative ; K14779
ATP-dependent RNA helicase DDX52/ROK1 [EC:3.6.4.13]
Length=652
Score = 143 bits (361), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 67/119 (56%), Positives = 92/119 (77%), Gaps = 0/119 (0%)
Query 1 SATLPPSVVLLAESIAYGAVHLSVGRASAAAPQIEQELVFCTTEAGKLWALKTLRLERRL 60
SATLPP V+ LA+S+ + VH+S+G +AAA IEQEL+FCT E GKL AL+TL L +
Sbjct 401 SATLPPDVLRLADSLLHNPVHISIGSPNAAATSIEQELLFCTNEEGKLLALRTLHLTGKF 460
Query 61 IPPCLIFVETQERASELLKEMLSDGMPVDLLHAAKSKQQRNATVDAFRIGKIWFLICTD 119
+PP LIFV+++ERA +L E++ DG+ V+ +HA K+K+QR+ TV+AFR G+IW LICTD
Sbjct 461 VPPVLIFVQSKERAKQLYCELVYDGIFVECIHADKTKKQRDDTVEAFRRGQIWVLICTD 519
> sce:YGL171W ROK1; ATP-dependent RNA helicase of the DEAD box
family; required for 18S rRNA synthesis (EC:3.6.1.-); K14779
ATP-dependent RNA helicase DDX52/ROK1 [EC:3.6.4.13]
Length=564
Score = 102 bits (254), Expect = 3e-22, Method: Composition-based stats.
Identities = 49/119 (41%), Positives = 75/119 (63%), Gaps = 0/119 (0%)
Query 1 SATLPPSVVLLAESIAYGAVHLSVGRASAAAPQIEQELVFCTTEAGKLWALKTLRLERRL 60
SAT+P +V +A+SI V + +G AA IEQ+L+FC E GKL A++ L E
Sbjct 312 SATIPSNVEEIAQSIMMDPVRVIIGHKEAANTNIEQKLIFCGNEEGKLIAIRQLVQEGEF 371
Query 61 IPPCLIFVETQERASELLKEMLSDGMPVDLLHAAKSKQQRNATVDAFRIGKIWFLICTD 119
PP +IF+E+ RA L E++ D + VD++HA ++ QR+ ++ F+ G++W LICTD
Sbjct 372 KPPIIIFLESITRAKALYHELMYDRINVDVIHAERTALQRDRIIERFKTGELWCLICTD 430
> mmu:78394 Ddx52, 2700029C06Rik, ROK1; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 52 (EC:3.6.4.13); K14779 ATP-dependent RNA
helicase DDX52/ROK1 [EC:3.6.4.13]
Length=598
Score = 101 bits (252), Expect = 4e-22, Method: Compositional matrix adjust.
Identities = 46/99 (46%), Positives = 72/99 (72%), Gaps = 1/99 (1%)
Query 20 VHLSVGRASAAAPQIEQELVFCTTEAGKLWALKTLRLERRLIPPCLIFVETQERASELLK 79
V +S+G ++A +EQEL+F +E GKL A++ L +++ PP L+FV++ ERA EL
Sbjct 373 VSVSIGARNSAVETVEQELLFVGSETGKLLAMREL-VKKGFKPPVLVFVQSIERAKELFH 431
Query 80 EMLSDGMPVDLLHAAKSKQQRNATVDAFRIGKIWFLICT 118
E++ +G+ VD++HA +++QQR+ TV +FR GKIW LICT
Sbjct 432 ELIYEGINVDVIHAERTQQQRDNTVHSFRAGKIWVLICT 470
> hsa:11056 DDX52, HUSSY19, ROK1; DEAD (Asp-Glu-Ala-Asp) box polypeptide
52 (EC:3.6.4.13); K14779 ATP-dependent RNA helicase
DDX52/ROK1 [EC:3.6.4.13]
Length=599
Score = 101 bits (251), Expect = 6e-22, Method: Compositional matrix adjust.
Identities = 45/99 (45%), Positives = 72/99 (72%), Gaps = 1/99 (1%)
Query 20 VHLSVGRASAAAPQIEQELVFCTTEAGKLWALKTLRLERRLIPPCLIFVETQERASELLK 79
+ +S+G ++A +EQEL+F +E GKL A++ L +++ PP L+FV++ ERA EL
Sbjct 372 ISVSIGARNSAVETVEQELLFVGSETGKLLAMREL-VKKGFNPPVLVFVQSIERAKELFH 430
Query 80 EMLSDGMPVDLLHAAKSKQQRNATVDAFRIGKIWFLICT 118
E++ +G+ VD++HA +++QQR+ TV +FR GKIW LICT
Sbjct 431 ELIYEGINVDVIHAERTQQQRDNTVHSFRAGKIWVLICT 469
> dre:407696 ddx52, MGC136495; DEAD (Asp-Glu-Ala-Asp) box polypeptide
52 (EC:3.6.4.13); K14779 ATP-dependent RNA helicase
DDX52/ROK1 [EC:3.6.4.13]
Length=606
Score = 98.2 bits (243), Expect = 6e-21, Method: Compositional matrix adjust.
Identities = 43/99 (43%), Positives = 73/99 (73%), Gaps = 1/99 (1%)
Query 20 VHLSVGRASAAAPQIEQELVFCTTEAGKLWALKTLRLERRLIPPCLIFVETQERASELLK 79
V +S+G ++AA +EQ+L+F +E GK+ A++ L +++ +PP L+FV++ +RA EL
Sbjct 374 VSVSIGARNSAAETVEQQLLFVGSENGKILAMRNL-IKQGFLPPVLVFVQSIDRARELYH 432
Query 80 EMLSDGMPVDLLHAAKSKQQRNATVDAFRIGKIWFLICT 118
E++ +G+ VD++HA +++QQR+ V +FR GKIW LICT
Sbjct 433 ELVYEGINVDVIHADRTQQQRDNVVSSFRSGKIWVLICT 471
> xla:446276 ddx52; DEAD (Asp-Glu-Ala-Asp) box polypeptide 52;
K14779 ATP-dependent RNA helicase DDX52/ROK1 [EC:3.6.4.13]
Length=614
Score = 97.8 bits (242), Expect = 7e-21, Method: Compositional matrix adjust.
Identities = 44/99 (44%), Positives = 70/99 (70%), Gaps = 1/99 (1%)
Query 20 VHLSVGRASAAAPQIEQELVFCTTEAGKLWALKTLRLERRLIPPCLIFVETQERASELLK 79
V +SVG ++A +EQ L+F +E GKL A++ L +++ PP L+FV++ ERA EL
Sbjct 378 VSVSVGARNSAVETVEQSLLFVGSETGKLLAMRDL-VKKGFAPPVLVFVQSIERAKELFH 436
Query 80 EMLSDGMPVDLLHAAKSKQQRNATVDAFRIGKIWFLICT 118
E++ +G+ VD++HA +++QQR+ + +FR GKIW LICT
Sbjct 437 ELIYEGINVDVIHAERTQQQRDNVIQSFREGKIWVLICT 475
> xla:379217 MGC53409; similar to ATP-dependent, RNA helicase;
K14779 ATP-dependent RNA helicase DDX52/ROK1 [EC:3.6.4.13]
Length=686
Score = 97.4 bits (241), Expect = 9e-21, Method: Compositional matrix adjust.
