bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0670_orf2
Length=149
Score E
Sequences producing significant alignments: (Bits) Value
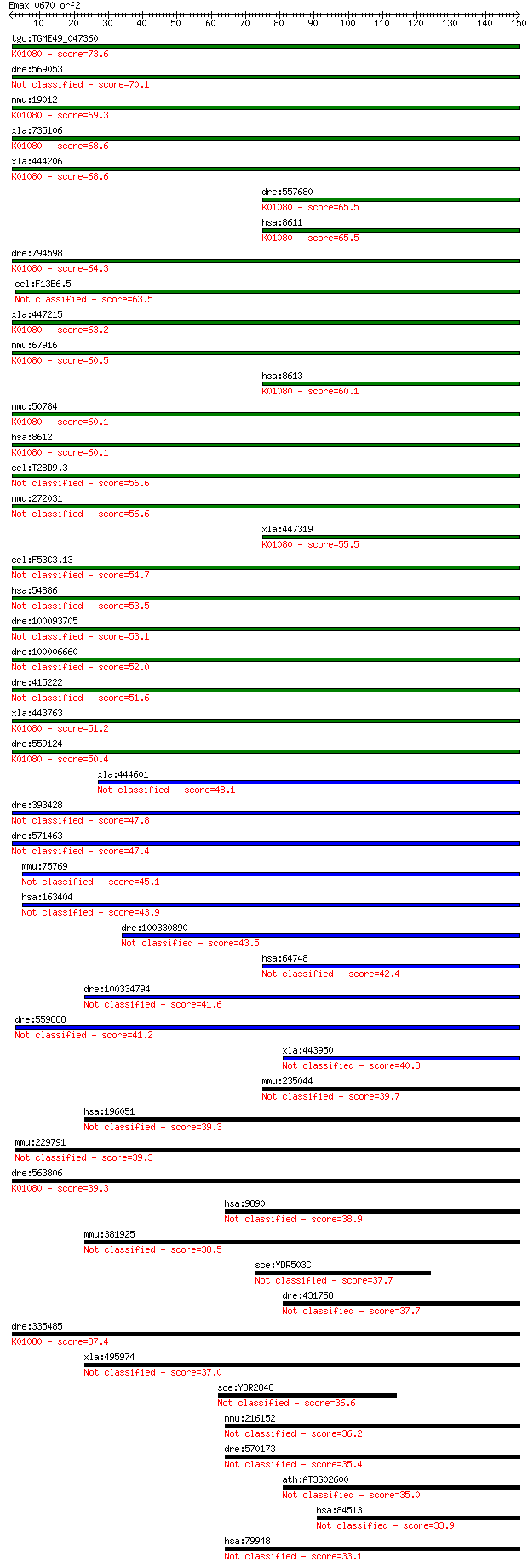
tgo:TGME49_047360 lipid phosphate phosphohydrolase 3, putative... 73.6 2e-13
dre:569053 hypothetical LOC569053 70.1 3e-12
mmu:19012 Ppap2a, Hic53, Hpic53, LPP-1, LPP1, mPAP; phosphatid... 69.3 4e-12
xla:735106 ppap2a, MGC114881; phosphatidic acid phosphatase ty... 68.6 7e-12
xla:444206 MGC80748 protein; K01080 phosphatidate phosphatase ... 68.6 8e-12
dre:557680 ppap2b, si:dkey-19f4.1; phosphatidic acid phosphata... 65.5 5e-11
hsa:8611 PPAP2A, LLP1a, LPP1, PAP-2a, PAP2, PAP2a2, PAP2alpha2... 65.5 6e-11
dre:794598 Lipid phosphate phosphohydrolase 1-like; K01080 pho... 64.3 1e-10
cel:F13E6.5 hypothetical protein 63.5 2e-10
xla:447215 ppap2b-b, dri42, lpp3, pap2b, ppap2bb, vcip; phosph... 63.2 3e-10
mmu:67916 Ppap2b, 1110003O22Rik, 2610002D05Rik, AV025606, D4Bw... 60.5 2e-09
hsa:8613 PPAP2B, Dri42, LPP3, MGC15306, PAP2B, VCIP; phosphati... 60.1 3e-09
mmu:50784 Ppap2c, Lpp2; phosphatidic acid phosphatase type 2C ... 60.1 3e-09
hsa:8612 PPAP2C, LPP2, PAP-2c, PAP2-g; phosphatidic acid phosp... 60.1 3e-09
cel:T28D9.3 hypothetical protein 56.6 3e-08
mmu:272031 E130309F12Rik, KIAA4247, Lppr1, PRG-3, mKIAA4247; R... 56.6 3e-08
xla:447319 ppap2b-a, dri42, lpp3, pap2b, ppap2b, ppap2ba, vcip... 55.5 7e-08
cel:F53C3.13 hypothetical protein 54.7 1e-07
hsa:54886 LPPR1, MGC26189, PRG-3; lipid phosphate phosphatase-... 53.5 2e-07
dre:100093705 zgc:165526 53.1 3e-07
dre:100006660 similar to phosphatidic acid phosphatase type 2d 52.0 7e-07
dre:415222 lppr1; zgc:86759 51.6 8e-07
xla:443763 ppap2c, MGC81169; phosphatidic acid phosphatase typ... 51.2 1e-06
dre:559124 MGC158309, ppap2b; zgc:158309 (EC:3.1.3.4); K01080 ... 50.4 2e-06
xla:444601 lppr5, MGC84075; lipid phosphate phosphatase-relate... 48.1 1e-05
dre:393428 MGC63577; zgc:63577 (EC:3.1.3.4) 47.8 1e-05
dre:571463 lipid phosphate phosphatase-related protein type 2-... 47.4 2e-05
mmu:75769 4833424O15Rik, Lppr5, PRG-5, Pap2d; RIKEN cDNA 48334... 45.1 8e-05
hsa:163404 LPPR5, PAP2, PAP2D, PRG5; lipid phosphate phosphata... 43.9 2e-04
dre:100330890 lipid phosphate phosphatase-related protein type... 43.5 2e-04
hsa:64748 LPPR2, DKFZp761E1121, FLJ13055, PRG4; lipid phosphat... 42.4 5e-04
dre:100334794 phosphatidic acid phosphatase type 2C-like 41.6 0.001
dre:559888 si:dkey-108c13.1 41.2 0.001
xla:443950 ppapdc1b, MGC80318; phosphatidic acid phosphatase t... 40.8 0.002
mmu:235044 Lppr2, MGC25492, PRG-4; cDNA sequence BC018242 (EC:... 39.7 0.003
hsa:196051 PPAPDC1A, DPPL2, MGC120299, MGC120300, PPAPDC1; pho... 39.3 0.004
mmu:229791 D3Bwg0562e, A330086D10, Lppr4, PRG-1, mKIAA0455; DN... 39.3 0.005
dre:563806 Lipid phosphate phosphohydrolase 2-like; K01080 pho... 39.3 0.005
hsa:9890 LPPR4, KIAA0455, LPR4, PHP1, PRG-1, PRG1, RP4-788L13.... 38.9 0.006
mmu:381925 Ppapdc1a, C030048B12Rik, Gm1090; phosphatidic acid ... 38.5 0.007
sce:YDR503C LPP1; Lipid phosphate phosphatase, catalyzes Mg(2+... 37.7 0.013
dre:431758 ppapdc1b, fc49c04, wu:fc49c04, zgc:92365; phosphati... 37.7 0.014
dre:335485 ppap2c, fj16e10, wu:fj16e10, zgc:66434; phosphatidi... 37.4 0.019
xla:495974 ppapdc1a; phosphatidic acid phosphatase type 2 doma... 37.0 0.026
sce:YDR284C DPP1, ZRG1; Diacylglycerol pyrophosphate (DGPP) ph... 36.6 0.034
mmu:216152 KIAA4076, Lppr3, Prg2; cDNA sequence BC005764 (EC:3... 36.2 0.037
dre:570173 lipid phosphate phosphatase-related protein type 3a... 35.4 0.069
ath:AT3G02600 LPP3; LPP3 (LIPID PHOSPHATE PHOSPHATASE 3); phos... 35.0 0.083
hsa:84513 PPAPDC1B, DPPL1, HTPAP; phosphatidic acid phosphatas... 33.9 0.22
hsa:79948 LPPR3, LPR3, PRG-2, PRG2; lipid phosphate phosphatas... 33.1 0.32
> tgo:TGME49_047360 lipid phosphate phosphohydrolase 3, putative
(EC:3.1.3.4); K01080 phosphatidate phosphatase [EC:3.1.3.4]
Length=320
Score = 73.6 bits (179), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 55/150 (36%), Positives = 81/150 (54%), Gaps = 13/150 (8%)
Query 2 FCGEDDIALPYMPASISAAGAFWISTMLPAFIIVVVEVLLWAVRATQEKDKSEREQQAVV 61
FC +D I P +P ++ A A I ++P +I+VV+ + W + ER + V
Sbjct 59 FCNDDSIRFPLLPQTVPAFEASLIILLIPLVLIIVVDTVSWVMFG-------ERFGRCVD 111
Query 62 VVLDRRIPETCVHLYTYC-GSLAMSVASVFLLTNSLKAAVGSLRPHFLDVCRPDWSRVSC 120
+ R P V LY G A ++ S + +T + K VG LRPHFL VC+PDWSR++C
