bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0653_orf2
Length=173
Score E
Sequences producing significant alignments: (Bits) Value
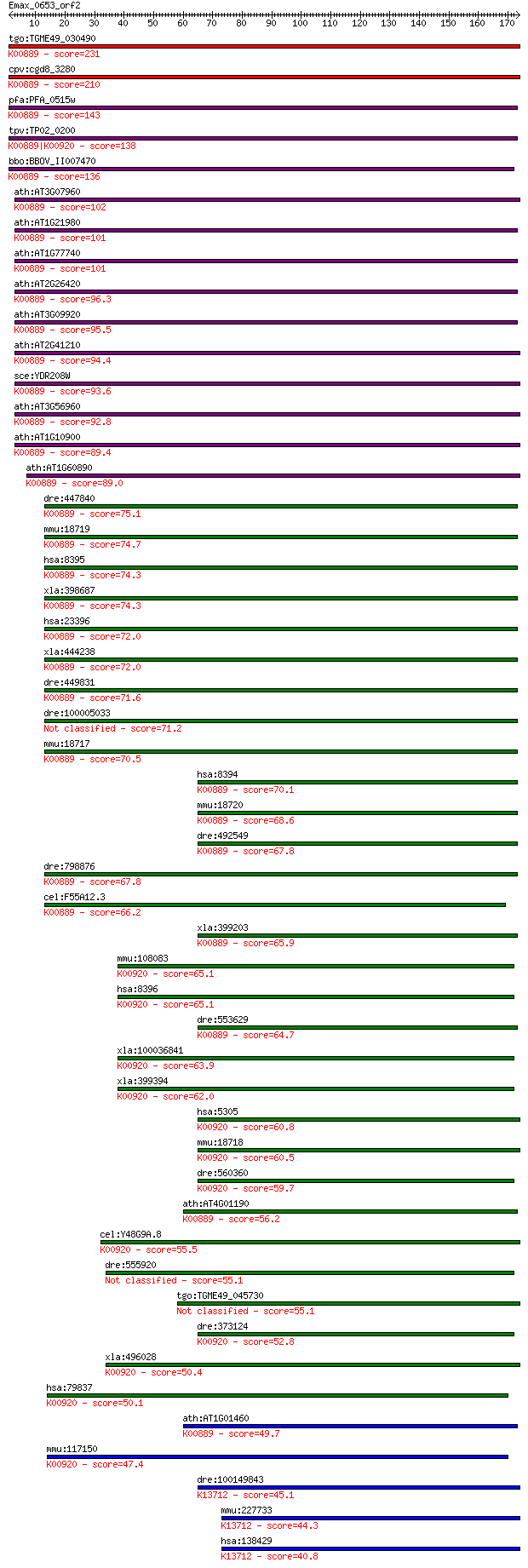
tgo:TGME49_030490 phosphatidylinositol-4-phosphate 5-kinase, p... 231 1e-60
cpv:cgd8_3280 phosphatidylinositol-4-phosphate 5-kinase ; K008... 210 2e-54
pfa:PFA_0515w PfPIP5K, NCS; phosphatidylinositol-4-phosphate-5... 143 2e-34
tpv:TP02_0200 phosphatidylinositol-4-phosphate 5-kinase; K0088... 138 1e-32
bbo:BBOV_II007470 18.m06619; 1-phosphatidylinositol-4-phosphat... 136 5e-32
ath:AT3G07960 phosphatidylinositol-4-phosphate 5-kinase family... 102 5e-22
ath:AT1G21980 ATPIP5K1 (PHOSPHATIDYLINOSITOL-4-PHOSPHATE 5-KIN... 101 1e-21
ath:AT1G77740 1-phosphatidylinositol-4-phosphate 5-kinase, put... 101 2e-21
ath:AT2G26420 PIP5K3; PIP5K3 (1-PHOSPHATIDYLINOSITOL-4-PHOSPHA... 96.3 4e-20
ath:AT3G09920 PIP5K9; PIP5K9 (PHOSPHATIDYL INOSITOL MONOPHOSPH... 95.5 9e-20
ath:AT2G41210 PIP5K5; PIP5K5 (PHOSPHATIDYLINOSITOL- 4-PHOSPHAT... 94.4 2e-19
sce:YDR208W MSS4; Mss4p (EC:2.7.1.68); K00889 1-phosphatidylin... 93.6 3e-19
ath:AT3G56960 PIP5K4; PIP5K4 (PHOSPHATIDYL INOSITOL MONOPHOSPH... 92.8 6e-19
ath:AT1G10900 phosphatidylinositol-4-phosphate 5-kinase family... 89.4 6e-18
ath:AT1G60890 phosphatidylinositol-4-phosphate 5-kinase family... 89.0 7e-18
dre:447840 fj29b05, fj93b05, wu:fj29b05, wu:fj93b05; zgc:92316... 75.1 1e-13
mmu:18719 Pip5k1b, AI844522, Pip5k1a, Pipk5a, Pipk5b, STM7; ph... 74.7 2e-13
hsa:8395 PIP5K1B, MSS4, STM7; phosphatidylinositol-4-phosphate... 74.3 2e-13
xla:398687 pip5k1c, MGC130766; phosphatidylinositol-4-phosphat... 74.3 2e-13
hsa:23396 PIP5K1C, KIAA0589, LCCS3, PIP5K-GAMMA, PIP5Kgamma, P... 72.0 9e-13
xla:444238 pip5k1b, MGC80809; phosphatidylinositol-4-phosphate... 72.0 1e-12
dre:449831 pip5k1b, zgc:101046; phosphatidylinositol-4-phospha... 71.6 1e-12
dre:100005033 si:ch211-243a15.1 71.2 2e-12
mmu:18717 Pip5k1c, AI115456, AI835305, Pip5kIgamma; phosphatid... 70.5 2e-12
hsa:8394 PIP5K1A; phosphatidylinositol-4-phosphate 5-kinase, t... 70.1 3e-12
mmu:18720 Pip5k1a, Pip5k1b, Pipk5a, Pipk5b; phosphatidylinosit... 68.6 9e-12
dre:492549 im:7140933; zgc:158838; K00889 1-phosphatidylinosit... 67.8 2e-11
dre:798876 si:ch211-197g15.13; K00889 1-phosphatidylinositol-4... 67.8 2e-11
cel:F55A12.3 ppk-1; PIP Kinase family member (ppk-1); K00889 1... 66.2 5e-11
xla:399203 pip5k1a, pip5k1c; phosphatidylinositol-4-phosphate ... 65.9 6e-11
mmu:108083 Pip4k2b, AI848124, PI5P4Kbeta, Pip5k2b, c11; phosph... 65.1 1e-10
hsa:8396 PIP4K2B, PI5P4KB, PIP5K2B, PIP5KIIB, PIP5KIIbeta; pho... 65.1 1e-10
dre:553629 pip5k1a, MGC110576, zgc:110576; phosphatidylinosito... 64.7 1e-10
xla:100036841 pip4k2a, pi5p4ka, pip5k2a, pip5kiia, pipk; phosp... 63.9 3e-10
xla:399394 pip4k2b, MGC81828, PI5P4KB, PIP5KIIB, PIP5KIIbeta, ... 62.0 9e-10
hsa:5305 PIP4K2A, FLJ13267, PI5P4KA, PIP5K2A, PIP5KII-alpha, P... 60.8 2e-09
mmu:18718 Pip4k2a, AW742916, Pip5k2a; phosphatidylinositol-5-p... 60.5 2e-09
dre:560360 similar to type II phosphatidylinositolphosphate ki... 59.7 4e-09
ath:AT4G01190 ATPIPK10 (Arabidopsis phosphatidylinositol phosp... 56.2 6e-08
cel:Y48G9A.8 ppk-2; PIP Kinase family member (ppk-2); K00920 1... 55.5 1e-07
dre:555920 novel protein similar to vertebrate phosphatidylino... 55.1 1e-07
tgo:TGME49_045730 phosphatidylinositol-4-phosphate 5-Kinase, p... 55.1 1e-07
dre:373124 pip5k2, Pip5k2c, cb770, pip4k2c, zgc:56046; phospha... 52.8 6e-07
xla:496028 pip4k2c, pip5k2c; phosphatidylinositol-5-phosphate ... 50.4 3e-06
hsa:79837 PIP4K2C, FLJ22055, PIP5K2C; phosphatidylinositol-5-p... 50.1 4e-06
ath:AT1G01460 PIPK11; PIPK11; 1-phosphatidylinositol-4-phospha... 49.7 5e-06
mmu:117150 Pip4k2c, Pip5k2c; phosphatidylinositol-5-phosphate ... 47.4 2e-05
dre:100149843 phosphatidylinositol-4-phosphate 5-kinase, type ... 45.1 1e-04
mmu:227733 Pip5kl1, BC028795; phosphatidylinositol-4-phosphate... 44.3 2e-04
hsa:138429 PIP5KL1, MGC46424, PIPKH, bA203J24.5; phosphatidyli... 40.8 0.002
> tgo:TGME49_030490 phosphatidylinositol-4-phosphate 5-kinase,
putative (EC:1.6.3.1 2.7.1.68); K00889 1-phosphatidylinositol-4-phosphate
5-kinase [EC:2.7.1.68]
Length=1313
Score = 231 bits (588), Expect = 1e-60, Method: Compositional matrix adjust.