Identities = 44/99 (44%), Positives = 69/99 (69%), Gaps = 1/99 (1%)
Query 20 VHLSVGRASAAAPQIEQELVFCTTEAGKLWALKTLRLERRLIPPCLIFVETQERASELLK 79
V +SVG ++A +EQ L+F +E GKL A++ L +++ PP L FV++ ERA EL
Sbjct 449 VSVSVGARNSAVETVEQSLLFVGSETGKLLAMRNL-IKKGFTPPVLAFVQSVERAKELFH 507
Query 80 EMLSDGMPVDLLHAAKSKQQRNATVDAFRIGKIWFLICT 118
E++ +G+ VD++HA +++QQR+ + +FR GKIW LICT
Sbjct 508 ELIYEGINVDVIHAERTQQQRDNVIQSFREGKIWVLICT 546
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 25/59 (42%), Positives = 38/59 (64%), Gaps = 1/59 (1%)
Query 20 VHLSVGRASAAAPQIEQELVFCTTEAGKLWALKTLRLERRLIPPCLIFVETQERASELL 78
V +SVG ++A +EQ L+F +E GKL A++ L +++ PP L FV++ ERA EL
Sbjct 378 VSVSVGARNSAVETVEQSLLFVGSETGKLLAMRNL-IKKGFTPPVLAFVQSVERAKELF 435
> ath:AT3G09720 DEAD/DEAH box helicase, putative; K14779 ATP-dependent
RNA helicase DDX52/ROK1 [EC:3.6.4.13]
Length=541
Score = 92.8 bits (229), Expect = 2e-19, Method: Composition-based stats.
Identities = 51/119 (42%), Positives = 72/119 (60%), Gaps = 1/119 (0%)
Query 1 SATLPPSVVLLAESIAYGAVHLSVGRASAAAPQIEQELVFCTTEAGKLWALKTLRLERRL 60
SATLP SV LA SI + AV + +GR + A+ ++Q+LVF +E GKL AL+ L
Sbjct 321 SATLPDSVEELARSIMHDAVRVIIGRKNTASETVKQKLVFAGSEEGKLLALRQ-SFAESL 379
Query 61 IPPCLIFVETQERASELLKEMLSDGMPVDLLHAAKSKQQRNATVDAFRIGKIWFLICTD 119
PP LIFV+++ERA EL E+ + + ++H+ +R VD FR G+ W LI TD
Sbjct 380 NPPVLIFVQSKERAKELYDELKCENIRAGVIHSDLPPGERENAVDQFRAGEKWVLIATD 438
> cpv:cgd6_3210 Rok1p, eIF4A-1-family RNA SFII helicase ; K14779
ATP-dependent RNA helicase DDX52/ROK1 [EC:3.6.4.13]
Length=480
Score = 89.7 bits (221), Expect = 2e-18, Method: Composition-based stats.
Identities = 45/119 (37%), Positives = 73/119 (61%), Gaps = 0/119 (0%)
Query 1 SATLPPSVVLLAESIAYGAVHLSVGRASAAAPQIEQELVFCTTEAGKLWALKTLRLERRL 60
SATLP V+ LA+SI V +++G AA+ I QELV T + K+ +L+ L + ++
Sbjct 273 SATLPQIVINLADSIMKSPVKVTLGHRLAASSTIIQELVCVTKDDAKIESLRQLIKQGKI 332
Query 61 IPPCLIFVETQERASELLKEMLSDGMPVDLLHAAKSKQQRNATVDAFRIGKIWFLICTD 119
+ P L+F +++ A L K+++ D + V+ +H+ K +R+ + FR GKIW LICTD
Sbjct 333 MLPTLVFTNSKDDAQRLFKKLMYDNLIVEAIHSDMPKVKRDNIIQRFRTGKIWILICTD 391
> tpv:TP03_0216 RNA helicase
Length=476
Score = 85.5 bits (210), Expect = 4e-17, Method: Composition-based stats.
Identities = 41/119 (34%), Positives = 70/119 (58%), Gaps = 0/119 (0%)
Query 1 SATLPPSVVLLAESIAYGAVHLSVGRASAAAPQIEQELVFCTTEAGKLWALKTLRLERRL 60
S+TL V+LL++S +H+++G+ + +EQEL+ T + GKL LK L + +L
Sbjct 244 SSTLQSEVLLLSKSHFNNPIHITIGKENVCCCNVEQELICVTNDKGKLLILKQLINDGKL 303
Query 61 IPPCLIFVETQERASELLKEMLSDGMPVDLLHAAKSKQQRNATVDAFRIGKIWFLICTD 119
+PP L+F+++ R ++L E+ + V + +QR + FRIG+IW L+CTD
Sbjct 304 LPPILVFLQSINRVNDLYNELSQLNLNVQKFTKQLTLKQRQNIIQKFRIGQIWILLCTD 362
> bbo:BBOV_IV006910 23.m06397; DEAD box RNA helicase (EC:3.6.1.-)
Length=457
Score = 82.0 bits (201), Expect = 4e-16, Method: Composition-based stats.
Identities = 39/119 (32%), Positives = 73/119 (61%), Gaps = 0/119 (0%)
Query 1 SATLPPSVVLLAESIAYGAVHLSVGRASAAAPQIEQELVFCTTEAGKLWALKTLRLERRL 60
S+T+ P V+ LA + AV ++VG+++ + QELV T E+GK+ L+ L + ++
Sbjct 230 SSTMQPQVLELAATFMPDAVKVAVGQSTRVCKNVRQELVCVTNESGKIPTLRQLIRDGKI 289
Query 61 IPPCLIFVETQERASELLKEMLSDGMPVDLLHAAKSKQQRNATVDAFRIGKIWFLICTD 119
PCL+F+++ +R +++ +M D + V+ L S +R+ ++ FR+ KIW L+CT+
Sbjct 290 KLPCLVFLQSIDRVTQVYNQMKDDNLLVERLTGQMSPSERDELINRFRLSKIWVLLCTN 348
> cel:R05D11.4 hypothetical protein; K14779 ATP-dependent RNA
helicase DDX52/ROK1 [EC:3.6.4.13]
Length=581
Score = 73.9 bits (180), Expect = 1e-13, Method: Composition-based stats.