Sbjct 112 LGFCR--PSGVVALYYQSFGGFAFALLSCYAITLTAKICVGRLRPHFLSVCQPDWSRIAC 169
Query 121 RDSSGLYDVYIPDFFCTG-DPHRVEEARRS 149
D++G +YI F C G D ++EAR S
Sbjct 170 SDANGF--LYIDKFECLGTDKAAIKEARVS 197
> dre:569053 hypothetical LOC569053
Length=303
Score = 70.1 bits (170), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 49/155 (31%), Positives = 65/155 (41%), Gaps = 34/155 (21%)
Query 2 FCGEDDIALPYMPASISAA--GAFWISTMLPAFIIVVVEVLLWAVRATQEKDKSEREQQA 59
FC +D I PY +IS G I L II +
Sbjct 61 FCNDDSIKYPYKEDTISYQLLGGVMIPVTLLTMII----------------------GEC 98
Query 60 VVVVLDRRIPETCVHLYTYC-----GSLAMSVASVFLLTNSLKAAVGSLRPHFLDVCRPD 114
+ V L+R + + Y C G+ A LT+ K ++G LRPHFLDVC+PD
Sbjct 99 LSVYLNRIKSNSFCNGYVACVYKAIGTFVFGAAISQSLTDIAKYSIGRLRPHFLDVCKPD 158
Query 115 WSRVSCRDSSGLYDVYIPDFFCTGDPHRVEEARRS 149
WS+++C + YI DF CTG V E R S
Sbjct 159 WSKINCTAGA-----YIEDFVCTGKESVVNEGRLS 188
> mmu:19012 Ppap2a, Hic53, Hpic53, LPP-1, LPP1, mPAP; phosphatidic
acid phosphatase type 2A (EC:3.1.3.4); K01080 phosphatidate
phosphatase [EC:3.1.3.4]
Length=284
Score = 69.3 bits (168), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 41/148 (27%), Positives = 64/148 (43%), Gaps = 20/148 (13%)
Query 2 FCGEDDIALPYMPASISAAGAFWISTMLPAFIIVVVEVLLWAVRATQEKDKSEREQQAVV 61
FC ++ + PY ++I + + LP F + + E L A +
Sbjct 40 FCTDNSVKYPYHDSTIPSRILAILGLGLPIFSMSIGESLSVYFNVLHSNSFVGNPYIATI 99
Query 62 VVLDRRIPETCVHLYTYCGSLAMSVASVFLLTNSLKAAVGSLRPHFLDVCRPDWSRVSCR 121
Y G+ V++ LT+ K +GSLRPHFL +C PDWS+++C
Sbjct 100 --------------YKAVGAFLFGVSASQSLTDIAKYTIGSLRPHFLAICNPDWSKINCS 145
Query 122 DSSGLYDVYIPDFFCTGDPHRVEEARRS 149
D YI D+ C G+ +V+E R S
Sbjct 146 DG------YIEDYICQGNEEKVKEGRLS 167
> xla:735106 ppap2a, MGC114881; phosphatidic acid phosphatase
type 2A (EC:3.1.3.4); K01080 phosphatidate phosphatase [EC:3.1.3.4]
Length=283
Score = 68.6 bits (166), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 45/148 (30%), Positives = 67/148 (45%), Gaps = 20/148 (13%)
Query 2 FCGEDDIALPYMPASISAAGAFWISTMLPAFIIVVVEVLLWAVRATQEKDKSEREQQAVV 61
FC +D + PY +IS M+P IIV++ +V + + + V
Sbjct 39 FCNDDSLWYPYKEDTISY--GLLGGIMIPFCIIVIILGEALSVFYSDLRSGAFIRNNYVA 96
Query 62 VVLDRRIPETCVHLYTYCGSLAMSVASVFLLTNSLKAAVGSLRPHFLDVCRPDWSRVSCR 121
+ Y G+ A LT+ K +G LRPHFLDVC+P+W++++C
Sbjct 97 TI------------YKAIGTFIFGAAVSQSLTDIAKYTIGRLRPHFLDVCKPNWAKINC- 143
Query 122 DSSGLYDVYIPDFFCTGDPHRVEEARRS 149
S G YI +F C GDP + E R S
Sbjct 144 -SLG----YIENFVCEGDPTKSSEGRLS 166
> xla:444206 MGC80748 protein; K01080 phosphatidate phosphatase
[EC:3.1.3.4]
Length=284
Score = 68.6 bits (166), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 42/148 (28%), Positives = 69/148 (46%), Gaps = 20/148 (13%)
Query 2 FCGEDDIALPYMPASISAAGAFWISTMLPAFIIVVVEVLLWAVRATQEKDKSEREQQAVV 61
FC +D I P+ +++++ + + +P +++ E L +V + + V
Sbjct 40 FCDDDSIKYPFHDSTVTSTVLYTVGFTVPICSMILGETL--SVVYNDLRSSAFIRNNYVA 97
Query 62 VVLDRRIPETCVHLYTYCGSLAMSVASVFLLTNSLKAAVGSLRPHFLDVCRPDWSRVSCR 121
+ Y G+ A+ LT+ K +G LRPHFLDVC+P+WS+++C
Sbjct 98 TI------------YKAIGTFIFGAAASQSLTDIAKYTIGRLRPHFLDVCKPNWSKINC- 144
Query 122 DSSGLYDVYIPDFFCTGDPHRVEEARRS 149
S G YI F C GDP + E R S
Sbjct 145 -SLG----YIETFVCEGDPTKSSEGRLS 167
> dre:557680 ppap2b, si:dkey-19f4.1; phosphatidic acid phosphatase
type 2B; K01080 phosphatidate phosphatase [EC:3.1.3.4]
Length=312
Score = 65.5 bits (158), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 34/75 (45%), Positives = 45/75 (60%), Gaps = 6/75 (8%)
Query 75 LYTYCGSLAMSVASVFLLTNSLKAAVGSLRPHFLDVCRPDWSRVSCRDSSGLYDVYIPDF 134
LY G A T+ K +VG +RPHFLDVCRP++S + C S G YI ++
Sbjct 126 LYRQVGVFIFGCAVSQSFTDIAKVSVGRMRPHFLDVCRPNYSTIDC--SLG----YITEY 179
Query 135 FCTGDPHRVEEARRS 149
CTGDP +V+EAR+S
Sbjct 180 TCTGDPSKVQEARKS 194
> hsa:8611 PPAP2A, LLP1a, LPP1, PAP-2a, PAP2, PAP2a2, PAP2alpha2,
PAPalpha1; phosphatidic acid phosphatase type 2A (EC:3.1.3.4);
K01080 phosphatidate phosphatase [EC:3.1.3.4]
Length=284
Score = 65.5 bits (158), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 31/75 (41%), Positives = 43/75 (57%), Gaps = 6/75 (8%)
Query 75 LYTYCGSLAMSVASVFLLTNSLKAAVGSLRPHFLDVCRPDWSRVSCRDSSGLYDVYIPDF 134
+Y G+ A+ LT+ K ++G LRPHFLDVC PDWS+++C D YI +
Sbjct 98 IYKAIGTFLFGAAASQSLTDIAKYSIGRLRPHFLDVCDPDWSKINCSDG------YIEYY 151
Query 135 FCTGDPHRVEEARRS 149
C G+ RV+E R S
Sbjct 152 ICRGNAERVKEGRLS 166
> dre:794598 Lipid phosphate phosphohydrolase 1-like; K01080 phosphatidate
phosphatase [EC:3.1.3.4]