Identities = 123/173 (71%), Positives = 143/173 (82%), Gaps = 0/173 (0%)
Query 1 HFGHESWNMVINIMVGIRLAGARAMSEPHRAVEPYDFLMKEKFSVLPKTGMVDKKRRKPS 60
HFGHE+W+ VINIMVGIRLA RA SEP RA+E YDF+MKEKFS+LP TGMV + S
Sbjct 893 HFGHENWDTVINIMVGIRLAAGRASSEPQRAIERYDFVMKEKFSILPNTGMVKSISDRRS 952
Query 61 LCAVRFIDYAPMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGS 120
L AVRF+DYAPMVFRRLR F I S Y+RSVGP+QLLGNL+LGNLSSLSELVSEG+SG+
Sbjct 953 LFAVRFVDYAPMVFRRLRERFHISSETYVRSVGPEQLLGNLLLGNLSSLSELVSEGRSGA 1012
Query 121 LFYYTTDGRFMIKTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALRL 173
LFYYT DG+FMIKT+SK TA F+RSIL DYY+HV +++LTR GLHA+RL
Sbjct 1013 LFYYTADGKFMIKTISKSTAMFLRSILLDYYEHVMANPDSLLTRFFGLHAIRL 1065
> cpv:cgd8_3280 phosphatidylinositol-4-phosphate 5-kinase ; K00889
1-phosphatidylinositol-4-phosphate 5-kinase [EC:2.7.1.68]
Length=828
Score = 210 bits (534), Expect = 2e-54, Method: Composition-based stats.
Identities = 99/183 (54%), Positives = 131/183 (71%), Gaps = 10/183 (5%)
Query 1 HFGHESWNMVINIMVGIRLAGARAMSEPHRAVEPYDFLMKEKFSVLPKTGMVDKKRR--- 57
+ GHESWN V+N+MVG+RLA R SEP+R V YDF+MKEKF ++P+T +
Sbjct 438 YIGHESWNTVLNMMVGMRLAIGRVYSEPNRDVADYDFIMKEKFFIIPRTSRESNTKSNVL 497
Query 58 -------KPSLCAVRFIDYAPMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLS 110
+ + V+FIDY PMVFR++R + I Y+RSVGP+QLLGN++LGNLSS++
Sbjct 498 IQKSNDYRGFMKPVQFIDYNPMVFRKIREICNISPESYVRSVGPEQLLGNMVLGNLSSMN 557
Query 111 ELVSEGKSGSLFYYTTDGRFMIKTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHA 170
EL SEGKSG+ FYYTTDGRF+IKTV K+TA F RSIL Y+ H+ + N+++TR+ GLHA
Sbjct 558 ELCSEGKSGAFFYYTTDGRFLIKTVGKKTAVFFRSILSKYFNHIKSNQNSLITRIYGLHA 617
Query 171 LRL 173
LR
Sbjct 618 LRF 620
> pfa:PFA_0515w PfPIP5K, NCS; phosphatidylinositol-4-phosphate-5-kinase
(EC:2.7.1.68); K00889 1-phosphatidylinositol-4-phosphate
5-kinase [EC:2.7.1.68]
Length=1710
Score = 143 bits (361), Expect = 2e-34, Method: Composition-based stats.
Identities = 67/172 (38%), Positives = 113/172 (65%), Gaps = 13/172 (7%)
Query 1 HFGHESWNMVINIMVGIRLAGARAMSEPHRAVEPYDFLMKEKFSVLPKTGMVDKKRRKPS 60
+FGHE W++V+N+M+GIR++ + S + ++ + LP + K
Sbjct 1345 YFGHERWDLVMNMMIGIRISSIKKFS----INDISNYFHHKDVIQLPTSNAQHK------ 1394
Query 61 LCAVRFIDYAPMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGS 120
V F +YAP++F+ +R +GI S Y+ SVGP+Q++ N++LGNLS+LSEL+SEGKSGS
Sbjct 1395 ---VIFKNYAPIIFKNIRNFYGIKSKEYLTSVGPEQVISNMVLGNLSTLSELLSEGKSGS 1451
Query 121 LFYYTTDGRFMIKTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALR 172
LFY+T++G+++IKTV + + +L YY+H+ +++LTRL G+H+++
Sbjct 1452 LFYFTSNGKYIIKTVCRNIHNLSKKLLPKYYEHIKKNPDSLLTRLYGIHSIK 1503
> tpv:TP02_0200 phosphatidylinositol-4-phosphate 5-kinase; K00889
1-phosphatidylinositol-4-phosphate 5-kinase [EC:2.7.1.68];
K00920 1-phosphatidylinositol-5-phosphate 4-kinase [EC:2.7.1.149]
Length=848
Score = 138 bits (347), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 73/211 (34%), Positives = 119/211 (56%), Gaps = 39/211 (18%)
Query 1 HFGHESWNMVINIMVGIRLAGARAMSEPHRAVEPYDFLMKEKFSV------------LPK 48
HFGHESWN V+N+M+G+ ++ ++ + + D+ +K F + +
Sbjct 454 HFGHESWNHVLNMMIGLSISARHVYAQVNAVLSDDDYNVKLCFHINENSHGLNVTKFITN 513
Query 49 TGMV-----------DKKRRKPSLCA----------------VRFIDYAPMVFRRLRALF 81
TG+ D + SL + F +YAPMVFR++R +
Sbjct 514 TGLSSSRFNSDYIINDNENGDNSLTGNNALISSNNMENVTRRIVFKEYAPMVFRQIRRIS 573
Query 82 GIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGSLFYYTTDGRFMIKTVSKETAF 141
G+ Y+ SV P+Q++GN++LGNLS++SELVSEGKSG+LFYYT +GR ++KT++K A
Sbjct 574 GLSEKDYMESVSPEQIVGNMVLGNLSTMSELVSEGKSGALFYYTINGRLILKTITKRCAK 633
Query 142 FMRSILFDYYKHVSTCTNTMLTRLCGLHALR 172
F++ L Y++H+ +++LTR CGL ++
Sbjct 634 FVKRWLKCYFEHLEKSPDSILTRFCGLFSIE 664
> bbo:BBOV_II007470 18.m06619; 1-phosphatidylinositol-4-phosphate
5-kinase (EC:2.7.1.68); K00889 1-phosphatidylinositol-4-phosphate
5-kinase [EC:2.7.1.68]
Length=662
Score = 136 bits (342), Expect = 5e-32, Method: Compositional matrix adjust.
Identities = 73/184 (39%), Positives = 112/184 (60%), Gaps = 13/184 (7%)
Query 1 HFGHESWNMVINIMVGIRLAGARAMSEPHRAVEPYDFLMKEKFSVLPKTGM--------V 52
+FGHE W+ VIN+MVG+ L + P F K+ FS+ P + V
Sbjct 294 YFGHERWDDVINMMVGLGLTARSKHEDITGDPCPDHFKEKKIFSISPSIAVDNMSSLNFV 353
Query 53 DKKRRKPSLCA-----VRFIDYAPMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLS 107
+ + P+ V F ++AP+VF+RLRAL + YI SVGP+ L+GN++LGNLS
Sbjct 354 ELESGVPTSSGDNSNRVVFTEHAPLVFKRLRALMNLSEEEYIHSVGPEHLVGNMVLGNLS 413
Query 108 SLSELVSEGKSGSLFYYTTDGRFMIKTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCG 167
+LSEL+SEG+SG+LFY+T +G+ ++KTV++ A F+ L YY+H+ N+++TR G
Sbjct 414 TLSELLSEGRSGALFYFTDNGKLVLKTVTRTCAEFVLQWLPHYYQHLKENPNSLITRFAG 473
Query 168 LHAL 171
L ++
Sbjct 474 LFSM 477
> ath:AT3G07960 phosphatidylinositol-4-phosphate 5-kinase family
protein; K00889 1-phosphatidylinositol-4-phosphate 5-kinase
[EC:2.7.1.68]
Length=715
Score = 102 bits (255), Expect = 5e-22, Method: Compositional matrix adjust.