Identities = 35/98 (35%), Positives = 65/98 (66%), Gaps = 3/98 (3%)
Query 24 VGRASAAAPQIEQELVFCTTEAGKLWALKTLRLERRLIPPCLIFVETQERASELLKEM-- 81
VG +++ ++Q+L +C TE GK A++ L L PP L+FV++++RA +L+K +
Sbjct 337 VGERNSSNTSVKQKLTYCGTEDGKKIAIRNL-LRTSFKPPALVFVQSKDRAVQLVKLLSA 395
Query 82 LSDGMPVDLLHAAKSKQQRNATVDAFRIGKIWFLICTD 119
+ + VD +++ KS ++R+ T++ FR G+IW L+CT+
Sbjct 396 IDSNLKVDSINSGKSDKERDETMERFRRGEIWVLVCTE 433
> mmu:72935 Ddx41, 2900024F02Rik, AA958953, ABS, AI324246; DEAD
(Asp-Glu-Ala-Asp) box polypeptide 41 (EC:3.6.4.13); K13116
ATP-dependent RNA helicase DDX41 [EC:3.6.4.13]
Length=622
Score = 65.1 bits (157), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 38/119 (31%), Positives = 64/119 (53%), Gaps = 3/119 (2%)
Query 1 SATLPPSVVLLAESIAYGAVHLSVGRASAAAPQIEQELVFCTTEAGKLWALKTLRLERRL 60
SAT+P + A+S V ++VGRA AA+ + QE+ + EA ++ L+ L+ +
Sbjct 375 SATMPKKIQNFAKSALVKPVTINVGRAGAASLDVIQEVEYVKEEAKMVYLLECLQ---KT 431
Query 61 IPPCLIFVETQERASELLKEMLSDGMPVDLLHAAKSKQQRNATVDAFRIGKIWFLICTD 119
PP LIF E + + + +L G+ +H K +++R ++AFR GK L+ TD
Sbjct 432 PPPVLIFAEKKADVDAIHEYLLLKGVEAVAIHGGKDQEERTKAIEAFREGKKDVLVATD 490
> hsa:51428 DDX41, ABS, MGC8828; DEAD (Asp-Glu-Ala-Asp) box polypeptide
41 (EC:3.6.4.13); K13116 ATP-dependent RNA helicase
DDX41 [EC:3.6.4.13]
Length=622
Score = 65.1 bits (157), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 38/119 (31%), Positives = 64/119 (53%), Gaps = 3/119 (2%)
Query 1 SATLPPSVVLLAESIAYGAVHLSVGRASAAAPQIEQELVFCTTEAGKLWALKTLRLERRL 60
SAT+P + A+S V ++VGRA AA+ + QE+ + EA ++ L+ L+ +
Sbjct 375 SATMPKKIQNFAKSALVKPVTINVGRAGAASLDVIQEVEYVKEEAKMVYLLECLQ---KT 431
Query 61 IPPCLIFVETQERASELLKEMLSDGMPVDLLHAAKSKQQRNATVDAFRIGKIWFLICTD 119
PP LIF E + + + +L G+ +H K +++R ++AFR GK L+ TD
Sbjct 432 PPPVLIFAEKKADVDAIHEYLLLKGVEAVAIHGGKDQEERTKAIEAFREGKKDVLVATD 490
> ath:AT5G51280 DEAD-box protein abstrakt, putative; K13116 ATP-dependent
RNA helicase DDX41 [EC:3.6.4.13]
Length=591
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 36/119 (30%), Positives = 62/119 (52%), Gaps = 3/119 (2%)
Query 1 SATLPPSVVLLAESIAYGAVHLSVGRASAAAPQIEQELVFCTTEAGKLWALKTLRLERRL 60
SAT+P + + A S V ++VGRA AA + QE+ + EA ++ L+ L+ +
Sbjct 340 SATMPTKIQIFARSALVKPVTVNVGRAGAANLDVIQEVEYVKQEAKIVYLLECLQ---KT 396
Query 61 IPPCLIFVETQERASELLKEMLSDGMPVDLLHAAKSKQQRNATVDAFRIGKIWFLICTD 119
PP LIF E + ++ + +L G+ +H K ++ R + +F+ GK L+ TD
Sbjct 397 SPPVLIFCENKADVDDIHEYLLLKGVEAVAIHGGKDQEDREYAISSFKAGKKDVLVATD 455
> ath:AT4G33370 DEAD-box protein abstrakt, putative
Length=542
Score = 62.8 bits (151), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 36/119 (30%), Positives = 62/119 (52%), Gaps = 3/119 (2%)
Query 1 SATLPPSVVLLAESIAYGAVHLSVGRASAAAPQIEQELVFCTTEAGKLWALKTLRLERRL 60
SAT+P + + A S V ++VGRA AA + QE+ + EA ++ L+ L+ +
Sbjct 291 SATMPAKIQIFATSALVKPVTVNVGRAGAANLDVIQEVEYVKQEAKIVYLLECLQ---KT 347
Query 61 IPPCLIFVETQERASELLKEMLSDGMPVDLLHAAKSKQQRNATVDAFRIGKIWFLICTD 119
PP LIF E + ++ + +L G+ +H K ++ R+ + F+ GK L+ TD
Sbjct 348 TPPVLIFCENKADVDDIHEYLLLKGVEAVAIHGGKDQEDRDYAISLFKAGKKDVLVATD 406
> cel:H27M09.1 hypothetical protein; K13116 ATP-dependent RNA
helicase DDX41 [EC:3.6.4.13]
Length=630
Score = 60.8 bits (146), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 37/119 (31%), Positives = 62/119 (52%), Gaps = 3/119 (2%)
Query 1 SATLPPSVVLLAESIAYGAVHLSVGRASAAAPQIEQELVFCTTEAGKLWALKTLRLERRL 60
SAT+P + A+S + ++VGRA AA+ + QEL F +E + L+ L+ +
Sbjct 384 SATMPRKIQFFAKSALVKPIVVNVGRAGAASLNVLQELEFVRSENKLVRVLECLQ---KT 440
Query 61 IPPCLIFVETQERASELLKEMLSDGMPVDLLHAAKSKQQRNATVDAFRIGKIWFLICTD 119
P LIF E + + + +L G+ V +H K + R+A ++AFR + L+ TD
Sbjct 441 SPKVLIFAEKKVDVDNIYEYLLVKGVEVASIHGGKDQSDRHAGIEAFRKNEKDVLVATD 499
> mmu:13207 Ddx5, 2600009A06Rik, G17P1, HUMP68, Hlr1, MGC118083,
p68; DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 (EC:3.6.4.13);
K12823 ATP-dependent RNA helicase DDX5/DBP2 [EC:3.6.4.13]
Length=615
Score = 58.5 bits (140), Expect = 5e-09, Method: Composition-based stats.
Identities = 39/123 (31%), Positives = 62/123 (50%), Gaps = 6/123 (4%)
Query 1 SATLPPSVVLLAESIAYGAVHLSVGRASAAAPQIEQELVFCTTEAGKLWALKTLRLERRL 60
SAT P V LAE +H+++G +A ++V + K K +RL +
Sbjct 279 SATWPKEVRQLAEDFLKDYIHINIGALELSANHNILQIVDVCHDVEK--DEKLIRLMEEI 336
Query 61 IP----PCLIFVETQERASELLKEMLSDGMPVDLLHAAKSKQQRNATVDAFRIGKIWFLI 116
+ ++FVET+ R EL ++M DG P +H KS+Q+R+ ++ F+ GK LI
Sbjct 337 MSEKENKTIVFVETKRRCDELTRKMRRDGWPAMGIHGDKSQQERDWVLNEFKHGKAPILI 396
Query 117 CTD 119
TD
Sbjct 397 ATD 399
> hsa:1655 DDX5, DKFZp434E109, DKFZp686J01190, G17P1, HLR1, HUMP68,
p68; DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 (EC:3.6.4.13);
K12823 ATP-dependent RNA helicase DDX5/DBP2 [EC:3.6.4.13]
Length=614
Score = 58.5 bits (140), Expect = 5e-09, Method: Composition-based stats.
Identities = 39/123 (31%), Positives = 62/123 (50%), Gaps = 6/123 (4%)
Query 1 SATLPPSVVLLAESIAYGAVHLSVGRASAAAPQIEQELVFCTTEAGKLWALKTLRLERRL 60
SAT P V LAE +H+++G +A ++V + K K +RL +
Sbjct 279 SATWPKEVRQLAEDFLKDYIHINIGALELSANHNILQIVDVCHDVEK--DEKLIRLMEEI 336
Query 61 IP----PCLIFVETQERASELLKEMLSDGMPVDLLHAAKSKQQRNATVDAFRIGKIWFLI 116
+ ++FVET+ R EL ++M DG P +H KS+Q+R+ ++ F+ GK LI
Sbjct 337 MSEKENKTIVFVETKRRCDELTRKMRRDGWPAMGIHGDKSQQERDWVLNEFKHGKAPILI 396
Query 117 CTD 119
TD
Sbjct 397 ATD 399
> xla:398649 ddx17, MGC80019; DEAD (Asp-Glu-Ala-Asp) box polypeptide
17; K13178 ATP-dependent RNA helicase DDX17 [EC:3.6.4.13]
Length=610
Score = 56.6 bits (135), Expect = 2e-08, Method: Composition-based stats.