Length=293
Score = 64.3 bits (155), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 46/148 (31%), Positives = 61/148 (41%), Gaps = 23/148 (15%)
Query 2 FCGEDDIALPYMPASISAAGAFWISTMLPAFIIVVVEVLLWAVRATQEKDKSEREQQAVV 61
FC +D I P+ +IS I L +IV E +R+ ++ + V
Sbjct 54 FCSDDSIRYPFKEDTISYQLLMGIMIPLALLLIVFGECFSIYLRS-----RASFSYEYVA 108
Query 62 VVLDRRIPETCVHLYTYCGSLAMSVASVFLLTNSLKAAVGSLRPHFLDVCRPDWSRVSCR 121
V Y GS A LT+ K +G LRPHFL VC+P WS + C+
Sbjct 109 CV------------YKAVGSFVFGAAVSQSLTDIAKYTIGRLRPHFLTVCKPHWSLIDCK 156
Query 122 DSSGLYDVYIPDFFCTGDPHRVEEARRS 149
YI +F CTGDP E R S
Sbjct 157 AG------YIENFTCTGDPTLTNEGRLS 178
> cel:F13E6.5 hypothetical protein
Length=304
Score = 63.5 bits (153), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 46/148 (31%), Positives = 62/148 (41%), Gaps = 20/148 (13%)
Query 3 CGEDDIALPYMPASISAAGAFWISTMLPAFIIVVVEVLLWAVRATQEKDKSEREQQAVVV 62
CG+ I P+ ++ I+ P I+ +VE +L K K
Sbjct 70 CGDISIQQPFKENTVGLKHLLVITLGSPFLIVALVEAIL------HFKSKGSNR------ 117
Query 63 VLDRRIPETCVHLYTYCGSLAMSVASVFLLTNSLKAAVGSLRPHFLDVCRPDWSRVSCRD 122
L + T + TY L M A F + LK VG LRPHF VC+PDWS+V C D
Sbjct 118 -LAKFFSATTI---TYLKYLLMYAACTFAM-EFLKCYVGRLRPHFFSVCKPDWSKVDCTD 172
Query 123 SSGLYDVYIPDFFCTG-DPHRVEEARRS 149
D D CT +P ++ AR S
Sbjct 173 KQSFIDS--SDLVCTNPNPRKIRTARTS 198
> xla:447215 ppap2b-b, dri42, lpp3, pap2b, ppap2bb, vcip; phosphatidic
acid phosphatase type 2B (EC:3.1.3.4); K01080 phosphatidate
phosphatase [EC:3.1.3.4]
Length=307
Score = 63.2 bits (152), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 47/149 (31%), Positives = 75/149 (50%), Gaps = 21/149 (14%)
Query 2 FCGEDDIALPYMPA-SISAAGAFWISTMLPAFIIVVVEVLLWAVRATQEKDKSEREQQAV 60
+CG++ I P +IS A + + I+V E + + +E+ +S Q
Sbjct 67 YCGDESIKYPANSGETISDAVLSAVGIFIAILAIIVGE--FFRIHYLKERPRS-FIQNPY 123
Query 61 VVVLDRRIPETCVHLYTYCGSLAMSVASVFLLTNSLKAAVGSLRPHFLDVCRPDWSRVSC 120
V L +++ C + SV+ F T+ K A+G LRPHFL+VC PD+S ++C
Sbjct 124 VAALYKQVG---------CFAFGCSVSQSF--TDIAKVAIGRLRPHFLNVCDPDFSTINC 172
Query 121 RDSSGLYDVYIPDFFCTGDPHRVEEARRS 149
S G YI ++ C G P++V EAR+S
Sbjct 173 --SLG----YIENYVCRGPPNKVMEARKS 195
> mmu:67916 Ppap2b, 1110003O22Rik, 2610002D05Rik, AV025606, D4Bwg0538e,
D4Bwg1535e, Lpp3, Ppab2b; phosphatidic acid phosphatase
type 2B (EC:3.1.3.4); K01080 phosphatidate phosphatase
[EC:3.1.3.4]
Length=312
Score = 60.5 bits (145), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 38/148 (25%), Positives = 67/148 (45%), Gaps = 19/148 (12%)
Query 2 FCGEDDIALPYMPASISAAGAFWISTMLPAFIIVVVEVLLWAVRATQEKDKSEREQQAVV 61
+C ++ I P + S + A + + I+ ++ + + +EK +S + V
Sbjct 67 YCNDESIKYP-LKVSETINDAVLCAVGIVIAILAIITGEFYRIYYLKEKSRSTTQNPYVA 125
Query 62 VVLDRRIPETCVHLYTYCGSLAMSVASVFLLTNSLKAAVGSLRPHFLDVCRPDWSRVSCR 121
LY G A T+ K ++G LRPHFL VC PD+S+++C
Sbjct 126 A------------LYKQVGCFLFGCAISQSFTDIAKVSIGRLRPHFLSVCDPDFSQINCS 173
Query 122 DSSGLYDVYIPDFFCTGDPHRVEEARRS 149
+ YI ++ C G+ +V+EAR+S
Sbjct 174 EG------YIQNYRCRGEDSKVQEARKS 195
> hsa:8613 PPAP2B, Dri42, LPP3, MGC15306, PAP2B, VCIP; phosphatidic
acid phosphatase type 2B (EC:3.1.3.4); K01080 phosphatidate
phosphatase [EC:3.1.3.4]
Length=311
Score = 60.1 bits (144), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 29/75 (38%), Positives = 42/75 (56%), Gaps = 6/75 (8%)
Query 75 LYTYCGSLAMSVASVFLLTNSLKAAVGSLRPHFLDVCRPDWSRVSCRDSSGLYDVYIPDF 134
LY G A T+ K ++G LRPHFL VC PD+S+++C + YI ++
Sbjct 126 LYKQVGCFLFGCAISQSFTDIAKVSIGRLRPHFLSVCNPDFSQINCSEG------YIQNY 179
Query 135 FCTGDPHRVEEARRS 149
C GD +V+EAR+S
Sbjct 180 RCRGDDSKVQEARKS 194
> mmu:50784 Ppap2c, Lpp2; phosphatidic acid phosphatase type 2C
(EC:3.1.3.4); K01080 phosphatidate phosphatase [EC:3.1.3.4]
Length=276
Score = 60.1 bits (144), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 48/153 (31%), Positives = 68/153 (44%), Gaps = 32/153 (20%)
Query 2 FCGEDDIALPYMPASISAAGAFWISTMLPAFIIVVVEVLLWAVRATQEKDKSEREQQAVV 61
+CG+D I PY P +I+ L A +I+ V+L ++ +A +
Sbjct 37 YCGDDSIRYPYRPDTITHG--------LMAGVIITATVILVSL------------GEAYL 76
Query 62 VVLDRRIPETCVH-----LYTYCGSLAMSVASVFLLTNSLKAAVGSLRPHFLDVCRPDWS 116
V DR + + +Y G+ A LT+ K +G LRP FL VC PDWS
Sbjct 77 VYTDRLYSRSNFNNYVAAIYKVLGTFLFGAAVSQSLTDLAKYMIGRLRPSFLAVCDPDWS 136
Query 117 RVSCRDSSGLYDVYIPDFFCTGDPHRVEEARRS 149
+V+C SG Y+ C G P V EAR S
Sbjct 137 QVNC---SG----YVQLEVCRGSPANVTEARLS 162
> hsa:8612 PPAP2C, LPP2, PAP-2c, PAP2-g; phosphatidic acid phosphatase