Identities = 67/174 (38%), Positives = 96/174 (55%), Gaps = 18/174 (10%)
Query 3 GHESWNMVINIMVGIRLAGARAMSEPHRAVEPYDFLMKEK-FSVLPKTGMVDKKRRKP-S 60
GH+++ +++N+ +GIR + R ++ F KEK ++ P G K P
Sbjct 325 GHKNYELMLNLQLGIRHSVGRPAPATSLDLKASAFDPKEKLWTKFPSEG---SKYTPPHQ 381
Query 61 LCAVRFIDYAPMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGS 120
C ++ DY P+VFR LR LF +D+ Y+ S I GN +L EL S GKSGS
Sbjct 382 SCEFKWKDYCPVVFRTLRKLFSVDAADYMLS----------ICGN-DALRELSSPGKSGS 430
Query 121 LFYYTTDGRFMIKTVSK-ETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALRL 173
FY T D R+MIKT+ K ET +R +L YY HV C NT++T+ GLH ++L
Sbjct 431 FFYLTNDDRYMIKTMKKAETKVLIR-MLPAYYNHVRACENTLVTKFFGLHCVKL 483
> ath:AT1G21980 ATPIP5K1 (PHOSPHATIDYLINOSITOL-4-PHOSPHATE 5-KINASE
1); 1-phosphatidylinositol-4-phosphate 5-kinase/ actin
filament binding / actin monomer binding / phosphatidylinositol
phosphate kinase; K00889 1-phosphatidylinositol-4-phosphate
5-kinase [EC:2.7.1.68]
Length=752
Score = 101 bits (252), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 67/173 (38%), Positives = 96/173 (55%), Gaps = 19/173 (10%)
Query 3 GHESWNMVINIMVGIRLAGARAMSEPHRAVEPYDFLMKEKF-SVLPKTGMVDKKRRKPSL 61
GH+ +++++N+ +GIR + + S R ++ DF KEKF + P G + P
Sbjct 351 GHKKYDLMLNLQLGIRYSVGKHAS-IVRDLKQTDFDPKEKFWTRFPPEGT----KTTPPH 405
Query 62 CAV--RFIDYAPMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSG 119
+V R+ DY P+VFRRLR LF +D Y+ + I GN +L EL S GKSG
Sbjct 406 QSVDFRWKDYCPLVFRRLRELFQVDPAKYMLA----------ICGN-DALRELSSPGKSG 454
Query 120 SLFYYTTDGRFMIKTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALR 172
S FY T D RFMIKTV K + +L YYKHV N+++TR G+H ++
Sbjct 455 SFFYLTQDDRFMIKTVKKSEVKVLLRMLPSYYKHVCQYENSLVTRFYGVHCVK 507
> ath:AT1G77740 1-phosphatidylinositol-4-phosphate 5-kinase, putative
/ PIP kinase, putative / PtdIns(4)P-5-kinase, putative
/ diphosphoinositide kinase, putative; K00889 1-phosphatidylinositol-4-phosphate
5-kinase [EC:2.7.1.68]
Length=754
Score = 101 bits (251), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 68/172 (39%), Positives = 93/172 (54%), Gaps = 17/172 (9%)
Query 3 GHESWNMVINIMVGIRLAGARAMSEPHRAVEPYDFLMKEKF-SVLPKTGMVDKKRRKPSL 61
GH+ +++++N+ GIR + + S R ++ DF EKF + P G K P L
Sbjct 353 GHKKYDLMLNLQHGIRYSVGKHAS-VVRDLKQSDFDPSEKFWTRFPPEG---SKTTPPHL 408
Query 62 CA-VRFIDYAPMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGS 120
R+ DY P+VFRRLR LF +D Y+ + I GN +L EL S GKSGS
Sbjct 409 SVDFRWKDYCPLVFRRLRELFTVDPADYMLA----------ICGN-DALRELSSPGKSGS 457
Query 121 LFYYTTDGRFMIKTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALR 172
FY T D RFMIKTV K + +L YYKHV NT++TR G+H ++
Sbjct 458 FFYLTQDDRFMIKTVKKSEVKVLLRMLPSYYKHVCQYENTLVTRFYGVHCIK 509
> ath:AT2G26420 PIP5K3; PIP5K3 (1-PHOSPHATIDYLINOSITOL-4-PHOSPHATE
5-KINASE 3); 1-phosphatidylinositol-4-phosphate 5-kinase;
K00889 1-phosphatidylinositol-4-phosphate 5-kinase [EC:2.7.1.68]
Length=705
Score = 96.3 bits (238), Expect = 4e-20, Method: Compositional matrix adjust.
Identities = 63/172 (36%), Positives = 96/172 (55%), Gaps = 17/172 (9%)
Query 3 GHESWNMVINIMVGIRLAGARAMSEPHRAVEPYDFLMKEK-FSVLPKTGMVDKKRRKPSL 61
GH+++++++N+ +GIR + + S R + DF K+K ++ P G K P L
Sbjct 325 GHKNYDLMLNLQLGIRYSVGKHAS-LLRELRHSDFDPKDKQWTRFPPEG---SKSTPPHL 380
Query 62 CA-VRFIDYAPMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGS 120
A ++ DY P+VFR LR LF ID Y+ + I GN SL E S GKSGS
Sbjct 381 SAEFKWKDYCPIVFRHLRDLFAIDQADYMLA----------ICGN-ESLREFASPGKSGS 429
Query 121 LFYYTTDGRFMIKTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALR 172
FY T D R+MIKT+ K + +L +YY+HVS N+++T+ G+H ++
Sbjct 430 AFYLTQDERYMIKTMKKSEIKVLLKMLPNYYEHVSKYKNSLVTKFFGVHCVK 481
> ath:AT3G09920 PIP5K9; PIP5K9 (PHOSPHATIDYL INOSITOL MONOPHOSPHATE
5 KINASE); 1-phosphatidylinositol-4-phosphate 5-kinase/
ATP binding / phosphatidylinositol phosphate kinase; K00889
1-phosphatidylinositol-4-phosphate 5-kinase [EC:2.7.1.68]
Length=815
Score = 95.5 bits (236), Expect = 9e-20, Method: Compositional matrix adjust.
Identities = 60/171 (35%), Positives = 90/171 (52%), Gaps = 14/171 (8%)
Query 3 GHESWNMVINIMVGIRLAGARAMSEPHRAVEPYDFLMKEKFSV-LPKTGMVDKKRRKPSL 61
GH S+++++++ +GIR + R V DF + F + P+ G
Sbjct 391 GHRSYDLMLSLQLGIRYTVGKITPIQRRQVRTADFGPRASFWMTFPRAGSTMTPPHHSE- 449
Query 62 CAVRFIDYAPMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGSL 121
++ DY PMVFR LR +F ID+ Y+ S I GN +L EL S GKSGS+
Sbjct 450 -DFKWKDYCPMVFRNLREMFKIDAADYMMS----------ICGN-DTLRELSSPGKSGSV 497
Query 122 FYYTTDGRFMIKTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALR 172
F+ + D RFMIKT+ K + +L DY+ HV T NT++T+ GLH ++
Sbjct 498 FFLSQDDRFMIKTLRKSEVKVLLRMLPDYHHHVKTYENTLITKFFGLHRIK 548
> ath:AT2G41210 PIP5K5; PIP5K5 (PHOSPHATIDYLINOSITOL- 4-PHOSPHATE
5-KINASE 5); 1-phosphatidylinositol-4-phosphate 5-kinase;
K00889 1-phosphatidylinositol-4-phosphate 5-kinase [EC:2.7.1.68]
Length=772
Score = 94.4 bits (233), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 61/173 (35%), Positives = 89/173 (51%), Gaps = 16/173 (9%)
Query 3 GHESWNMVINIMVGIRLAGARAMSEPHRAVEPYDFLMKEK-FSVLPKTGMVDKKRRKP-S 60
GH ++ +++N+ +GIR + R ++P F K+K + P+ G K P
Sbjct 379 GHRNYELMLNLQLGIRHSVGRQAPAASLDLKPSAFDPKDKIWRRFPREGT---KYTPPHQ 435
Query 61 LCAVRFIDYAPMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGS 120
++ DY P+VFR LR LF +D Y+ S I GN +L EL S GKSGS
Sbjct 436 STEFKWKDYCPLVFRSLRKLFKVDPADYMLS----------ICGN-DALRELSSPGKSGS 484
Query 121 LFYYTTDGRFMIKTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALRL 173
FY T D R+MIKT+ K + +L YY HV N+++ R GLH ++L
Sbjct 485 FFYLTNDDRYMIKTMKKSETKVLLGMLAAYYNHVRAFENSLVIRFFGLHCVKL 537
> sce:YDR208W MSS4; Mss4p (EC:2.7.1.68); K00889 1-phosphatidylinositol-4-phosphate
5-kinase [EC:2.7.1.68]
Length=779
Score = 93.6 bits (231), Expect = 3e-19, Method: Composition-based stats.