Identities = 40/123 (32%), Positives = 59/123 (47%), Gaps = 6/123 (4%)
Query 1 SATLPPSVVLLAESIAYGAVHLSVGRASAAAPQIEQELVFCTTEAGKLWALKTLRLERRL 60
SAT P V LAE +++G +A ++V E+ K K ++L +
Sbjct 267 SATWPKEVRQLAEDFLRDYSQINIGNLELSANHNILQIVDVCQESEK--DHKLIQLMEEI 324
Query 61 IP----PCLIFVETQERASELLKEMLSDGMPVDLLHAAKSKQQRNATVDAFRIGKIWFLI 116
+ +IFVET+ R EL + M DG P +H KS+Q+R+ + FR GK LI
Sbjct 325 MAEKENKTIIFVETKRRCDELTRRMRRDGWPAMCIHGDKSQQERDWVLCEFRTGKAPILI 384
Query 117 CTD 119
TD
Sbjct 385 ATD 387
> tgo:TGME49_010360 DEAD/DEAH box helicase, putative (EC:5.99.1.3);
K13116 ATP-dependent RNA helicase DDX41 [EC:3.6.4.13]
Length=657
Score = 55.8 bits (133), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 35/119 (29%), Positives = 62/119 (52%), Gaps = 3/119 (2%)
Query 1 SATLPPSVVLLAESIAYGAVHLSVGRASAAAPQIEQELVFCTTEAGKLWALKTLRLERRL 60
SAT+P + A+S + ++VGRA AA + QE+ + E + L L+ +
Sbjct 402 SATMPRKIQEFAKSALIDPLVVNVGRAGAANLDVVQEVEYVKQENKLPYLLHCLQ---KT 458
Query 61 IPPCLIFVETQERASELLKEMLSDGMPVDLLHAAKSKQQRNATVDAFRIGKIWFLICTD 119
PP LIF E ++ ++ + +L G+ +H ++++R+ V AFR G+ L+ TD
Sbjct 459 APPVLIFCENKKDVDDIQEYLLLKGVDAAAVHGGLAQEERSEAVRAFREGRKDVLVGTD 517
> tpv:TP03_0478 RNA helicase; K11594 ATP-dependent RNA helicase
[EC:3.6.4.13]
Length=741
Score = 55.8 bits (133), Expect = 3e-08, Method: Composition-based stats.
Identities = 34/119 (28%), Positives = 58/119 (48%), Gaps = 0/119 (0%)
Query 1 SATLPPSVVLLAESIAYGAVHLSVGRASAAAPQIEQELVFCTTEAGKLWALKTLRLERRL 60
SAT P + LA ++L+VGR + I+Q L++ + + +K LR L
Sbjct 481 SATFPKEIQQLAREFLSDYIYLAVGRVGSTNEFIKQRLLYADQDQKIKYLIKLLRDNTNL 540
Query 61 IPPCLIFVETQERASELLKEMLSDGMPVDLLHAAKSKQQRNATVDAFRIGKIWFLICTD 119
LIFVET++RA + +LS+ +H +S++ R + F+ G ++ TD
Sbjct 541 GGLVLIFVETKKRADLIEGYLLSENFKAVNIHGDRSQEDREKALSLFKAGVRPIMVATD 599
> xla:399382 ddx5, MGC81559; DEAD (Asp-Glu-Ala-Asp) box polypeptide
5; K12823 ATP-dependent RNA helicase DDX5/DBP2 [EC:3.6.4.13]
Length=608
Score = 55.5 bits (132), Expect = 4e-08, Method: Composition-based stats.
Identities = 38/123 (30%), Positives = 61/123 (49%), Gaps = 6/123 (4%)
Query 1 SATLPPSVVLLAESIAYGAVHLSVGRASAAAPQIEQELVFCTTEAGKLWALKTLRLERRL 60
SAT P V LAE VH+++G +A ++V + K K +RL +
Sbjct 277 SATWPKEVRQLAEDFLRDYVHINIGALELSANHNILQIVDVCNDGEK--DDKLVRLMEEI 334
Query 61 IP----PCLIFVETQERASELLKEMLSDGMPVDLLHAAKSKQQRNATVDAFRIGKIWFLI 116
+ ++FVET+ R +L + + DG P +H KS+Q+R+ ++ F+ GK LI
Sbjct 335 MSEKENKTIVFVETKRRCDDLTRRLRRDGWPAMGIHGDKSQQERDWVLNEFKHGKSPILI 394
Query 117 CTD 119
TD
Sbjct 395 ATD 397
> mmu:432554 Gm12183, OTTMUSG00000005521; predicted gene 12183;
K12823 ATP-dependent RNA helicase DDX5/DBP2 [EC:3.6.4.13]
Length=670
Score = 55.5 bits (132), Expect = 4e-08, Method: Composition-based stats.
Identities = 37/123 (30%), Positives = 61/123 (49%), Gaps = 6/123 (4%)
Query 1 SATLPPSVVLLAESIAYGAVHLSVGRASAAAPQIEQELVFCTTEAGK----LWALKTLRL 56
SAT P V LAE +H+++G +A ++V + K + ++ +
Sbjct 334 SATWPKEVRQLAEDFLKDYIHINIGALELSANHNILQIVDVCHDVEKDEKLILLMEEIMS 393
Query 57 ERRLIPPCLIFVETQERASELLKEMLSDGMPVDLLHAAKSKQQRNATVDAFRIGKIWFLI 116
E+ ++FVET+ R EL ++M DG P +H KS+Q+R+ + F+ GK LI
Sbjct 394 EKE--NKTIVFVETKRRCDELTRKMRRDGWPAMGIHGDKSQQERDWVLSEFKHGKASILI 451
Query 117 CTD 119
TD
Sbjct 452 ATD 454
> bbo:BBOV_I004920 19.m02284; DEAD/DEAH box helicase and helicase
conserved C-terminal domain containing protein (EC:3.6.1.-);
K12811 ATP-dependent RNA helicase DDX46/PRP5 [EC:3.6.4.13]
Length=994
Score = 55.1 bits (131), Expect = 5e-08, Method: Composition-based stats.
Identities = 36/118 (30%), Positives = 56/118 (47%), Gaps = 1/118 (0%)
Query 1 SATLPPSVVLLAESIAYGAVHLSVGRASAAAPQIEQELVFCTTEAGKLWALKTLRLERRL 60
SAT PP++ LA+ I + + VG + +A Q++Q V E K++AL L E
Sbjct 576 SATFPPTIEALAKKILTKPLQIIVGESGKSASQVDQH-VMVLPERQKMYALLKLLGEWHE 634
Query 61 IPPCLIFVETQERASELLKEMLSDGMPVDLLHAAKSKQQRNATVDAFRIGKIWFLICT 118
++FV Q A + E++ G +LH + + R T+ FR G LI T
Sbjct 635 HGSIIVFVNRQLDADSMYAELIKHGYDCAVLHGGQDQTDREFTLQDFRDGTKGILIAT 692
> xla:379390 MGC53795; similar to DEAD/H (Asp-Glu-Ala-Asp/His)
box polypeptide 5 (RNA helicase, 68kDa); K12823 ATP-dependent
RNA helicase DDX5/DBP2 [EC:3.6.4.13]
Length=607
Score = 55.1 bits (131), Expect = 5e-08, Method: Composition-based stats.