type 2C (EC:3.1.3.4); K01080 phosphatidate phosphatase
[EC:3.1.3.4]
Length=288
Score = 60.1 bits (144), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 45/149 (30%), Positives = 64/149 (42%), Gaps = 23/149 (15%)
Query 2 FCGEDDIALPYMPASISAAGAFWISTMLPAFIIVVVEVLLWAVRATQEKDKSEREQQAVV 61
+CG+D I PY P +I+ + A +I+V + V + +S+
Sbjct 37 YCGDDSIRYPYRPDTITHG--LMAGVTITATVILVSAGEAYLVYTDRLYSRSDFNNYVAA 94
Query 62 VVLDRRIPETCVHLYTYCGSLAMSVASVFLLTNSLKAAVGSLRPHFLDVCRPDWSRVSCR 121
V Y G+ A LT+ K +G LRP+FL VC PDWSRV+C
Sbjct 95 V-------------YKVLGTFLFGAAVSQSLTDLAKYMIGRLRPNFLAVCDPDWSRVNC- 140
Query 122 DSSGLYDVYIP-DFFCTGDPHRVEEARRS 149
VY+ + C G+P V EAR S
Sbjct 141 ------SVYVQLEKVCRGNPADVTEARLS 163
> cel:T28D9.3 hypothetical protein
Length=358
Score = 56.6 bits (135), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 46/148 (31%), Positives = 64/148 (43%), Gaps = 11/148 (7%)
Query 2 FCGEDDIALPYMPASISAAGAFWISTMLPAFIIVVVEVLLWAVRATQEKDKSEREQQAVV 61
FC +D I Y +I+A + +L A ++ VE + + R + +
Sbjct 73 FCDDDSIRYEYRKDTITAVQLMLYNLVLNAATVLFVEYYRMQKVESNINNPRYRWRNNHL 132
Query 62 VVLDRRIPETCVHLYTYCGSLAMSVASVFLLTNSLKAAVGSLRPHFLDVCRPDWSRVSCR 121
VL V L TY G + L K VG LRPHFLDVC+ + +C
Sbjct 133 HVL-------FVRLLTYFGYSQIGFVMNIALNIVTKHVVGRLRPHFLDVCK--LANDTC- 182
Query 122 DSSGLYDVYIPDFFCTGDPHRVEEARRS 149
+G YI D+ CTG P V EAR+S
Sbjct 183 -VTGDSHRYITDYTCTGPPELVLEARKS 209
> mmu:272031 E130309F12Rik, KIAA4247, Lppr1, PRG-3, mKIAA4247;
RIKEN cDNA E130309F12 gene
Length=325
Score = 56.6 bits (135), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 41/158 (25%), Positives = 71/158 (44%), Gaps = 21/158 (13%)
Query 2 FCGEDDIALPYMPAS-----ISAAGAFWISTMLPAFIIVVVEVLLWAVRATQEKDKSERE 56
FC + D+ PY P + IS + + P II + E+ ++ +++T+E +E +
Sbjct 49 FCQDGDLMKPY-PGTEEESFISPLVLYCVLAATPTAIIFIGEISMYFIKSTRESLIAEEK 107
Query 57 QQAV-----VVVLDRRIPETCVHLYTYCGSLAMSVASVFLLTNSLKAAVGSLRPHFLDVC 111
+ L RRI + G A + + + N+ + G L P+FL VC
Sbjct 108 MILTGDCCYLSPLLRRIIR-------FIGVFAFGLFATDIFVNAGQVVTGHLTPYFLTVC 160
Query 112 RPDWSRVSCRDSSGLYDVYIPDFFCTGDPHRVEEARRS 149
+P+++ CR + CTGD +E+ARRS
Sbjct 161 QPNYTSTDCRAHQQFIN---NGNICTGDLEVIEKARRS 195
> xla:447319 ppap2b-a, dri42, lpp3, pap2b, ppap2b, ppap2ba, vcip;
phosphatidic acid phosphatase type 2B (EC:3.1.3.4); K01080
phosphatidate phosphatase [EC:3.1.3.4]
Length=307
Score = 55.5 bits (132), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 31/75 (41%), Positives = 41/75 (54%), Gaps = 6/75 (8%)
Query 75 LYTYCGSLAMSVASVFLLTNSLKAAVGSLRPHFLDVCRPDWSRVSCRDSSGLYDVYIPDF 134
LY G + T+ K A+G LRPHFL VC PD+S + C S G YI ++
Sbjct 127 LYKQVGCFVFGCSVSQSFTDIAKVAIGRLRPHFLKVCDPDFSIIDC--SLG----YIENY 180
Query 135 FCTGDPHRVEEARRS 149
C G P++V EAR+S
Sbjct 181 ECRGPPNKVMEARKS 195
> cel:F53C3.13 hypothetical protein
Length=385
Score = 54.7 bits (130), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 41/149 (27%), Positives = 69/149 (46%), Gaps = 10/149 (6%)
Query 2 FCGEDDIALPYMPASISAAGAFWISTMLPAFIIVVVEVLLWAVRATQEKDKSEREQQAVV 61
+C ++ I P+ + ++ + ++P +I+ E+ R + K E E +
Sbjct 41 YCDDESIRYPFRDSKVTRQMLIVVGLLIPILLILATELF----RTLAWEKKCETEFKTYH 96
Query 62 VVLDRRIPETCVHLYTYCGSLAMSVASVFLLTNSLKAAVGSLRPHFLDVCRPDWSRVSCR 121
V + + V LY + G + V L+ + K +G RPHF+DVCRPD +C
Sbjct 97 V-RNHSVHRLVVRLYCFIGYFFVGVCFNQLMVDIAKYTIGRQRPHFMDVCRPDIGYQTCS 155
Query 122 DSSGLYDVYIPDFFC-TGDPHRVEEARRS 149
D+YI DF C T D ++ EA+ S
Sbjct 156 QP----DLYITDFKCTTTDTKKIHEAQLS 180
> hsa:54886 LPPR1, MGC26189, PRG-3; lipid phosphate phosphatase-related
protein type 1
Length=325
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 40/161 (24%), Positives = 72/161 (44%), Gaps = 27/161 (16%)
Query 2 FCGEDDIALPYMPAS-----ISAAGAFWISTMLPAFIIVVVEVLLWAVRATQEKDKSERE 56
FC + D+ PY P + I+ + + P II + E+ ++ +++T+E S
Sbjct 49 FCQDGDLMKPY-PGTEEESFITPLVLYCVLAATPTAIIFIGEISMYFIKSTRE---SLIA 104
Query 57 QQAVVVV--------LDRRIPETCVHLYTYCGSLAMSVASVFLLTNSLKAAVGSLRPHFL 108
Q+ ++ L RRI + G A + + + N+ + G L P+FL
Sbjct 105 QEKTILTGECCYLNPLLRRIIR-------FTGVFAFGLFATDIFVNAGQVVTGHLTPYFL 157
Query 109 DVCRPDWSRVSCRDSSGLYDVYIPDFFCTGDPHRVEEARRS 149
VC+P+++ C+ + CTGD +E+ARRS
Sbjct 158 TVCKPNYTSADCQAHHQFIN---NGNICTGDLEVIEKARRS 195
> dre:100093705 zgc:165526
Length=301
Score = 53.1 bits (126), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 40/156 (25%), Positives = 73/156 (46%), Gaps = 17/156 (10%)
Query 2 FCGEDDIALPYM----PASISAAGAFWISTMLPAFIIVVVEVLLWAVRATQEKDKSEREQ 57
FC E PY+ ++I A + + T +P +I V E +L+ ++ T KD RE+