Identities = 58/171 (33%), Positives = 89/171 (52%), Gaps = 16/171 (9%)
Query 3 GHESWNMVINIMVGIRLAGARAMSEPHRAVEPYDFLMKEKFSVLPKTGMVDKKRRKPSLC 62
GH ++ + N++ GIR+A +R S + + P DF +K + + S
Sbjct 382 GHVNFIIAYNMLTGIRVAVSRC-SGIMKPLTPADFRFTKKLAF----DYHGNELTPSSQY 436
Query 63 AVRFIDYAPMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGSLF 122
A +F DY P VFR LRALFG+D Y+ S+ +L SEL S GKSGS F
Sbjct 437 AFKFKDYCPEVFRELRALFGLDPADYLVSLTSKYIL-----------SELNSPGKSGSFF 485
Query 123 YYTTDGRFMIKTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALRL 173
YY+ D +++IKT+ +R + +YY HV NT++ + GLH +++
Sbjct 486 YYSRDYKYIIKTIHHSEHIHLRKHIQEYYNHVRDNPNTLICQFYGLHRVKM 536
> ath:AT3G56960 PIP5K4; PIP5K4 (PHOSPHATIDYL INOSITOL MONOPHOSPHATE
5 KINASE 4); 1-phosphatidylinositol-4-phosphate 5-kinase;
K00889 1-phosphatidylinositol-4-phosphate 5-kinase [EC:2.7.1.68]
Length=779
Score = 92.8 bits (229), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 66/178 (37%), Positives = 93/178 (52%), Gaps = 26/178 (14%)
Query 3 GHESWNMVINIMVGIRLA-GARA----MSEPHRAVEPYDFLMKEK-FSVLPKTGMVDKKR 56
GH ++ +++N+ +GIR A G +A + H A +P KEK ++ P G K
Sbjct 386 GHRNYELMLNLQLGIRHAVGKQAPVVSLDLKHSAFDP-----KEKVWTRFPPEGT---KY 437
Query 57 RKPSLCA-VRFIDYAPMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSE 115
P + ++ DY P+VFR LR LF +D Y+ S I GN +L EL S
Sbjct 438 TPPHQSSEFKWKDYCPLVFRSLRKLFKVDPADYMLS----------ICGN-DALRELSSP 486
Query 116 GKSGSLFYYTTDGRFMIKTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALRL 173
GKSGS FY T D R+MIKT+ K + +L YY HV NT++ R GLH ++L
Sbjct 487 GKSGSFFYLTNDDRYMIKTMKKSETKVLLRMLAAYYNHVRAFENTLVIRFYGLHCVKL 544
> ath:AT1G10900 phosphatidylinositol-4-phosphate 5-kinase family
protein; K00889 1-phosphatidylinositol-4-phosphate 5-kinase
[EC:2.7.1.68]
Length=754
Score = 89.4 bits (220), Expect = 6e-18, Method: Compositional matrix adjust.
Identities = 57/174 (32%), Positives = 88/174 (50%), Gaps = 18/174 (10%)
Query 3 GHESWNMVINIMVGIRLAGARAMSEPHRAVEPYDFLMKEKFSVL-PKTGMVDKKRRKPSL 61
G ++ +++N+ +GIR + P R V DF + + P+ G P
Sbjct 329 GEHNYYLMLNLQLGIRYTVGKITPVPRREVRASDFGKNARTKMFFPRDG----SNFTPPH 384
Query 62 CAVRFI--DYAPMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSG 119
+V F DY PMVFR LR +F +D+ Y+ S+ D L+E+ S GKSG
Sbjct 385 KSVDFSWKDYCPMVFRNLRQMFKLDAAEYMMSICGDD-----------GLTEISSPGKSG 433
Query 120 SLFYYTTDGRFMIKTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALRL 173
S+FY + D RF+IKT+ K + +L YY+HV NT++T+ G+H + L
Sbjct 434 SIFYLSHDDRFVIKTLKKSELQVLLRMLPKYYEHVGDHENTLITKFFGVHRITL 487
> ath:AT1G60890 phosphatidylinositol-4-phosphate 5-kinase family
protein; K00889 1-phosphatidylinositol-4-phosphate 5-kinase
[EC:2.7.1.68]
Length=769
Score = 89.0 bits (219), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 56/172 (32%), Positives = 89/172 (51%), Gaps = 20/172 (11%)
Query 7 WN--MVINIMVGIRLAGARAMSEPHRAVEPYDFLMKEKFSVL-PKTGMVDKKRRKPSLCA 63
WN +++N+ +GIR + P R V DF + + + P+ G + P +
Sbjct 344 WNHYLMLNLQLGIRYTVGKITPVPPREVRASDFSERARIMMFFPRNG----SQYTPPHKS 399
Query 64 VRFI--DYAPMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGSL 121
+ F DY PMVFR LR +F +D+ Y+ S+ D L E+ S GKSGS+
Sbjct 400 IDFDWKDYCPMVFRNLREMFKLDAADYMMSICGDD-----------GLREISSPGKSGSI 448
Query 122 FYYTTDGRFMIKTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALRL 173
FY + D RF+IKT+ + + +L YY+HV NT++T+ G+H ++L
Sbjct 449 FYLSHDDRFVIKTLKRSELKVLLRMLPRYYEHVGDYENTLITKFFGVHRIKL 500
> dre:447840 fj29b05, fj93b05, wu:fj29b05, wu:fj93b05; zgc:92316
(EC:2.7.1.68); K00889 1-phosphatidylinositol-4-phosphate
5-kinase [EC:2.7.1.68]
Length=527
Score = 75.1 bits (183), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 51/160 (31%), Positives = 77/160 (48%), Gaps = 15/160 (9%)
Query 13 IMVGIRLAGARAMSEPHRAVEPYDFLMKEKFSVLPKTGMVDKKRRKPSLCAVRFIDYAPM 72
I +GI S+P R V DF + E + + + P RF YAP+
Sbjct 33 IQLGIGYTVGNLTSKPDRDVLMQDFYVVESVFLPSEGSNLTPAHHYPDF---RFKTYAPL 89
Query 73 VFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGSLFYYTTDGRFMI 132
FR R LFGI Y+ S+ + L+ EL + G SGSLFY T+D F+I
Sbjct 90 AFRYFRELFGIKPDDYLYSICKEPLI------------ELSNPGASGSLFYLTSDDEFII 137
Query 133 KTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALR 172
KTV + A F++ +L YY +++ T+L + GL+ ++
Sbjct 138 KTVQHKEAEFLQKLLPGYYMNLNQNPRTLLPKFYGLYCVQ 177
> mmu:18719 Pip5k1b, AI844522, Pip5k1a, Pipk5a, Pipk5b, STM7;
phosphatidylinositol-4-phosphate 5-kinase, type 1 beta (EC:2.7.1.68);
K00889 1-phosphatidylinositol-4-phosphate 5-kinase
[EC:2.7.1.68]
Length=539
Score = 74.7 bits (182), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 50/160 (31%), Positives = 77/160 (48%), Gaps = 15/160 (9%)
Query 13 IMVGIRLAGARAMSEPHRAVEPYDFLMKEKFSVLPKTGMVDKKRRKPSLCAVRFIDYAPM 72
I +GI S+P R V DF + E + + + P RF YAP+
Sbjct 33 IQLGIGYTVGNLTSKPERDVLMQDFYVVESVFLPSEGSNLTPAHHYPDF---RFKTYAPL 89
Query 73 VFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGSLFYYTTDGRFMI 132
FR R LFGI Y+ S+ + L+ EL + G SGSLF+ T+D F+I
Sbjct 90 AFRYFRELFGIKPDDYLYSICSEPLI------------ELSNPGASGSLFFLTSDDEFII 137
Query 133 KTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALR 172
KTV + A F++ +L YY +++ T+L + GL+ ++
Sbjct 138 KTVQHKEAEFLQKLLPGYYMNLNQNPRTLLPKFYGLYCMQ 177
> hsa:8395 PIP5K1B, MSS4, STM7; phosphatidylinositol-4-phosphate
5-kinase, type I, beta (EC:2.7.1.68); K00889 1-phosphatidylinositol-4-phosphate
5-kinase [EC:2.7.1.68]
Length=540
Score = 74.3 bits (181), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 50/160 (31%), Positives = 77/160 (48%), Gaps = 15/160 (9%)
Query 13 IMVGIRLAGARAMSEPHRAVEPYDFLMKEKFSVLPKTGMVDKKRRKPSLCAVRFIDYAPM 72
I +GI S+P R V DF + E + + + P RF YAP+
Sbjct 33 IQLGIGYTVGNLTSKPERDVLMQDFYVVESVFLPSEGSNLTPAHHYPDF---RFKTYAPL 89
Query 73 VFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGSLFYYTTDGRFMI 132
FR R LFGI Y+ S+ + L+ EL + G SGSLF+ T+D F+I
Sbjct 90 AFRYFRELFGIKPDDYLYSICSEPLI------------ELSNPGASGSLFFVTSDDEFII 137
Query 133 KTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALR 172
KTV + A F++ +L YY +++ T+L + GL+ ++
Sbjct 138 KTVQHKEAEFLQKLLPGYYMNLNQNPRTLLPKFYGLYCMQ 177
> xla:398687 pip5k1c, MGC130766; phosphatidylinositol-4-phosphate
5-kinase, type I, gamma (EC:2.7.1.68); K00889 1-phosphatidylinositol-4-phosphate
5-kinase [EC:2.7.1.68]
Length=240
Score = 74.3 bits (181), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 50/160 (31%), Positives = 77/160 (48%), Gaps = 15/160 (9%)
Query 13 IMVGIRLAGARAMSEPHRAVEPYDFLMKEKFSVLPKTGMVDKKRRKPSLCAVRFIDYAPM 72
I +GI S+P R V DF + E + + + R P RF YAP+
Sbjct 33 IQLGIGYTVGNLTSKPDRDVLMQDFYVVESVFLPSEGSNLTPAHRYPDF---RFKTYAPL 89
Query 73 VFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGSLFYYTTDGRFMI 132
FR R LFGI Y+ S+ + L+ EL + G SGS+F+ T+D F+I
Sbjct 90 AFRYFRELFGIKPDDYLYSLCSEPLI------------ELSNPGASGSIFFVTSDDEFII 137
Query 133 KTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALR 172
KTV + A F++ +L YY +++ T+L + GL+ +
Sbjct 138 KTVQHKEAEFLQKLLPGYYMNLNQNPRTLLPKFYGLYCFQ 177
> hsa:23396 PIP5K1C, KIAA0589, LCCS3, PIP5K-GAMMA, PIP5Kgamma,
PIPKIg_v4; phosphatidylinositol-4-phosphate 5-kinase, type
I, gamma (EC:2.7.1.68); K00889 1-phosphatidylinositol-4-phosphate
5-kinase [EC:2.7.1.68]
Length=640
Score = 72.0 bits (175), Expect = 9e-13, Method: Composition-based stats.