Identities = 38/123 (30%), Positives = 61/123 (49%), Gaps = 6/123 (4%)
Query 1 SATLPPSVVLLAESIAYGAVHLSVGRASAAAPQIEQELVFCTTEAGKLWALKTLRLERRL 60
SAT P V LAE VH+++G +A ++V + K K +RL +
Sbjct 275 SATWPKEVRQLAEDFLKEYVHINIGALELSANHNILQIVDVCNDGEK--DDKLVRLMEEI 332
Query 61 IP----PCLIFVETQERASELLKEMLSDGMPVDLLHAAKSKQQRNATVDAFRIGKIWFLI 116
+ ++FVET+ R +L + + DG P +H KS+Q+R+ ++ F+ GK LI
Sbjct 333 MSEKENKTIVFVETKRRCDDLTRRLRRDGWPAMGIHGDKSQQERDWVLNEFKHGKSPILI 392
Query 117 CTD 119
TD
Sbjct 393 ATD 395
> pfa:PF10_0209 RNA helicase, putative
Length=680
Score = 55.1 bits (131), Expect = 5e-08, Method: Composition-based stats.
Identities = 29/85 (34%), Positives = 46/85 (54%), Gaps = 0/85 (0%)
Query 35 EQELVFCTTEAGKLWALKTLRLERRLIPPCLIFVETQERASELLKEMLSDGMPVDLLHAA 94
+QEL++ E KL L L + + P LIFV++ +A+ + + + LL +
Sbjct 456 KQELLYVNNEEEKLLVLNNLIKNKEIHIPVLIFVDSIIKANMIYTNLHKSVSYIALLTSE 515
Query 95 KSKQQRNATVDAFRIGKIWFLICTD 119
KSK++R F+ G IW+LICTD
Sbjct 516 KSKEERKIIFQKFQQGHIWYLICTD 540
> tgo:TGME49_098020 DEAD-box ATP-dependent RNA helicase, putative
(EC:2.7.11.25); K12858 ATP-dependent RNA helicase DDX23/PRP28
[EC:3.6.4.13]
Length=1158
Score = 55.1 bits (131), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 39/119 (32%), Positives = 59/119 (49%), Gaps = 3/119 (2%)
Query 1 SATLPPSVVLLAESIAYGAVHLSVGRASAAAPQIEQELVFCTTEAGKLWALKTLRLERRL 60
SAT+PP+V LA ++S+G A IEQ + F EA K L+ + LE
Sbjct 949 SATMPPAVERLARKYLRQPSYISIGDPGAGKRAIEQRVEFVP-EARKKQRLQDV-LEN-A 1005
Query 61 IPPCLIFVETQERASELLKEMLSDGMPVDLLHAAKSKQQRNATVDAFRIGKIWFLICTD 119
PP ++FV ++ A L K + G LH K+++ R A + +F+ G L+ TD
Sbjct 1006 TPPVMVFVNQKKSADALAKVLGKLGYSACSLHGGKAQENREAALSSFKEGSHDVLVATD 1064
> dre:556764 similar to Probable RNA-dependent helicase p72 (DEAD-box
protein p72) (DEAD-box protein 17); K13178 ATP-dependent
RNA helicase DDX17 [EC:3.6.4.13]
Length=671
Score = 55.1 bits (131), Expect = 6e-08, Method: Composition-based stats.
Identities = 40/123 (32%), Positives = 59/123 (47%), Gaps = 6/123 (4%)
Query 1 SATLPPSVVLLAESIAYGAVHLSVGRASAAAPQIEQELVFCTTEAGKLWALKTLRLERRL 60
SAT P V LAE V +++G +A ++V E K K ++L +
Sbjct 280 SATWPKEVRQLAEDFLQDYVQINIGALELSANHNILQIVDVCMENEK--DNKLIQLMEEI 337
Query 61 IP----PCLIFVETQERASELLKEMLSDGMPVDLLHAAKSKQQRNATVDAFRIGKIWFLI 116
+ +IFVET++R EL + M DG P +H KS+ +R+ + FR GK LI
Sbjct 338 MAEKENKTIIFVETKKRCDELTRRMRRDGWPAMCIHGDKSQPERDWVLTEFRSGKAPILI 397
Query 117 CTD 119
TD
Sbjct 398 ATD 400
> pfa:PFE0925c snrnp protein, putative; K12858 ATP-dependent RNA
helicase DDX23/PRP28 [EC:3.6.4.13]
Length=1123
Score = 54.7 bits (130), Expect = 7e-08, Method: Composition-based stats.
Identities = 35/119 (29%), Positives = 56/119 (47%), Gaps = 3/119 (2%)
Query 1 SATLPPSVVLLAESIAYGAVHLSVGRASAAAPQIEQELVFCTTEAGKLWALKTLRLERRL 60
SAT+PPSV L+ ++S+G A IEQ+L F T K + L +
Sbjct 910 SATMPPSVERLSRKYLRAPAYISIGDPGAGKRSIEQKLEFLTEGKKKQKLQEILEMYE-- 967
Query 61 IPPCLIFVETQERASELLKEMLSDGMPVDLLHAAKSKQQRNATVDAFRIGKIWFLICTD 119
PP ++FV ++ A + K + LH K+++ R T+ AF+ + L+ TD
Sbjct 968 -PPIIVFVNQKKVADIISKSITKMKYKAVALHGGKAQEIREQTLSAFKNAEFDILVATD 1025
> mmu:67040 Ddx17, 2610007K22Rik, A430025E01Rik, AI047725, C80929,
Gm926, MGC79147, p72; DEAD (Asp-Glu-Ala-Asp) box polypeptide
17 (EC:3.6.4.13); K13178 ATP-dependent RNA helicase DDX17
[EC:3.6.4.13]
Length=650
Score = 54.7 bits (130), Expect = 7e-08, Method: Composition-based stats.
Identities = 39/123 (31%), Positives = 59/123 (47%), Gaps = 6/123 (4%)
Query 1 SATLPPSVVLLAESIAYGAVHLSVGRASAAAPQIEQELVFCTTEAGKLWALKTLRLERRL 60
SAT P V LAE ++VG +A ++V E+ K K ++L +
Sbjct 277 SATWPKEVRQLAEDFLRDYTQINVGNLELSANHNILQIVDVCMESEK--DHKLIQLMEEI 334
Query 61 IP----PCLIFVETQERASELLKEMLSDGMPVDLLHAAKSKQQRNATVDAFRIGKIWFLI 116
+ +IFVET+ R +L + M DG P +H KS+ +R+ ++ FR GK LI
Sbjct 335 MAEKENKTIIFVETKRRCDDLTRRMRRDGWPAMCIHGDKSQPERDWVLNEFRSGKAPILI 394
Query 117 CTD 119
TD
Sbjct 395 ATD 397
> dre:322206 ddx5, wu:fa56a07, wu:fb11e01, wu:fb16c10, wu:fb53b05;
DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 (EC:3.6.1.-);
K12823 ATP-dependent RNA helicase DDX5/DBP2 [EC:3.6.4.13]
Length=518
Score = 54.3 bits (129), Expect = 1e-07, Method: Composition-based stats.