Sbjct 25 FCYETAYTKPYLGPEDTSAIPPALLYAVVTGVPTLMITVTETVLFLIQYT-SKDLDSREK 83
Query 58 QAVV---VVLDRRIPETCVHLYTYCGSLAMSVASVFLLTNSLKAAVGSLRPHFLDVCRPD 114
V L+ + T + + G + + + N+ + G+L PHFL VC+P+
Sbjct 84 TMVTGDCCYLNPLVRRT----FRFLGVYLFGLFTTDIFVNAGQVVTGNLAPHFLTVCKPN 139
Query 115 WSRVSCRDSSGLYDVYIP-DFFCTGDPHRVEEARRS 149
++ + C+ + YI CTG+ + AR++
Sbjct 140 FTALGCQQALR----YISHQEACTGNEDDILRARKT 171
> dre:100006660 similar to phosphatidic acid phosphatase type
2d
Length=309
Score = 52.0 bits (123), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 38/159 (23%), Positives = 75/159 (47%), Gaps = 23/159 (14%)
Query 2 FCGEDDIALPYM----PASISAAGAFWISTMLPAFIIVVVEVLLWAVRATQEKDKSEREQ 57
FC ++ PY+ ++I A + + +PA +I V E +L+ ++ E D RE+
Sbjct 33 FCHDNAYTKPYLGPEESSAIPPAILYAVVAGVPALVITVTESVLFLLQYVSE-DLDNREK 91
Query 58 QAVVVVLDRRIPETCVHL-------YTYCGSLAMSVASVFLLTNSLKAAVGSLRPHFLDV 110
++V+ D C +L + + G A + + + N+ + G+L P+FL V
Sbjct 92 --IIVMGD------CCYLNPLVRRTFRFLGVYAFGLFATDIFVNAGQVVTGNLSPYFLTV 143
Query 111 CRPDWSRVSCRDSSGLYDVYIPDFFCTGDPHRVEEARRS 149
C+P+++ + C+ + CTG+ + AR+S
Sbjct 144 CKPNYTALGCQQVVRFIN---QQDACTGNEDDILHARKS 179
> dre:415222 lppr1; zgc:86759
Length=333
Score = 51.6 bits (122), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 41/160 (25%), Positives = 71/160 (44%), Gaps = 25/160 (15%)
Query 2 FCGEDDIALPY----MPASISAAGAFWISTMLPAFIIVVVEVLLWAVRATQEKDKSEREQ 57
FC + D+ PY + I + + P II V E+ ++ +++T E + Q
Sbjct 48 FCNDADLLKPYPGPEESSFIQPLILYCVVAAAPTAIIFVGEISMYIMKSTGE---ALLAQ 104
Query 58 QAVVVV--------LDRRIPETCVHLYTYCGSLAMSVASVFLLTNSLKAAVGSLRPHFLD 109
+ +V L RRI + G A + + + N+ + G+L P+FL+
Sbjct 105 EKTIVTGECCYLNPLIRRIIR-------FIGVFAFGLFATDIFVNAGQVVTGNLAPYFLN 157
Query 110 VCRPDWSRVSCRDSSGLYDVYIPDFFCTGDPHRVEEARRS 149
VC+P+++ + C S + CTG+ VE ARRS
Sbjct 158 VCKPNYTGLDCHFS---HQFIANGNICTGNQVVVERARRS 194
> xla:443763 ppap2c, MGC81169; phosphatidic acid phosphatase type
2C (EC:3.1.3.4); K01080 phosphatidate phosphatase [EC:3.1.3.4]
Length=257
Score = 51.2 bits (121), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 37/148 (25%), Positives = 61/148 (41%), Gaps = 22/148 (14%)
Query 2 FCGEDDIALPYMPASISAAGAFWISTMLPAFIIVVVEVLLWAVRATQEKDKSEREQQAVV 61
+C ++ I PY +I+ + + +I++ ++ V + + +SE
Sbjct 13 YCNDESIRYPYREDTIT--NGLMATVTISCTVIIISSGEMYMVFSKRLYSRSECNNYIAA 70
Query 62 VVLDRRIPETCVHLYTYCGSLAMSVASVFLLTNSLKAAVGSLRPHFLDVCRPDWSRVSCR 121
LY G+ A LT+ K +G RP+FL VC PDWS V+C
Sbjct 71 -------------LYKVVGTYLFGAAVSQSLTDLAKYMIGRPRPNFLAVCDPDWSTVNCS 117
Query 122 DSSGLYDVYIPDFFCTGDPHRVEEARRS 149
Y+ DF C G+ V ++R S
Sbjct 118 R-------YVTDFTCRGNYANVTDSRLS 138
> dre:559124 MGC158309, ppap2b; zgc:158309 (EC:3.1.3.4); K01080
phosphatidate phosphatase [EC:3.1.3.4]
Length=323
Score = 50.4 bits (119), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 47/153 (30%), Positives = 61/153 (39%), Gaps = 28/153 (18%)
Query 2 FCGEDDIALPYM-----PASISAAGAFWISTMLPAFIIVVVEVLLWAVRATQEKDKSERE 56
FCG+ I PY+ P S+ AG I+ I V E + VR +D R
Sbjct 85 FCGDTSITYPYIESEAIPDSVLIAGGIIIT----GLTIAVGEC--YRVRF---RDVHSR- 134
Query 57 QQAVVVVLDRRIPETCVHLYTYCGSLAMSVASVFLLTNSLKAAVGSLRPHFLDVCRPDWS 116
R + +C LY GS LTN K +VG LRPHFL C +
Sbjct 135 ------AFVRNLYVSC--LYKELGSFLFGCCVGQSLTNMAKLSVGRLRPHFLSACNVTYE 186
Query 117 RVSCRDSSGLYDVYIPDFFCTGDPHRVEEARRS 149
++C + YI C VEEAR+S
Sbjct 187 SLNCTPGT-----YISHVVCKSSKKIVEEARKS 214
> xla:444601 lppr5, MGC84075; lipid phosphate phosphatase-related
protein type 5
Length=305
Score = 48.1 bits (113), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 34/129 (26%), Positives = 63/129 (48%), Gaps = 17/129 (13%)
Query 27 TMLPAFIIVVVEVLLWAVR-ATQEKDKSEREQQA----VVVVLDRRIPETCVHLYTYCGS 81
T +P +I+V E +++ ++ AT++ + E+ + L RR + G
Sbjct 62 TGVPVLVIIVGETVVFCLQVATRDFENQEKTLLTGDCCYINPLVRRTVR-------FLGI 114
Query 82 LAMSVASVFLLTNSLKAAVGSLRPHFLDVCRPDWSRVSCRDSSGLYDVYIPD-FFCTGDP 140
+ + + N+ + G+L PHFL VC+P+++ + CR + +I D CTG P
Sbjct 115 YTFGLFATDIFVNAGQVVTGNLAPHFLTVCKPNYTALGCRQ----FTQFITDANACTGIP 170
Query 141 HRVEEARRS 149
V +ARR+
Sbjct 171 DLVIKARRT 179
> dre:393428 MGC63577; zgc:63577 (EC:3.1.3.4)
Length=360
Score = 47.8 bits (112), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 39/161 (24%), Positives = 68/161 (42%), Gaps = 31/161 (19%)
Query 2 FCGEDDIALPY----MPASISAAGAFWISTMLPAFIIVVVEVLLWAVRATQEKDKSEREQ 57
FC + + PY +++ + + +P I+ EV+++ +RA E Q
Sbjct 64 FCFDKAFSKPYPGPDETSNVPPVLVYSLIAAIPTITILAGEVMVFFMRA-------EGTQ 116