Identities = 53/162 (32%), Positives = 77/162 (47%), Gaps = 19/162 (11%)
Query 13 IMVGIRLAGARAMSEPHRAVEPYDFLMKEKFSVLPKTGMVDKKRRKPS--LCAVRFIDYA 70
I +GI S+P R V DF + E P G P+ RF YA
Sbjct 83 IQLGIGYTVGHLSSKPERDVLMQDFYVVESI-FFPSEG----SNLTPAHHFQDFRFKTYA 137
Query 71 PMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGSLFYYTTDGRF 130
P+ FR R LFGI Y+ S+ + L+ EL + G SGSLFY T+D F
Sbjct 138 PVAFRYFRELFGIRPDDYLYSLCNEPLI------------ELSNPGASGSLFYVTSDDEF 185
Query 131 MIKTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALR 172
+IKTV + A F++ +L YY +++ T+L + GL+ ++
Sbjct 186 IIKTVMHKEAEFLQKLLPGYYMNLNQNPRTLLPKFYGLYCVQ 227
> xla:444238 pip5k1b, MGC80809; phosphatidylinositol-4-phosphate
5-kinase, type I, beta (EC:2.7.1.68); K00889 1-phosphatidylinositol-4-phosphate
5-kinase [EC:2.7.1.68]
Length=537
Score = 72.0 bits (175), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 52/161 (32%), Positives = 78/161 (48%), Gaps = 17/161 (10%)
Query 13 IMVGIRLAGARAMSEPHRAVEPYDFLMKEKFSVLPKTGM-VDKKRRKPSLCAVRFIDYAP 71
I +GI S+P R V DF + E LP G + P RF YAP
Sbjct 33 IQLGIGYTVGNLTSKPDRDVLMQDFYVVESV-FLPSEGSNLTPAHHYPDF---RFKTYAP 88
Query 72 MVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGSLFYYTTDGRFM 131
+ FR R LFGI Y+ S+ + L+ EL + G SGS+F+ T+D F+
Sbjct 89 LAFRYFRELFGIKPDDYLYSLCSEPLI------------ELSNPGASGSIFFVTSDDEFI 136
Query 132 IKTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALR 172
IKTV + A F++ +L YY +++ T+L + GL+ ++
Sbjct 137 IKTVQHKEAEFLQKLLPGYYMNLNQNPRTLLPKFYGLYCVQ 177
> dre:449831 pip5k1b, zgc:101046; phosphatidylinositol-4-phosphate
5-kinase, type I, beta (EC:2.7.1.68); K00889 1-phosphatidylinositol-4-phosphate
5-kinase [EC:2.7.1.68]
Length=521
Score = 71.6 bits (174), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 49/160 (30%), Positives = 76/160 (47%), Gaps = 15/160 (9%)
Query 13 IMVGIRLAGARAMSEPHRAVEPYDFLMKEKFSVLPKTGMVDKKRRKPSLCAVRFIDYAPM 72
I +GI S+P R V DF + E + + + P RF +YAP+
Sbjct 33 IQLGIGYTVGNLTSKPDRDVLMQDFYVVESVFLPSEGSNLTPAHHYPDF---RFKNYAPL 89
Query 73 VFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGSLFYYTTDGRFMI 132
FR R LFGI Y+ S+ + L+ EL + G S S FY T+D F+I
Sbjct 90 AFRYFRELFGIKPDDYLYSICKEPLI------------ELSNPGASSSWFYLTSDDEFII 137
Query 133 KTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALR 172
KTV + A F++ +L YY +++ T+L + GL+ ++
Sbjct 138 KTVQHKEAEFLQKLLPGYYMNLNQNPRTLLPKFYGLYCIQ 177
> dre:100005033 si:ch211-243a15.1
Length=802
Score = 71.2 bits (173), Expect = 2e-12, Method: Composition-based stats.
Identities = 49/160 (30%), Positives = 75/160 (46%), Gaps = 15/160 (9%)
Query 13 IMVGIRLAGARAMSEPHRAVEPYDFLMKEKFSVLPKTGMVDKKRRKPSLCAVRFIDYAPM 72
I +GI S+P R V DF + E + + P RF YAP+
Sbjct 126 IQLGIGYTVGNLSSKPERDVLMQDFYVVESIFFPSEGSNLTPAHHYPDF---RFKTYAPV 182
Query 73 VFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGSLFYYTTDGRFMI 132
FR R LFGI Y+ S+ + L+ EL + G SGS+FY T D F++
Sbjct 183 AFRYFRELFGIRPDDYLYSLCNEPLI------------ELSNPGASGSIFYVTRDDEFIL 230
Query 133 KTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALR 172
KTV + A F++ +L YY +++ T+L + GL+ ++
Sbjct 231 KTVMHKEAEFLQKLLPGYYMNLNQNPRTLLPKFFGLYCVQ 270
> mmu:18717 Pip5k1c, AI115456, AI835305, Pip5kIgamma; phosphatidylinositol-4-phosphate
5-kinase, type 1 gamma (EC:2.7.1.68);
K00889 1-phosphatidylinositol-4-phosphate 5-kinase [EC:2.7.1.68]
Length=635
Score = 70.5 bits (171), Expect = 2e-12, Method: Composition-based stats.
Identities = 52/162 (32%), Positives = 77/162 (47%), Gaps = 19/162 (11%)
Query 13 IMVGIRLAGARAMSEPHRAVEPYDFLMKEKFSVLPKTGMVDKKRRKPS--LCAVRFIDYA 70
I +GI S+P R V DF + E P G P+ RF YA
Sbjct 83 IQLGIGYTVGNLSSKPERDVLMQDFYVVESI-FFPSEG----SNLTPAHHFQDFRFKTYA 137
Query 71 PMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGSLFYYTTDGRF 130
P+ FR R LFGI Y+ S+ + L+ EL + G SGS+FY T+D F
Sbjct 138 PVAFRYFRELFGIRPDDYLYSLCNEPLI------------ELSNPGASGSVFYVTSDDEF 185
Query 131 MIKTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALR 172
+IKTV + A F++ +L YY +++ T+L + GL+ ++
Sbjct 186 IIKTVMHKEAEFLQKLLPGYYMNLNQNPRTLLPKFYGLYCVQ 227
> hsa:8394 PIP5K1A; phosphatidylinositol-4-phosphate 5-kinase,
type I, alpha (EC:2.7.1.68); K00889 1-phosphatidylinositol-4-phosphate
5-kinase [EC:2.7.1.68]
Length=522
Score = 70.1 bits (170), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 40/108 (37%), Positives = 60/108 (55%), Gaps = 12/108 (11%)
Query 65 RFIDYAPMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGSLFYY 124
RF YAP+ FR R LFGI Y+ S+ + L+ EL S G SGSLFY
Sbjct 126 RFKTYAPVAFRYFRELFGIRPDDYLYSLCSEPLI------------ELCSSGASGSLFYV 173
Query 125 TTDGRFMIKTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALR 172
++D F+IKTV + A F++ +L YY +++ T+L + GL+ ++
Sbjct 174 SSDDEFIIKTVQHKEAEFLQKLLPGYYMNLNQNPRTLLPKFYGLYCVQ 221
> mmu:18720 Pip5k1a, Pip5k1b, Pipk5a, Pipk5b; phosphatidylinositol-4-phosphate
5-kinase, type 1 alpha (EC:2.7.1.68); K00889
1-phosphatidylinositol-4-phosphate 5-kinase [EC:2.7.1.68]
Length=546
Score = 68.6 bits (166), Expect = 9e-12, Method: Compositional matrix adjust.