Identities = 38/123 (30%), Positives = 60/123 (48%), Gaps = 6/123 (4%)
Query 1 SATLPPSVVLLAESIAYGAVHLSVGRASAAAPQIEQELVFCTTEAGKLWALKTLRLERRL 60
SAT P V LAE + ++VG +A ++V + K K +RL +
Sbjct 281 SATWPKEVRQLAEDFLKEYIQINVGALQLSANHNILQIVDVCNDGEK--EDKLIRLLEEI 338
Query 61 IP----PCLIFVETQERASELLKEMLSDGMPVDLLHAAKSKQQRNATVDAFRIGKIWFLI 116
+ +IFVET+ R +L + M DG P +H K++Q+R+ ++ F+ GK LI
Sbjct 339 MSEKENKTIIFVETKRRCDDLTRRMRRDGWPAMGIHGDKNQQERDWVLNEFKYGKAPILI 398
Query 117 CTD 119
TD
Sbjct 399 ATD 401
> hsa:10521 DDX17, DKFZp761H2016, P72, RH70; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 17 (EC:3.6.4.13); K13178 ATP-dependent
RNA helicase DDX17 [EC:3.6.4.13]
Length=731
Score = 53.9 bits (128), Expect = 1e-07, Method: Composition-based stats.
Identities = 39/123 (31%), Positives = 59/123 (47%), Gaps = 6/123 (4%)
Query 1 SATLPPSVVLLAESIAYGAVHLSVGRASAAAPQIEQELVFCTTEAGKLWALKTLRLERRL 60
SAT P V LAE ++VG +A ++V E+ K K ++L +
Sbjct 356 SATWPKEVRQLAEDFLRDYTQINVGNLELSANHNILQIVDVCMESEK--DHKLIQLMEEI 413
Query 61 IP----PCLIFVETQERASELLKEMLSDGMPVDLLHAAKSKQQRNATVDAFRIGKIWFLI 116
+ +IFVET+ R +L + M DG P +H KS+ +R+ ++ FR GK LI
Sbjct 414 MAEKENKTIIFVETKRRCDDLTRRMRRDGWPAMCIHGDKSQPERDWVLNEFRSGKAPILI 473
Query 117 CTD 119
TD
Sbjct 474 ATD 476
> pfa:PFE1390w RNA helicase-1; K13116 ATP-dependent RNA helicase
DDX41 [EC:3.6.4.13]
Length=665
Score = 53.9 bits (128), Expect = 1e-07, Method: Composition-based stats.
Identities = 34/119 (28%), Positives = 60/119 (50%), Gaps = 3/119 (2%)
Query 1 SATLPPSVVLLAESIAYGAVHLSVGRASAAAPQIEQELVFCTTEAGKLWALKTLRLERRL 60
SAT+P + A+S + ++VGRA AA + QE+ + E + L+ L+ +
Sbjct 411 SATMPKKIQEFAKSTLVNPIIINVGRAGAANLDVIQEVEYVKEEFKLSYLLEVLQ---KT 467
Query 61 IPPCLIFVETQERASELLKEMLSDGMPVDLLHAAKSKQQRNATVDAFRIGKIWFLICTD 119
PP LIF E ++ ++ + +L G+ +H + +R ++ FR GK L+ TD
Sbjct 468 GPPVLIFCENKKDVDDVHEYLLLKGVNAVAIHGNLGQSERQEAINLFREGKKDILVGTD 526
> sce:YDR243C PRP28; Prp28p (EC:3.6.1.-); K12858 ATP-dependent
RNA helicase DDX23/PRP28 [EC:3.6.4.13]
Length=588
Score = 53.9 bits (128), Expect = 1e-07, Method: Composition-based stats.
Identities = 31/120 (25%), Positives = 62/120 (51%), Gaps = 3/120 (2%)
Query 1 SATLPPSVVLLAESIAYGAVHLSVGRASAAAPQIEQELVFCTTEAGKLWALKTLRLERRL 60
+AT+ P + +A V+ ++G + + P I+Q + + + K LK + +
Sbjct 378 TATMTPVIEKIAAGYMQKPVYATIGVETGSEPLIQQVVEYADNDEDKFKKLKPI--VAKY 435
Query 61 IPPCLIFVETQERASELLKEMLSD-GMPVDLLHAAKSKQQRNATVDAFRIGKIWFLICTD 119
PP +IF+ ++ A L ++ + M V +LH +KS++QR ++ FR K+ +I T+
Sbjct 436 DPPIIIFINYKQTADWLAEKFQKETNMKVTILHGSKSQEQREHSLQLFRTNKVQIMIATN 495
> cel:F58E10.3 hypothetical protein; K12823 ATP-dependent RNA
helicase DDX5/DBP2 [EC:3.6.4.13]
Length=561
Score = 53.5 bits (127), Expect = 2e-07, Method: Composition-based stats.
Identities = 39/121 (32%), Positives = 55/121 (45%), Gaps = 2/121 (1%)
Query 1 SATLPPSVVLLAESIAYGAVHLSVGRASAAAPQIEQELVFCTTEAGKLWALKTLRLERRL 60
SAT P V LA A L+VG AA ++V E K L L
Sbjct 314 SATWPKEVRALASDFQKDAAFLNVGSLELAANHNITQVVDILEEHAKQAKLMELLNHIMN 373
Query 61 IPPC--LIFVETQERASELLKEMLSDGMPVDLLHAAKSKQQRNATVDAFRIGKIWFLICT 118
C +IFVET+ +A EL + M DG P +H K++ +R+ + F+ GK ++ T
Sbjct 374 QKECKTIIFVETKRKADELTRAMRRDGWPTLCIHGDKNQGERDWVLQEFKAGKTPIMLAT 433
Query 119 D 119
D
Sbjct 434 D 434
> bbo:BBOV_IV001840 21.m02846; DEAD/DEAH box helicase (EC:3.6.1.-);
K11594 ATP-dependent RNA helicase [EC:3.6.4.13]
Length=609
Score = 53.5 bits (127), Expect = 2e-07, Method: Composition-based stats.
Identities = 35/119 (29%), Positives = 56/119 (47%), Gaps = 2/119 (1%)
Query 1 SATLPPSVVLLAESIAYGAVHLSVGRASAAAPQIEQELVFCTTEAGKLWALKTLRLERRL 60
SAT P + LA ++L+VGR + I Q L++ E + +K LR
Sbjct 347 SATFPKEIQQLARDFLRDYLYLAVGRVGSTNEFIRQRLLYADQEQKLHYLVKLLRENTNG 406
Query 61 IPPCLIFVETQERASELLKEMLSDGMPVDLLHAAKSKQQRNATVDAFRIGKIWFLICTD 119
+ LIFVET+ RA + +L + +H +S+Q R + F+ G+ L+ TD
Sbjct 407 L--VLIFVETKRRADMIESYLLKENFMAVNIHGDRSQQDREEALRLFKTGERPILVATD 463
> ath:AT2G33730 DEAD box RNA helicase, putative; K12858 ATP-dependent
RNA helicase DDX23/PRP28 [EC:3.6.4.13]
Length=733
Score = 53.1 bits (126), Expect = 2e-07, Method: Composition-based stats.