Query 58 QAVVVVLD--------RRIPETCVHLYTYCGSLAMSVASVFLLTNSLKAAVGSLRPHFLD 109
+ +V D RRI + G V + + N+ + G+ PHFL
Sbjct 117 EKTIVTADCCYFNPLLRRI-------IRFLGVYTFGVFTTTIFANAGQVVTGNQTPHFLS 169
Query 110 VCRPDWSRVSCRDSSGLYDVYIPD-FFCTGDPHRVEEARRS 149
CRP+++ + C + YI + CTG+P V AR+S
Sbjct 170 ACRPNYTALGCHSNLQ----YITERKACTGNPLIVASARKS 206
> dre:571463 lipid phosphate phosphatase-related protein type
2-like
Length=336
Score = 47.4 bits (111), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 39/156 (25%), Positives = 69/156 (44%), Gaps = 21/156 (13%)
Query 2 FCGEDDIALPY-MPASISAAGAFWISTMLPAFIIVVVEVLLWAVRATQEKDKSEREQQAV 60
FC + + PY P S A + +++ A I V +LL + + K ++ +E+ V
Sbjct 33 FCYDKTFSKPYPGPEDDSKAPPVLVYSLVTA--IPTVTILLGELASFFTKPEASQEKTIV 90
Query 61 VV------VLDRRIPETCVHLYTYCGSLAMSVASVFLLTNSLKAAVGSLRPHFLDVCRPD 114
L RRI + G + + + + N+ + G+ PHFL CRP+
Sbjct 91 TADCCYFNPLLRRIVR-------FLGVYSFGLFTTTIFANAGQVVTGNQTPHFLSTCRPN 143
Query 115 WSRVSCRDSSGLYDVYIPD-FFCTGDPHRVEEARRS 149
++ + C YI + CTG+P+ + AR+S
Sbjct 144 YTALGCHSPMQ----YITERRACTGNPYLIASARKS 175
> mmu:75769 4833424O15Rik, Lppr5, PRG-5, Pap2d; RIKEN cDNA 4833424O15
gene
Length=316
Score = 45.1 bits (105), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 37/147 (25%), Positives = 66/147 (44%), Gaps = 16/147 (10%)
Query 5 EDDIALPYMPASISAAGAFWISTMLPAFIIVVVEVLLWAVRATQEKDKSEREQQAVVVVL 64
ED A+P + AAG +P +I+V E AV Q + Q+ ++
Sbjct 58 EDSSAVPPVLLYSLAAG-------VPVLVIIVGET---AVFCLQLATRDFENQEKTILTG 107
Query 65 DR-RIPETCVHLYTYCGSLAMSVASVFLLTNSLKAAVGSLRPHFLDVCRPDWSRVSCRDS 123
D I + G A + + + N+ + G+L PHFL +C+P+++ + C+
Sbjct 108 DCCYINPLVRRTVRFLGIYAFGLFATDIFVNAGQVVTGNLAPHFLALCKPNYTALGCQQ- 166
Query 124 SGLYDVYIP-DFFCTGDPHRVEEARRS 149
Y +I + CTG+P + AR++
Sbjct 167 ---YTQFISGEEACTGNPDLIMRARKT 190
> hsa:163404 LPPR5, PAP2, PAP2D, PRG5; lipid phosphate phosphatase-related
protein type 5
Length=316
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 36/147 (24%), Positives = 65/147 (44%), Gaps = 16/147 (10%)
Query 5 EDDIALPYMPASISAAGAFWISTMLPAFIIVVVEVLLWAVRATQEKDKSEREQQAVVVVL 64
ED A+P + AAG +P +I+V E AV Q + Q+ ++
Sbjct 58 EDSSAVPPVLLYSLAAG-------VPVLVIIVGET---AVFCLQLATRDFENQEKTILTG 107
Query 65 DR-RIPETCVHLYTYCGSLAMSVASVFLLTNSLKAAVGSLRPHFLDVCRPDWSRVSCRDS 123
D I + G + + + N+ + G+L PHFL +C+P+++ + C+
Sbjct 108 DCCYINPLVRRTVRFLGIYTFGLFATDIFVNAGQVVTGNLAPHFLALCKPNYTALGCQQ- 166
Query 124 SGLYDVYIP-DFFCTGDPHRVEEARRS 149
Y +I + CTG+P + AR++
Sbjct 167 ---YTQFISGEEACTGNPDLIMRARKT 190
> dre:100330890 lipid phosphate phosphatase-related protein type
2-like
Length=299
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 34/125 (27%), Positives = 55/125 (44%), Gaps = 27/125 (21%)
Query 34 IVVVEVLLWAVRATQEKDKSEREQQAVVVVLD--------RRIPETCVHLYTYCGSLAMS 85
I+ EV+++ +RA E Q+ +V D RRI + G
Sbjct 39 ILAGEVMVFFMRA-------EGTQEKTIVTADCCYFNPLLRRI-------IRFLGVYTFG 84
Query 86 VASVFLLTNSLKAAVGSLRPHFLDVCRPDWSRVSCRDSSGLYDVYIPD-FFCTGDPHRVE 144
V + + N+ + G+ PHFL CRP+++ + C + YI + CTG+P V
Sbjct 85 VFTTTIFANAGQVVTGNQTPHFLSACRPNYTALGCHSNLQ----YITERKACTGNPLIVA 140
Query 145 EARRS 149
AR+S
Sbjct 141 SARKS 145
> hsa:64748 LPPR2, DKFZp761E1121, FLJ13055, PRG4; lipid phosphate
phosphatase-related protein type 2 (EC:3.1.3.4)
Length=427
Score = 42.4 bits (98), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 25/81 (30%), Positives = 38/81 (46%), Gaps = 8/81 (9%)
Query 75 LYTYCGSLAMSVASVFLLTNSLKAAVGSLRPHFLDVCRPDWSRVSCRDSSGLYDVYIPDF 134
L + G + + + + N+ + G+ PHFL VCRP+++ + C S D PD
Sbjct 101 LVRFLGVYSFGLFTTTIFANAGQVVTGNPTPHFLSVCRPNYTALGCLPPSP--DRPGPDR 158
Query 135 F------CTGDPHRVEEARRS 149
F C G P V ARR+
Sbjct 159 FVTDQGACAGSPSLVAAARRA 179
> dre:100334794 phosphatidic acid phosphatase type 2C-like
Length=282
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 36/127 (28%), Positives = 54/127 (42%), Gaps = 40/127 (31%)
Query 23 FWISTMLPAFIIVVVEVLLWAVRATQEKDKSEREQQAVVVVLDRRIPETCVHLYTYCGSL 82
F IS + P +I VV+++ Q D++E I E C+ + SL
Sbjct 62 FAISFLTPLAVIFVVKII-------QRTDRTE-------------IKEACLAV-----SL 96
Query 83 AMSVASVFLLTNSLKAAVGSLRPHFLDVCRPDWSRVSCRDSSGLYDVYIPDFFCTGDPHR 142
A+++ VF TN++K VG RP + C PD + CTG+P
Sbjct 97 ALALNGVF--TNTIKLIVGRPRPDYYQRCFPDGQMNA-------------KMLCTGEPDL 141
Query 143 VEEARRS 149
V E R+S
Sbjct 142 VSEGRKS 148
> dre:559888 si:dkey-108c13.1
Length=775
Score = 41.2 bits (95), Expect = 0.001, Method: Composition-based stats.