Identities = 39/108 (36%), Positives = 60/108 (55%), Gaps = 12/108 (11%)
Query 65 RFIDYAPMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGSLFYY 124
RF YAP+ FR R LFGI Y+ S+ + L+ EL + G SGSLFY
Sbjct 123 RFKTYAPVAFRYFRELFGIRPDDYLYSLCSEPLI------------ELSNSGASGSLFYV 170
Query 125 TTDGRFMIKTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALR 172
++D F+IKTV + A F++ +L YY +++ T+L + GL+ ++
Sbjct 171 SSDDEFIIKTVQHKEAEFLQKLLPGYYMNLNQNPRTLLPKFYGLYCVQ 218
> dre:492549 im:7140933; zgc:158838; K00889 1-phosphatidylinositol-4-phosphate
5-kinase [EC:2.7.1.68]
Length=584
Score = 67.8 bits (164), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 39/108 (36%), Positives = 60/108 (55%), Gaps = 12/108 (11%)
Query 65 RFIDYAPMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGSLFYY 124
RF YAP+ FR R LFGI Y+ S+ D L+ EL + G SGS+FY
Sbjct 113 RFKTYAPIAFRYFRELFGIRPDDYLYSLCNDPLI------------ELPNPGASGSIFYV 160
Query 125 TTDGRFMIKTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALR 172
T+D F+IKTV + A F++ +L Y+ +++ T+L + GL+ ++
Sbjct 161 TSDDEFIIKTVMHKEAEFLQKLLPGYFMNLNQNKRTLLPKFYGLYCVQ 208
> dre:798876 si:ch211-197g15.13; K00889 1-phosphatidylinositol-4-phosphate
5-kinase [EC:2.7.1.68]
Length=682
Score = 67.8 bits (164), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 50/160 (31%), Positives = 75/160 (46%), Gaps = 15/160 (9%)
Query 13 IMVGIRLAGARAMSEPHRAVEPYDFLMKEKFSVLPKTGMVDKKRRKPSLCAVRFIDYAPM 72
I +GI S+P R V DF + E + + P RF YAP+
Sbjct 88 IQLGIGYTVGNLSSKPERDVLMQDFYVVESIFFPSEGSNLTPAHHFPDF---RFKTYAPV 144
Query 73 VFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGSLFYYTTDGRFMI 132
FR R LFGI Y+ S+ + L+ EL + G SGS+FY T D F+I
Sbjct 145 AFRYFRELFGIRPDDYLYSLCNEPLI------------ELSNPGASGSVFYLTKDDEFII 192
Query 133 KTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALR 172
KTV + A F++ +L YY +++ T+L + GL+ ++
Sbjct 193 KTVMHKEAEFLQKLLPGYYMNLNQNPRTLLPKFFGLYCVQ 232
> cel:F55A12.3 ppk-1; PIP Kinase family member (ppk-1); K00889
1-phosphatidylinositol-4-phosphate 5-kinase [EC:2.7.1.68]
Length=611
Score = 66.2 bits (160), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 51/158 (32%), Positives = 74/158 (46%), Gaps = 19/158 (12%)
Query 13 IMVGIRLAGARAMSEPHRAVEPYDFLMKEKFSVL--PKTGMVDKKRRKPSLCAVRFIDYA 70
I +GI + S P+R V DF EK ++ P G S RF YA
Sbjct 92 IQLGISNSIGSLASLPNRDVLLQDF---EKVDIVAFPAAGSTITPSH--SFGDFRFRTYA 146
Query 71 PMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGSLFYYTTDGRF 130
P+ FR R LF I ++RS+ L EL + G SGS+FY + D +F
Sbjct 147 PIAFRYFRNLFHIKPADFLRSIC------------TEPLKELSNAGASGSIFYVSQDDQF 194
Query 131 MIKTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGL 168
+IKTV + A F++ +L YY +++ T+L + GL
Sbjct 195 IIKTVQHKEADFLQKLLPGYYMNLNQNPRTLLPKFFGL 232
> xla:399203 pip5k1a, pip5k1c; phosphatidylinositol-4-phosphate
5-kinase, type I, alpha (EC:2.7.1.68); K00889 1-phosphatidylinositol-4-phosphate
5-kinase [EC:2.7.1.68]
Length=570
Score = 65.9 bits (159), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 38/108 (35%), Positives = 59/108 (54%), Gaps = 12/108 (11%)
Query 65 RFIDYAPMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGSLFYY 124
RF YAP+ FR R LFGI Y+ S+ + L+ EL + G SGS+FY
Sbjct 116 RFKTYAPVAFRYFRELFGIRPDDYLYSLCNEPLI------------ELSNPGASGSVFYV 163
Query 125 TTDGRFMIKTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALR 172
+ D F+IKTV + A F++ +L YY +++ T+L + GL+ ++
Sbjct 164 SGDDEFIIKTVQHKEAEFLQKLLPGYYMNLNQNPRTLLPKFYGLYCVQ 211
> mmu:108083 Pip4k2b, AI848124, PI5P4Kbeta, Pip5k2b, c11; phosphatidylinositol-5-phosphate
4-kinase, type II, beta (EC:2.7.1.149);
K00920 1-phosphatidylinositol-5-phosphate 4-kinase
[EC:2.7.1.149]
Length=416
Score = 65.1 bits (157), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 47/136 (34%), Positives = 67/136 (49%), Gaps = 14/136 (10%)
Query 38 LMKEKFSVLPKTGMVDKKRRKPSLCA-VRFIDYAPMVFRRLRALFGIDSLCYIRSVGPDQ 96
LM + F K + + K +L + +F +Y PMVFR LR FGID Y SV
Sbjct 66 LMPDDFKAYSKIKVDNHLFNKENLPSRFKFKEYCPMVFRNLRERFGIDDQDYQNSV---- 121
Query 97 LLGNLILGNLSSLSELVSEGKSGSLFYYTTDGRFMIKTVSKETAFFMRSILFDYYKHVST 156
S+ S+G+ G+ F T D RF+IKTVS E M +IL Y++ +
Sbjct 122 --------TRSAPINSDSQGRCGTRFLTTYDRRFVIKTVSSEDVAEMHNILKKYHQFIVE 173
Query 157 C-TNTMLTRLCGLHAL 171
C NT+L + G++ L
Sbjct 174 CHGNTLLPQFLGMYRL 189
> hsa:8396 PIP4K2B, PI5P4KB, PIP5K2B, PIP5KIIB, PIP5KIIbeta; phosphatidylinositol-5-phosphate
4-kinase, type II, beta (EC:2.7.1.149);
K00920 1-phosphatidylinositol-5-phosphate 4-kinase
[EC:2.7.1.149]
Length=416
Score = 65.1 bits (157), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 47/136 (34%), Positives = 67/136 (49%), Gaps = 14/136 (10%)
Query 38 LMKEKFSVLPKTGMVDKKRRKPSLCA-VRFIDYAPMVFRRLRALFGIDSLCYIRSVGPDQ 96
LM + F K + + K +L + +F +Y PMVFR LR FGID Y SV
Sbjct 66 LMPDDFKAYSKIKVDNHLFNKENLPSRFKFKEYCPMVFRNLRERFGIDDQDYQNSV---- 121
Query 97 LLGNLILGNLSSLSELVSEGKSGSLFYYTTDGRFMIKTVSKETAFFMRSILFDYYKHVST 156
S+ S+G+ G+ F T D RF+IKTVS E M +IL Y++ +
Sbjct 122 --------TRSAPINSDSQGRCGTRFLTTYDRRFVIKTVSSEDVAEMHNILKKYHQFIVE 173
Query 157 C-TNTMLTRLCGLHAL 171
C NT+L + G++ L
Sbjct 174 CHGNTLLPQFLGMYRL 189
> dre:553629 pip5k1a, MGC110576, zgc:110576; phosphatidylinositol-4-phosphate
5-kinase, type I, alpha (EC:2.7.1.68); K00889
1-phosphatidylinositol-4-phosphate 5-kinase [EC:2.7.1.68]
Length=559
Score = 64.7 bits (156), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 36/108 (33%), Positives = 61/108 (56%), Gaps = 12/108 (11%)
Query 65 RFIDYAPMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGSLFYY 124
+F YAP+ FR R +FGI Y+ S+ + L+ EL + G SGSLFY
Sbjct 115 KFKTYAPIAFRYFREMFGIRPDDYLYSLCNEPLI------------ELSNPGASGSLFYV 162
Query 125 TTDGRFMIKTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALR 172
++D F+IKTV + A F++++L Y+ +++ T+L + GL+ ++
Sbjct 163 SSDDEFIIKTVQHKEAEFLQTLLPGYFMNLNQNMRTLLPKFYGLYCVQ 210
> xla:100036841 pip4k2a, pi5p4ka, pip5k2a, pip5kiia, pipk; phosphatidylinositol-5-phosphate
4-kinase, type II, alpha (EC:2.7.1.149);
K00920 1-phosphatidylinositol-5-phosphate 4-kinase
[EC:2.7.1.149]