Identities = 35/119 (29%), Positives = 58/119 (48%), Gaps = 2/119 (1%)
Query 1 SATLPPSVVLLAESIAYGAVHLSVGRASAAAPQIEQELVFCTTEAGKLWALKTLRLERRL 60
SAT+PP V LA V +++G A I Q ++ E+ K + L+ L L+
Sbjct 518 SATMPPGVERLARKYLRNPVVVTIGTAGKTTDLISQHVIM-MKESEKFFRLQKL-LDELG 575
Query 61 IPPCLIFVETQERASELLKEMLSDGMPVDLLHAAKSKQQRNATVDAFRIGKIWFLICTD 119
++FV T++ + K + G V LH KS++QR +++ FR + L+ TD
Sbjct 576 EKTAIVFVNTKKNCDSIAKNLDKAGYRVTTLHGGKSQEQREISLEGFRAKRYNVLVATD 634
> cpv:cgd4_3000 Dbp1p, eIF4a-1 family RNA SFII helicase (DEXDC+HELICc)
; K11594 ATP-dependent RNA helicase [EC:3.6.4.13]
Length=702
Score = 52.8 bits (125), Expect = 3e-07, Method: Composition-based stats.
Identities = 32/119 (26%), Positives = 58/119 (48%), Gaps = 2/119 (1%)
Query 1 SATLPPSVVLLAESIAYGAVHLSVGRASAAAPQIEQELVFCTTEAGKLWALKTLRLERRL 60
SAT P + LA+ + + L+VGR A + I Q +V+ + +K L +
Sbjct 401 SATFPREIQQLAKDFLHNYIFLTVGRVGATSGSIVQRVVYAEEDHKPRLLVKLLLEQGEG 460
Query 61 IPPCLIFVETQERASELLKEMLSDGMPVDLLHAAKSKQQRNATVDAFRIGKIWFLICTD 119
+ ++FVE + RA ++ ++ P +H +S+Q+R + FR G+ L+ TD
Sbjct 461 L--TVVFVEMKRRADQIEDFLIDQNFPAVSIHGDRSQQEREHALRLFRSGQRPILVATD 517
> ath:AT3G58570 DEAD box RNA helicase, putative; K11594 ATP-dependent
RNA helicase [EC:3.6.4.13]
Length=646
Score = 52.0 bits (123), Expect = 4e-07, Method: Composition-based stats.
Identities = 33/124 (26%), Positives = 56/124 (45%), Gaps = 5/124 (4%)
Query 1 SATLPPSVVLLAESIAYGAVHLSVGRASAAAPQIEQELVFCTTEAGKLWALKTLRLERR- 59
SAT P + LA + L+VGR ++ I Q + F + + L +R
Sbjct 340 SATFPREIQRLASDFLSNYIFLAVGRVGSSTDLIVQRVEFVHDSDKRSHLMDLLHAQREN 399
Query 60 ----LIPPCLIFVETQERASELLKEMLSDGMPVDLLHAAKSKQQRNATVDAFRIGKIWFL 115
L+FVET++ A L + +G P +H +S+Q+R + +F+ G+ L
Sbjct 400 GNQGKQALTLVFVETKKGADSLENWLCINGFPATTIHGDRSQQEREVALRSFKTGRTPIL 459
Query 116 ICTD 119
+ TD
Sbjct 460 VATD 463
> cpv:cgd7_4600 abstrakt protein SF II helicase + Znknuckle C2HC
(PA) ; K13116 ATP-dependent RNA helicase DDX41 [EC:3.6.4.13]
Length=570
Score = 52.0 bits (123), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 34/119 (28%), Positives = 61/119 (51%), Gaps = 3/119 (2%)
Query 1 SATLPPSVVLLAESIAYGAVHLSVGRASAAAPQIEQELVFCTTEAGKLWALKTLRLERRL 60
SAT+P A++ V ++VGRA AA ++ QE + E + L L+ +
Sbjct 317 SATMPRKTQEFAQTALIDPVVVNVGRAGAANLRVIQEFEYVRQERRLVSLLSCLQ---KT 373
Query 61 IPPCLIFVETQERASELLKEMLSDGMPVDLLHAAKSKQQRNATVDAFRIGKIWFLICTD 119
P LIF E ++ E+ + +L G+ +H +++QR +++ FR G++ L+ TD
Sbjct 374 APRVLIFSENKKDVDEIHEYLLLKGVNAVAIHGGLTQEQRFRSIEKFRNGEMDVLVGTD 432
> pfa:PF14_0437 helicase, putative
Length=527
Score = 51.6 bits (122), Expect = 6e-07, Method: Composition-based stats.
Identities = 38/126 (30%), Positives = 66/126 (52%), Gaps = 12/126 (9%)
Query 1 SATLPPSVVLLAESIAY-GAVHLSVGRASAAAPQIEQELVFCTTEAGKLWALKTLRLER- 58
SAT P V LA+ + + ++VG + A + ++ ++ E K+ LK+L L+R
Sbjct 298 SATWPKEVQALAKDLCKEQPIQVNVGSLTLTACRSIKQEIYLLEEHEKIGNLKSL-LQRI 356
Query 59 -----RLIPPCLIFVETQERASELLKEMLSDGMPVDLLHAAKSKQQRNATVDAFRIGKIW 113
R+I +FVET++ A + K + DGMP +H K +++R ++ F+ GK
Sbjct 357 FKDNDRII----VFVETKKNADFITKALRLDGMPALCIHGDKKQEERRWVLNEFKTGKSP 412
Query 114 FLICTD 119
+I TD
Sbjct 413 IMIATD 418
> cpv:cgd2_1010 hypothetical protein ; K12823 ATP-dependent RNA
helicase DDX5/DBP2 [EC:3.6.4.13]
Length=586
Score = 50.1 bits (118), Expect = 2e-06, Method: Composition-based stats.
Identities = 41/124 (33%), Positives = 59/124 (47%), Gaps = 6/124 (4%)
Query 1 SATLPPSVVLLAESIAYG-AVHLSVGRASA--AAPQIEQELVFCTTEAGKLWALKTL--R 55
SAT P V LA + +H++VG A A+ I+Q V E+ K LK +
Sbjct 353 SATWPKEVQKLARDLCKEIPIHINVGSVDALKASHNIKQ-YVNVVEESEKKAKLKMFLGQ 411
Query 56 LERRLIPPCLIFVETQERASELLKEMLSDGMPVDLLHAAKSKQQRNATVDAFRIGKIWFL 115
+ P LIF ET+ A L KE+ DG P +H K +++R ++ FR G +
Sbjct 412 VMVESAPKVLIFCETKRGADILTKELRLDGWPALCIHGDKKQEERTWVLNEFRTGASPIM 471
Query 116 ICTD 119
I TD
Sbjct 472 IATD 475
> cpv:cgd3_3690 U5 snRNP 100 kD protein ; K12858 ATP-dependent
RNA helicase DDX23/PRP28 [EC:3.6.4.13]
Length=529
Score = 50.1 bits (118), Expect = 2e-06, Method: Composition-based stats.