Identities = 39/158 (24%), Positives = 74/158 (46%), Gaps = 24/158 (15%)
Query 3 CGEDDIALPYMPAS---ISAAGAFWISTMLPAFIIVVVEVLLWAVRATQE-KDKSEREQQ 58
C + +++PY+ + I F ++ PA I++ E +L+ A + K+E
Sbjct 55 CNDRSLSMPYIEPTKEVIPFLMLFSLAFAGPAVTIMIGEGILYCCVARRNIAIKTEANIN 114
Query 59 AVVVVLD----RRIPETCVHLYTYCGSLAMSVASVFLLTNSLKAAVGSLRPHFLDVCRPD 114
A + R + VH++ C + L+T+ ++ A G P+FL VC+P+
Sbjct 115 AAGCNFNSYIRRAVRFVGVHVFGLCITA--------LITDIIQLATGYHAPYFLTVCKPN 166
Query 115 WS--RVSCRDSSGLYDVYIPDFFCTG-DPHRVEEARRS 149
++ +SC ++S +I D C+G DP + R+S
Sbjct 167 YTTLNISCDENS-----FIVDDICSGPDPAAINSGRKS 199
> xla:443950 ppapdc1b, MGC80318; phosphatidic acid phosphatase
type 2 domain containing 1B (EC:3.1.3.4)
Length=226
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 27/69 (39%), Positives = 38/69 (55%), Gaps = 15/69 (21%)
Query 81 SLAMSVASVFLLTNSLKAAVGSLRPHFLDVCRPDWSRVSCRDSSGLYDVYIPDFFCTGDP 140
SL++++ +F TN++K VG RP FL C PD ++S GL+ CTGDP
Sbjct 53 SLSLALNGIF--TNTVKLIVGRPRPDFLFRCFPDG-----QESPGLH--------CTGDP 97
Query 141 HRVEEARRS 149
V E R+S
Sbjct 98 ELVIEGRKS 106
> mmu:235044 Lppr2, MGC25492, PRG-4; cDNA sequence BC018242 (EC:3.1.3.4)
Length=343
Score = 39.7 bits (91), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 22/79 (27%), Positives = 37/79 (46%), Gaps = 4/79 (5%)
Query 75 LYTYCGSLAMSVASVFLLTNSLKAAVGSLRPHFLDVCRPDWSRVSC---RDSSGLYDVYI 131
L + G + + + + N+ + G+ PHFL VCRP+++ + C D ++
Sbjct 126 LVRFLGVYSFGLFTTTIFANAGQVVTGNPTPHFLSVCRPNYTALGCPPPSPDRPGPDRFV 185
Query 132 PD-FFCTGDPHRVEEARRS 149
D C G P V ARR+
Sbjct 186 TDQSACAGSPSLVAAARRA 204
> hsa:196051 PPAPDC1A, DPPL2, MGC120299, MGC120300, PPAPDC1; phosphatidic
acid phosphatase type 2 domain containing 1A (EC:3.1.3.4)
Length=271
Score = 39.3 bits (90), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 36/127 (28%), Positives = 53/127 (41%), Gaps = 40/127 (31%)
Query 23 FWISTMLPAFIIVVVEVLLWAVRATQEKDKSEREQQAVVVVLDRRIPETCVHLYTYCGSL 82
F IS + P +I VV+++ + DK+E ++ + V SL
Sbjct 55 FAISFLTPLAVICVVKII-------RRTDKTEIKEAFLAV------------------SL 89
Query 83 AMSVASVFLLTNSLKAAVGSLRPHFLDVCRPDWSRVSCRDSSGLYDVYIPDFFCTGDPHR 142
A+++ V TN++K VG RP F C PD V + CTGDP
Sbjct 90 ALALNGV--CTNTIKLIVGRPRPDFFYRCFPD-------------GVMNSEMHCTGDPDL 134
Query 143 VEEARRS 149
V E R+S
Sbjct 135 VSEGRKS 141
> mmu:229791 D3Bwg0562e, A330086D10, Lppr4, PRG-1, mKIAA0455;
DNA segment, Chr 3, Brigham & Women's Genetics 0562 expressed
(EC:3.1.3.4)
Length=766
Score = 39.3 bits (90), Expect = 0.005, Method: Composition-based stats.
Identities = 37/159 (23%), Positives = 74/159 (46%), Gaps = 26/159 (16%)
Query 3 CGEDDIALPYMPASISAAGAFWISTML---PAFIIVVVEVLLWAVRATQEKDKSEREQQA 59
C + +++PY+ + A + ++ PA I+V E +L+ ++ ++ + E
Sbjct 104 CYDRSLSMPYIEPTQEAIPFLMLLSLAFAGPAITIMVGEGILYCC-LSKRRNGAGLEPNI 162
Query 60 VV------VVLDRRIPETCVHLYTYCGSLAMSVASVFLLTNSLKAAVGSLRPHFLDVCRP 113
L R + VH++ C S L+T+ ++ + G P+FL VC+P
Sbjct 163 NAGGCNFNSFLRRAVRFVGVHVFGLC--------STALITDIIQLSTGYQAPYFLTVCKP 214
Query 114 DWS--RVSCRDSSGLYDVYIPDFFCTG-DPHRVEEARRS 149
+++ VSC+++S YI + C+G D + R+S
Sbjct 215 NYTSLNVSCKENS-----YIVEDICSGSDLTVINSGRKS 248
> dre:563806 Lipid phosphate phosphohydrolase 2-like; K01080 phosphatidate
phosphatase [EC:3.1.3.4]
Length=365
Score = 39.3 bits (90), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 37/148 (25%), Positives = 57/148 (38%), Gaps = 26/148 (17%)
Query 2 FCGEDDIALPYMPASISAAGAFWISTMLPAFIIVVVEVLLWAVRATQEKDKSEREQQAVV 61
+C ++ I PY P +IS ++ II+ E L V + S+ Q A
Sbjct 72 YCNDETIQYPYRPDTISHKMMAAVTISCSVIIIISGEAYL--VYTKRLYSNSDFNQYAAA 129
Query 62 VVLDRRIPETCVHLYTYCGSLAMSVASVFLLTNSLKAAVGSLRPHFLDVCRPDWSRVSCR 121
LY G+ LT+ K +G LRP+F+ VC P
Sbjct 130 -------------LYKVVGTFLFGACVSQSLTDMAKYTIGRLRPNFMSVCAP-------- 168
Query 122 DSSGLYDVYIPDFFCTGDPHRVEEARRS 149
+ + Y+ + CTG+ V E+R S
Sbjct 169 ---AVCEGYMLEINCTGNARNVTESRLS 193
> hsa:9890 LPPR4, KIAA0455, LPR4, PHP1, PRG-1, PRG1, RP4-788L13.1;
lipid phosphate phosphatase-related protein type 4 (EC:3.1.3.4)
Length=705
Score = 38.9 bits (89), Expect = 0.006, Method: Composition-based stats.
Identities = 26/89 (29%), Positives = 46/89 (51%), Gaps = 16/89 (17%)
Query 64 LDRRIPETCVHLYTYCGSLAMSVASVFLLTNSLKAAVGSLRPHFLDVCRPDWS--RVSCR 121
L R + VH++ C S L+T+ ++ + G P+FL VC+P+++ VSC+
Sbjct 172 LRRAVRFVGVHVFGLC--------STALITDIIQLSTGYQAPYFLTVCKPNYTSLNVSCK 223
Query 122 DSSGLYDVYIPDFFCTG-DPHRVEEARRS 149
++S YI + C+G D + R+S
Sbjct 224 ENS-----YIVEDICSGSDLTVINSGRKS 247
> mmu:381925 Ppapdc1a, C030048B12Rik, Gm1090; phosphatidic acid
phosphatase type 2 domain containing 1A (EC:3.1.3.4)
Length=271
Score = 38.5 bits (88), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 36/127 (28%), Positives = 53/127 (41%), Gaps = 40/127 (31%)
Query 23 FWISTMLPAFIIVVVEVLLWAVRATQEKDKSEREQQAVVVVLDRRIPETCVHLYTYCGSL 82
F IS + P +I VV+++ + DK+E ++ + V SL
Sbjct 55 FAISFLTPLAVICVVKII-------RRTDKTEIKEAFLAV------------------SL 89
Query 83 AMSVASVFLLTNSLKAAVGSLRPHFLDVCRPDWSRVSCRDSSGLYDVYIPDFFCTGDPHR 142
A+++ V TN++K VG RP F C PD V + CTGDP
Sbjct 90 ALALNGV--CTNTIKLIVGRPRPDFFYRCFPD-------------GVMNSEMRCTGDPDL 134
Query 143 VEEARRS 149
V E R+S
Sbjct 135 VSEGRKS 141
> sce:YDR503C LPP1; Lipid phosphate phosphatase, catalyzes Mg(2+)-independent
dephosphorylation of phosphatidic acid (PA),
lysophosphatidic acid, and diacylglycerol pyrophosphate; involved
in control of the cellular levels of phosphatidylinositol
and PA (EC:3.1.3.4 3.1.3.-)
Length=274
Score = 37.7 bits (86), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 20/51 (39%), Positives = 30/51 (58%), Gaps = 2/51 (3%)
Query 73 VHLYTYCGSLAMSVASVFLLTNSLKAAVGSLRPHFLDVCRPDWSRVSCRDS 123
+H C L +S+ + LT +LK +G+LRP F+D C PD ++S DS
Sbjct 114 MHTSILCLMLIISINAA--LTGALKLIIGNLRPDFVDRCIPDLQKMSDSDS 162
> dre:431758 ppapdc1b, fc49c04, wu:fc49c04, zgc:92365; phosphatidic
acid phosphatase type 2 domain containing 1B (EC:3.1.3.4)
Length=266
Score = 37.7 bits (86), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 25/71 (35%), Positives = 34/71 (47%), Gaps = 15/71 (21%)
Query 81 SLAMSVASVF--LLTNSLKAAVGSLRPHFLDVCRPDWSRVSCRDSSGLYDVYIPDFFCTG 138
SLA+++ V + TN++K AVG RP F C PD P+ C+G
Sbjct 89 SLAVTLTLVLNGVFTNAVKLAVGRPRPDFFYRCFPDGQMN-------------PELHCSG 135
Query 139 DPHRVEEARRS 149
DP V E R+S
Sbjct 136 DPDVVMEGRKS 146
> dre:335485 ppap2c, fj16e10, wu:fj16e10, zgc:66434; phosphatidic
acid phosphatase type 2C (EC:3.1.3.4); K01080 phosphatidate
phosphatase [EC:3.1.3.4]
Length=273
Score = 37.4 bits (85), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 36/148 (24%), Positives = 56/148 (37%), Gaps = 26/148 (17%)
Query 2 FCGEDDIALPYMPASISAAGAFWISTMLPAFIIVVVEVLLWAVRATQEKDKSEREQQAVV 61
FC ++ I P +I+ ++ II E L + + A+
Sbjct 37 FCQDESIGYPVKTDTITNVTLAAVTITCTILIICSGEAYLVYSKKIHSNSSFNQYVSAIY 96
Query 62 VVLDRRIPETCVHLYTYCGSLAMSVASVFLLTNSLKAAVGSLRPHFLDVCRPDWSRVSCR 121
VL + + G+++ S LT+ K +G RP+FL VC P C+
Sbjct 97 KVLGA---------FLFGGAVSQS------LTDLAKYTIGRPRPNFLAVCAPK----VCK 137
Query 122 DSSGLYDVYIPDFFCTGDPHRVEEARRS 149
L + CTG+P V EAR S
Sbjct 138 GFVNLNN-------CTGNPADVTEARLS 158
> xla:495974 ppapdc1a; phosphatidic acid phosphatase type 2 domain
containing 1A (EC:3.1.3.4)
Length=218
Score = 37.0 bits (84), Expect = 0.026, Method: Compositional matrix adjust.