Length=415
Score = 63.9 bits (154), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 47/136 (34%), Positives = 69/136 (50%), Gaps = 14/136 (10%)
Query 38 LMKEKFSVLPKTGMVDKKRRKPSLCA-VRFIDYAPMVFRRLRALFGIDSLCYIRSVGPDQ 96
LM + F K + + K +L + +F +Y PMVFR LR FGID DQ
Sbjct 67 LMPDDFKAYSKIKVDNHLFNKENLPSRFKFKEYCPMVFRNLRERFGID----------DQ 116
Query 97 LLGNLILGNLSSLSELVSEGKSGSLFYYTTDGRFMIKTVSKETAFFMRSILFDYYKHVST 156
N + + SE ++G+ GS F T D RF+IKT+S E M +IL Y++ +
Sbjct 117 DYQNSLTRSAPVNSE--NQGRFGSRFLTTYDRRFVIKTISSEDVAEMHNILKKYHQFIVE 174
Query 157 C-TNTMLTRLCGLHAL 171
C NT+L + G++ L
Sbjct 175 CHGNTLLPQFLGMYRL 190
> xla:399394 pip4k2b, MGC81828, PI5P4KB, PIP5KIIB, PIP5KIIbeta,
pip5k2b; phosphatidylinositol-5-phosphate 4-kinase, type II,
beta (EC:2.7.1.149); K00920 1-phosphatidylinositol-5-phosphate
4-kinase [EC:2.7.1.149]
Length=417
Score = 62.0 bits (149), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 47/138 (34%), Positives = 68/138 (49%), Gaps = 18/138 (13%)
Query 38 LMKEKFSVLPKTGMVDKKRRKPSLCA-VRFIDYAPMVFRRLRALFGIDSLCYIRSVGPDQ 96
LM + F K + + K +L + +F +Y PMVFR LR FGID Y S
Sbjct 69 LMPDDFKAYSKIKVDNHLFNKENLPSRFKFKEYCPMVFRNLRERFGIDDQDYQNS----- 123
Query 97 LLGNLILGNLSSLSELVSE--GKSGSLFYYTTDGRFMIKTVSKETAFFMRSILFDYYKHV 154
L+ + + SE G+ GS F T D RF+IKT+S E M +IL Y++ +
Sbjct 124 ---------LTRSAPVNSENLGRFGSRFLTTYDRRFVIKTISGEDVAEMHNILKKYHQFI 174
Query 155 STC-TNTMLTRLCGLHAL 171
C NT+L + G++ L
Sbjct 175 VECHGNTLLPQFLGMYRL 192
> hsa:5305 PIP4K2A, FLJ13267, PI5P4KA, PIP5K2A, PIP5KII-alpha,
PIP5KIIA, PIPK; phosphatidylinositol-5-phosphate 4-kinase,
type II, alpha (EC:2.7.1.149); K00920 1-phosphatidylinositol-5-phosphate
4-kinase [EC:2.7.1.149]
Length=406
Score = 60.8 bits (146), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 35/110 (31%), Positives = 58/110 (52%), Gaps = 13/110 (11%)
Query 65 RFIDYAPMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGSLFYY 124
+F +Y PMVFR LR FGID + S+ L N S+ +SG+ F+
Sbjct 89 KFKEYCPMVFRNLRERFGIDDQDFQNSLTRSAPLPN------------DSQARSGARFHT 136
Query 125 TTDGRFMIKTVSKETAFFMRSILFDYYKHVSTCTN-TMLTRLCGLHALRL 173
+ D R++IKT++ E M +IL Y++++ C T+L + G++ L +
Sbjct 137 SYDKRYIIKTITSEDVAEMHNILKKYHQYIVECHGITLLPQFLGMYRLNV 186
> mmu:18718 Pip4k2a, AW742916, Pip5k2a; phosphatidylinositol-5-phosphate
4-kinase, type II, alpha (EC:2.7.1.149); K00920 1-phosphatidylinositol-5-phosphate
4-kinase [EC:2.7.1.149]
Length=405
Score = 60.5 bits (145), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 35/110 (31%), Positives = 58/110 (52%), Gaps = 13/110 (11%)
Query 65 RFIDYAPMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGSLFYY 124
+F +Y PMVFR LR FGID + S+ L N S+ +SG+ F+
Sbjct 89 KFKEYCPMVFRNLRERFGIDDQDFQNSLTRSAPLPN------------DSQARSGARFHT 136
Query 125 TTDGRFMIKTVSKETAFFMRSILFDYYKHVSTCTN-TMLTRLCGLHALRL 173
+ D R++IKT++ E M +IL Y++++ C T+L + G++ L +
Sbjct 137 SYDKRYVIKTITSEDVAEMHNILKKYHQYIVECHGVTLLPQFLGMYRLNV 186
> dre:560360 similar to type II phosphatidylinositolphosphate
kinase-alpha; K00920 1-phosphatidylinositol-5-phosphate 4-kinase
[EC:2.7.1.149]
Length=404
Score = 59.7 bits (143), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 36/108 (33%), Positives = 61/108 (56%), Gaps = 13/108 (12%)
Query 65 RFIDYAPMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGSLFYY 124
+F +Y P+VFR LR F ID DQ N + + +SE ++G+SG+ F+
Sbjct 88 KFKEYCPLVFRNLRERFVID----------DQDFQNSLTRSAPLVSE--AQGRSGARFHT 135
Query 125 TTDGRFMIKTVSKETAFFMRSILFDYYKHVSTCT-NTMLTRLCGLHAL 171
+ D R++IKT+S E M +IL Y++++ C +T+L + G++ L
Sbjct 136 SYDKRYVIKTISSEDVAEMHNILKKYHQYIVECHGSTLLPQFLGMYRL 183
> ath:AT4G01190 ATPIPK10 (Arabidopsis phosphatidylinositol phosphate
kinase 10); 1-phosphatidylinositol-4-phosphate 5-kinase;
K00889 1-phosphatidylinositol-4-phosphate 5-kinase [EC:2.7.1.68]
Length=401
Score = 56.2 bits (134), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 32/113 (28%), Positives = 59/113 (52%), Gaps = 12/113 (10%)
Query 60 SLCAVRFIDYAPMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSG 119
S+ + DY P+ F ++ L GID Y+ S+ D+ L + +S GK G
Sbjct 25 SITEFDWKDYCPVGFGLIQELEGIDHDDYLLSICTDETL------------KKISSGKIG 72
Query 120 SLFYYTTDGRFMIKTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALR 172
++F+ + D RF+IK + K +L YY+H++ +++ TR+ G H+++
Sbjct 73 NVFHISNDNRFLIKILRKSEIKVTLEMLPRYYRHINYHRSSLFTRIFGAHSVK 125
> cel:Y48G9A.8 ppk-2; PIP Kinase family member (ppk-2); K00920
1-phosphatidylinositol-5-phosphate 4-kinase [EC:2.7.1.149]
Length=401
Score = 55.5 bits (132), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 42/146 (28%), Positives = 70/146 (47%), Gaps = 15/146 (10%)
Query 32 VEPYDFLMKEKFSVLPKTGMVDKKRRK---PSLCAVRFIDYAPMVFRRLRALFGIDSLCY 88
V P LM + F K + + K PS V+ +Y P VFR LR FG+D+ Y
Sbjct 49 VPPPGLLMPDDFKAYSKVKIDNHNFNKDIMPSHYKVK--EYCPNVFRNLREQFGVDNFEY 106
Query 89 IRSVGPDQLLGNLILGNLSSLSELVSEGKSGSLFYYTTDGRFMIKTVSKETAFFMRSILF 148
+RS+ + +L+ G S S F+ + D +F+IK++ E + S+L
Sbjct 107 LRSLTSYEPEPDLLDG---------SAKDSTPRFFISYDKKFVIKSMDSEAVAELHSVLR 157
Query 149 DYYKH-VSTCTNTMLTRLCGLHALRL 173
+Y+++ V T+L + GL+ L +
Sbjct 158 NYHQYVVEKQGKTLLPQYLGLYRLTI 183
> dre:555920 novel protein similar to vertebrate phosphatidylinositol-4-phosphate
5-kinase, type II, alpha (PIP5K2A)
Length=413
Score = 55.1 bits (131), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 41/139 (29%), Positives = 67/139 (48%), Gaps = 18/139 (12%)
Query 34 PYDFLMKEKFSVLPKTGMVDKKRRKPSLCAVRFIDYAPMVFRRLRALFGIDSLCYIRSVG 93
P DF K V + K PS +F +Y P+VFR LR F ID
Sbjct 67 PDDFKANSKIKV---DNHLFNKENMPS--HFKFKEYCPLVFRNLRERFSID--------- 112
Query 94 PDQLLGNLILGNLSSLSELVSEGKSGSLFYYTTDGRFMIKTVSKETAFFMRSILFDYYKH 153
DQ N + S+ ++G+SG+ F+ + + R++IK ++ E M +IL Y+++
Sbjct 113 -DQEYQNSLTRRAPIPSD--AQGRSGARFHTSHNKRYVIKIITSEDVAEMHNILKKYHQY 169
Query 154 VSTCT-NTMLTRLCGLHAL 171
+ C NT+L + G++ L
Sbjct 170 IVECHGNTLLPQFLGMYRL 188
> tgo:TGME49_045730 phosphatidylinositol-4-phosphate 5-Kinase,
putative (EC:2.7.1.68)
Length=2153
Score = 55.1 bits (131), Expect = 1e-07, Method: Composition-based stats.