Identities = 28/119 (23%), Positives = 58/119 (48%), Gaps = 0/119 (0%)
Query 1 SATLPPSVVLLAESIAYGAVHLSVGRASAAAPQIEQELVFCTTEAGKLWALKTLRLERRL 60
SAT+ + +A+ +++++G A I+Q L F + K + TL +
Sbjct 336 SATMQKELENIAKRYLNSPINVTIGDIGAGKKSIQQILNFISENKKKSTLINTLNNKELA 395
Query 61 IPPCLIFVETQERASELLKEMLSDGMPVDLLHAAKSKQQRNATVDAFRIGKIWFLICTD 119
+PP ++F+ ++ + +E++S G LH K ++ R +++ F+ G L+ TD
Sbjct 396 VPPIIVFLNQKKMVDIVCREIVSHGFKATSLHGGKMQEVRENSLNLFKSGVFDILVSTD 454
> tgo:TGME49_026250 ATP-dependent RNA helicase, putative ; K11594
ATP-dependent RNA helicase [EC:3.6.4.13]
Length=734
Score = 50.1 bits (118), Expect = 2e-06, Method: Composition-based stats.
Identities = 32/119 (26%), Positives = 58/119 (48%), Gaps = 2/119 (1%)
Query 1 SATLPPSVVLLAESIAYGAVHLSVGRASAAAPQIEQELVFCTTEAGKLWALKTLRLERRL 60
SAT P + +LA+ ++L+VGR + I Q L + + +K LR +
Sbjct 453 SATFPREIQMLAKDFLEDYIYLAVGRVGSTNEFIRQRLQYADEDQKLKLLVKLLRETEKG 512
Query 61 IPPCLIFVETQERASELLKEMLSDGMPVDLLHAAKSKQQRNATVDAFRIGKIWFLICTD 119
+ +IFVET+ +A + ++ D P +H +++Q+R + F+ K L+ TD
Sbjct 513 L--TIIFVETKRKADMIEDYLVDDDFPAVSIHGDRTQQEREEALRLFKAAKCPILVATD 569
> bbo:BBOV_II004470 18.m06373; p68-like protein; K12823 ATP-dependent
RNA helicase DDX5/DBP2 [EC:3.6.4.13]
Length=529
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 36/120 (30%), Positives = 56/120 (46%), Gaps = 1/120 (0%)
Query 1 SATLPPSVVLLAESIAYGA-VHLSVGRASAAAPQIEQELVFCTTEAGKLWALKTLRLERR 59
SAT P V LA + VH++VG + VF E K LK + +
Sbjct 293 SATWPREVQSLAHDLCREEPVHINVGSLDLKTCHNVSQEVFVIEEHEKRSQLKKILGQIG 352
Query 60 LIPPCLIFVETQERASELLKEMLSDGMPVDLLHAAKSKQQRNATVDAFRIGKIWFLICTD 119
LIF +T++ A + KE+ DG P +H K +++RN ++ F+ GK ++ TD
Sbjct 353 QGTKILIFTDTKKTADSITKELRLDGWPALSIHGDKKQEERNWVLNEFKSGKHPIMVATD 412
> hsa:9416 DDX23, MGC8416, PRPF28, U5-100K, U5-100KD, prp28; DEAD
(Asp-Glu-Ala-Asp) box polypeptide 23 (EC:3.6.4.13); K12858
ATP-dependent RNA helicase DDX23/PRP28 [EC:3.6.4.13]
Length=820
Score = 50.1 bits (118), Expect = 2e-06, Method: Composition-based stats.
Identities = 36/119 (30%), Positives = 57/119 (47%), Gaps = 2/119 (1%)
Query 1 SATLPPSVVLLAESIAYGAVHLSVGRASAAAPQIEQELVFCTTEAGKLWALKTLRLERRL 60
+AT+PP+V LA S + +G A ++EQ+ VF +E+ K L + LE+
Sbjct 606 TATMPPAVERLARSYLRRPAVVYIGSAGKPHERVEQK-VFLMSESEKRKKLLAI-LEQGF 663
Query 61 IPPCLIFVETQERASELLKEMLSDGMPVDLLHAAKSKQQRNATVDAFRIGKIWFLICTD 119
PP +IFV ++ L K + G LH K ++QR + + G L+ TD
Sbjct 664 DPPIIIFVNQKKGCDVLAKSLEKMGYNACTLHGGKGQEQREFALSNLKAGAKDILVATD 722
> mmu:74351 Ddx23, 3110082M05Rik, 4921506D17Rik; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 23 (EC:3.6.1.-); K12858 ATP-dependent
RNA helicase DDX23/PRP28 [EC:3.6.4.13]
Length=819
Score = 50.1 bits (118), Expect = 2e-06, Method: Composition-based stats.
Identities = 36/119 (30%), Positives = 57/119 (47%), Gaps = 2/119 (1%)
Query 1 SATLPPSVVLLAESIAYGAVHLSVGRASAAAPQIEQELVFCTTEAGKLWALKTLRLERRL 60
+AT+PP+V LA S + +G A ++EQ+ VF +E+ K L + LE+
Sbjct 605 TATMPPAVERLARSYLRRPAVVYIGSAGKPHERVEQK-VFLMSESEKRKKLLAI-LEQGF 662
Query 61 IPPCLIFVETQERASELLKEMLSDGMPVDLLHAAKSKQQRNATVDAFRIGKIWFLICTD 119
PP +IFV ++ L K + G LH K ++QR + + G L+ TD
Sbjct 663 DPPIIIFVNQKKGCDVLAKSLEKMGYNACTLHGGKGQEQREFALSNLKAGAKDILVATD 721
> tgo:TGME49_036650 DEAD/DEAH box helicase, putative (EC:5.99.1.3);
K12823 ATP-dependent RNA helicase DDX5/DBP2 [EC:3.6.4.13]
Length=550
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 39/122 (31%), Positives = 59/122 (48%), Gaps = 5/122 (4%)
Query 1 SATLPPSVVLLAESI-AYGAVHLSVGRASAAAPQIEQELVFCTTEAGKLWALKTLRLERR 59
SAT P V LA + VH++VG A Q ++ V E K L + L RR
Sbjct 314 SATWPKEVQNLARDLCKEEPVHINVGSLDLQACQNIKQEVMVVQEYEKRGQL--MSLLRR 371
Query 60 LI--PPCLIFVETQERASELLKEMLSDGMPVDLLHAAKSKQQRNATVDAFRIGKIWFLIC 117
++ LIF ET+ A L ++M +G P LH K +++R +D F+ G+ ++
Sbjct 372 IMDGSKILIFAETKRGADNLTRDMRVEGWPALSLHGDKKQEERTWVLDEFKNGRNPIMVA 431
Query 118 TD 119
TD
Sbjct 432 TD 433
> ath:AT2G42520 DEAD box RNA helicase, putative; K11594 ATP-dependent
RNA helicase [EC:3.6.4.13]
Length=633
Score = 50.1 bits (118), Expect = 2e-06, Method: Composition-based stats.
Identities = 33/124 (26%), Positives = 55/124 (44%), Gaps = 5/124 (4%)
Query 1 SATLPPSVVLLAESIAYGAVHLSVGRASAAAPQIEQELVFCTTEAGKLWALKTLRLERR- 59
SAT P + LA + L+VGR ++ I Q + F + + L +R
Sbjct 353 SATFPREIQRLAADFLANYIFLAVGRVGSSTDLIVQRVEFVLDSDKRSHLMDLLHAQREN 412
Query 60 ----LIPPCLIFVETQERASELLKEMLSDGMPVDLLHAAKSKQQRNATVDAFRIGKIWFL 115
L+FVET+ A L + +G P +H +++Q+R + AF+ G+ L
Sbjct 413 GIQGKQALTLVFVETKRGADSLENWLCINGFPATSIHGDRTQQEREVALKAFKSGRTPIL 472
Query 116 ICTD 119
+ TD
Sbjct 473 VATD 476
Lambda K H
0.322 0.134 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2022937320
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40