Identities = 34/127 (26%), Positives = 52/127 (40%), Gaps = 40/127 (31%)
Query 23 FWISTMLPAFIIVVVEVLLWAVRATQEKDKSEREQQAVVVVLDRRIPETCVHLYTYCGSL 82
F IS + P +I VV+++L D++E ++ + V SL
Sbjct 2 FAISFLTPLAVIFVVKIIL-------RTDRTEVKEACLAV------------------SL 36
Query 83 AMSVASVFLLTNSLKAAVGSLRPHFLDVCRPDWSRVSCRDSSGLYDVYIPDFFCTGDPHR 142
A+++ V TN++K VG RP F C PD + + CTGD
Sbjct 37 ALALNGV--CTNTIKLIVGRPRPDFFYRCFPD-------------GISNEEMHCTGDASL 81
Query 143 VEEARRS 149
V E R+S
Sbjct 82 VSEGRKS 88
> sce:YDR284C DPP1, ZRG1; Diacylglycerol pyrophosphate (DGPP)
phosphatase, zinc-regulated vacuolar membrane-associated lipid
phosphatase, dephosphorylates DGPP to phosphatidate (PA)
and Pi, then PA to diacylglycerol; involved in lipid signaling
and cell metabolism (EC:3.1.3.4 3.1.3.-)
Length=289
Score = 36.6 bits (83), Expect = 0.034, Method: Compositional matrix adjust.
Identities = 20/52 (38%), Positives = 28/52 (53%), Gaps = 2/52 (3%)
Query 62 VVLDRRIPETCVHLYTYCGSLAMSVASVFLLTNSLKAAVGSLRPHFLDVCRP 113
++ DRR LYT L+++ S TN +K +G LRP FLD C+P
Sbjct 85 ILADRR--HLIFILYTSLLGLSLAWFSTSFFTNFIKNWIGRLRPDFLDRCQP 134
> mmu:216152 KIAA4076, Lppr3, Prg2; cDNA sequence BC005764 (EC:3.1.3.4)
Length=454
Score = 36.2 bits (82), Expect = 0.037, Method: Composition-based stats.
Identities = 25/89 (28%), Positives = 43/89 (48%), Gaps = 16/89 (17%)
Query 64 LDRRIPETCVHLYTYCGSLAMSVASVFLLTNSLKAAVGSLRPHFLDVCRPDWSRV--SCR 121
L R + VH++ C + L+T+ ++ A G P FL VC+P+++ + SC
Sbjct 125 LRRTVRFVGVHVFGLCATA--------LVTDVIQLATGYHTPFFLTVCKPNYTLLGTSCE 176
Query 122 DSSGLYDVYIPDFFCTG-DPHRVEEARRS 149
+ YI C+G D H + AR++
Sbjct 177 SNP-----YITQDICSGHDTHAILSARKT 200
> dre:570173 lipid phosphate phosphatase-related protein type
3a-like
Length=694
Score = 35.4 bits (80), Expect = 0.069, Method: Composition-based stats.
Identities = 26/89 (29%), Positives = 42/89 (47%), Gaps = 16/89 (17%)
Query 64 LDRRIPETCVHLYTYCGSLAMSVASVFLLTNSLKAAVGSLRPHFLDVCRPDWS--RVSCR 121
L R + VH++ C + L+T+ ++ A G P FL VC+P+++ VSC
Sbjct 125 LRRTVRFVGVHVFGLCATA--------LVTDVIQLATGYHTPFFLTVCKPNYTIPGVSCD 176
Query 122 DSSGLYDVYIPDFFCTG-DPHRVEEARRS 149
+ YI C G D H + AR++
Sbjct 177 KNP-----YITKDICAGQDQHAILSARKT 200
> ath:AT3G02600 LPP3; LPP3 (LIPID PHOSPHATE PHOSPHATASE 3); phosphatidate
phosphatase (EC:3.1.3.4)
Length=333
Score = 35.0 bits (79), Expect = 0.083, Method: Compositional matrix adjust.
Identities = 24/69 (34%), Positives = 31/69 (44%), Gaps = 9/69 (13%)
Query 81 SLAMSVASVFLLTNSLKAAVGSLRPHFLDVCRPDWSRVSCRDSSGLYDVYIPDFFCTGDP 140
L SV +LT+++K AVG RP F C P D LYD + D C GD
Sbjct 121 GLLYSVLVTAVLTDAIKNAVGRPRPDFFWRCFP--------DGKALYDS-LGDVICHGDK 171
Query 141 HRVEEARRS 149
+ E +S
Sbjct 172 SVIREGHKS 180
> hsa:84513 PPAPDC1B, DPPL1, HTPAP; phosphatidic acid phosphatase
type 2 domain containing 1B (EC:3.1.3.4)
Length=264
Score = 33.9 bits (76), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 21/59 (35%), Positives = 25/59 (42%), Gaps = 13/59 (22%)
Query 91 LLTNSLKAAVGSLRPHFLDVCRPDWSRVSCRDSSGLYDVYIPDFFCTGDPHRVEEARRS 149
+ TN++K VG RP F C PD S D CTGD V E R+S
Sbjct 102 VFTNTIKLIVGRPRPDFFYRCFPDGLAHS-------------DLMCTGDKDVVNEGRKS 147
> hsa:79948 LPPR3, LPR3, PRG-2, PRG2; lipid phosphate phosphatase-related
protein type 3 (EC:3.1.3.4)
Length=746
Score = 33.1 bits (74), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 25/89 (28%), Positives = 42/89 (47%), Gaps = 16/89 (17%)
Query 64 LDRRIPETCVHLYTYCGSLAMSVASVFLLTNSLKAAVGSLRPHFLDVCRPDWS--RVSCR 121
L R + VH++ C + L+T+ ++ A G P FL VC+P+++ SC
Sbjct 127 LRRTVRFVGVHVFGLC--------ATALVTDVIQLATGYHTPFFLTVCKPNYTLLGTSCE 178
Query 122 DSSGLYDVYIPDFFCTG-DPHRVEEARRS 149
+ YI C+G D H + AR++
Sbjct 179 -----VNPYITQDICSGHDIHAILSARKT 202
Lambda K H
0.325 0.137 0.430
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3068761412
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40