Identities = 37/116 (31%), Positives = 52/116 (44%), Gaps = 12/116 (10%)
Query 58 KPSLCAVRFIDYAPMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGK 117
+P C F D AP F R L G+ Y S+ S E S K
Sbjct 1019 RPFFCV--FEDLAPDTFECARELAGVTEQDYRSSMCRTDF----------SFIEFESNSK 1066
Query 118 SGSLFYYTTDGRFMIKTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALRL 173
SG F ++ DG+++IKT+ M IL Y H+ + N++LTRL GLH + +
Sbjct 1067 SGQFFLFSHDGKYLIKTIGLREVKQMLKILPAYVDHLLSNPNSLLTRLFGLHRVEI 1122
> dre:373124 pip5k2, Pip5k2c, cb770, pip4k2c, zgc:56046; phosphatidylinositol-4-phosphate
5-kinase, type II (EC:2.7.1.149);
K00920 1-phosphatidylinositol-5-phosphate 4-kinase [EC:2.7.1.149]
Length=416
Score = 52.8 bits (125), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 30/108 (27%), Positives = 53/108 (49%), Gaps = 15/108 (13%)
Query 65 RFIDYAPMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGSLFYY 124
F +Y P VFR LR FGI+ L Y S L+ + + +G+ L +
Sbjct 94 EFKEYCPQVFRNLRERFGIEDLDYQAS--------------LARSAPMKGDGQGEGLLFT 139
Query 125 TTDGRFMIKTVSKETAFFMRSILFDYYKHVSTC-TNTMLTRLCGLHAL 171
+ D ++K +S E M +IL +Y++H+ C +T+L + G++ +
Sbjct 140 SYDRTLIVKQISSEEVADMHNILSEYHQHIVKCHGSTLLPQFLGMYRI 187
> xla:496028 pip4k2c, pip5k2c; phosphatidylinositol-5-phosphate
4-kinase, type II, gamma (EC:2.7.1.149); K00920 1-phosphatidylinositol-5-phosphate
4-kinase [EC:2.7.1.149]
Length=419
Score = 50.4 bits (119), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 45/141 (31%), Positives = 61/141 (43%), Gaps = 20/141 (14%)
Query 34 PYDFLMKEKFSVLPKTGMVDKKRRKPSLCAVRFIDYAPMVFRRLRALFGIDSLCYIRSVG 93
P DF K V T + + PS +F DY P VFR LR FGID + S
Sbjct 76 PDDFKANSKIKV---TNHLFNRENLPS--HFKFKDYCPQVFRNLRERFGIDDQDFQVS-- 128
Query 94 PDQLLGNLILGNLSSLSELVSEGKSGSLFYYTTDGRFMIKTVSKETAFFMRSILFDYYKH 153
L S E SEG G F + D +IK +S E M IL Y++H
Sbjct 129 ---------LTRSSPYCE--SEGHDGR-FLLSYDKTLVIKEISSEDVADMHIILSHYHQH 176
Query 154 VSTC-TNTMLTRLCGLHALRL 173
+ C +T+L + G++ L +
Sbjct 177 IVKCHESTLLPQFLGMYRLSV 197
> hsa:79837 PIP4K2C, FLJ22055, PIP5K2C; phosphatidylinositol-5-phosphate
4-kinase, type II, gamma (EC:2.7.1.149); K00920 1-phosphatidylinositol-5-phosphate
4-kinase [EC:2.7.1.149]
Length=421
Score = 50.1 bits (118), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 45/158 (28%), Positives = 69/158 (43%), Gaps = 17/158 (10%)
Query 14 MVGIRLAGARAMSEPHRAVEPYDFLMKEKFSVLPKTGMVDKKRRKPSLCA-VRFIDYAPM 72
+VG+ L G V P L+ + F K + + + +L + +F +Y P
Sbjct 47 LVGVFLWGVAHSINELSQVPPPVMLLPDDFKASSKIKVNNHLFHRENLPSHFKFKEYCPQ 106
Query 73 VFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGSLFYYTTDGRFMI 132
VFR LR FGID Y+ S L+ SEG G F + D +I
Sbjct 107 VFRNLRDRFGIDDQDYLVS--------------LTRNPPSESEGSDGR-FLISYDRTLVI 151
Query 133 KTVSKETAFFMRSILFDYYKHVSTC-TNTMLTRLCGLH 169
K VS E M S L +Y++++ C NT+L + G++
Sbjct 152 KEVSSEDIADMHSNLSNYHQYIVKCHGNTLLPQFLGMY 189
> ath:AT1G01460 PIPK11; PIPK11; 1-phosphatidylinositol-4-phosphate
5-kinase/ phosphatidylinositol phosphate kinase; K00889
1-phosphatidylinositol-4-phosphate 5-kinase [EC:2.7.1.68]
Length=427
Score = 49.7 bits (117), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 29/113 (25%), Positives = 57/113 (50%), Gaps = 12/113 (10%)
Query 60 SLCAVRFIDYAPMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSG 119
++ + DY P+ FR ++ L I+ Y++S+ D+ L L S K G
Sbjct 33 TITEFEWKDYCPLGFRLIQELEDINHDEYMKSICNDETLRKL------------STSKVG 80
Query 120 SLFYYTTDGRFMIKTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALR 172
++F + D RF+IK + K + +L Y++H+ +T+L++ G H+++
Sbjct 81 NMFLLSKDDRFLIKILRKSEIKVILEMLPGYFRHIHKYRSTLLSKNYGAHSVK 133
> mmu:117150 Pip4k2c, Pip5k2c; phosphatidylinositol-5-phosphate
4-kinase, type II, gamma (EC:2.7.1.149); K00920 1-phosphatidylinositol-5-phosphate
4-kinase [EC:2.7.1.149]
Length=421
Score = 47.4 bits (111), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 45/158 (28%), Positives = 70/158 (44%), Gaps = 17/158 (10%)
Query 14 MVGIRLAGARAMSEPHRAVEPYDFLMKEKFSVLPKTGMVDKKRRKPSLCA-VRFIDYAPM 72
+VG+ L G V P L+ + F K + + + +L + +F +Y P
Sbjct 47 LVGVFLWGVAHSINELSQVPPPVMLLPDDFKASSKIKVNNHFFHRENLPSHFKFKEYCPQ 106
Query 73 VFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGSLFYYTTDGRFMI 132
VFR LR F ID Y+ S+ S SE +EG G F + D +I
Sbjct 107 VFRNLRDRFAIDDHDYLVSL------------TRSPPSE--TEGSDGR-FLISYDRTLVI 151
Query 133 KTVSKETAFFMRSILFDYYKHVSTCT-NTMLTRLCGLH 169
K VS E M S L +Y++++ C NT+L + G++
Sbjct 152 KEVSSEDIADMHSNLSNYHQYIVKCHGNTLLPQFLGMY 189
> dre:100149843 phosphatidylinositol-4-phosphate 5-kinase, type
I, alpha-like; K13712 phosphatidylinositol-4-phosphate 5-kinase-like
protein 1 [EC:2.7.1.68]
Length=388
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 31/109 (28%), Positives = 50/109 (45%), Gaps = 11/109 (10%)
Query 65 RFIDYAPMVFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGSLFYY 124
R YA VF LR G+ Y +S L + + +S KS + F+
Sbjct 85 RMETYAGPVFASLRRSLGMTEKEYQQS-----------LSSEGCYLQFISNSKSKADFFL 133
Query 125 TTDGRFMIKTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALRL 173
T D RF +KT SK F+ S L Y +H+ +++L + G+H +++
Sbjct 134 TNDKRFFLKTQSKREVRFLLSNLRIYMEHLEKYPHSLLVKFLGVHRIKI 182
> mmu:227733 Pip5kl1, BC028795; phosphatidylinositol-4-phosphate
5-kinase-like 1 (EC:2.7.1.68); K13712 phosphatidylinositol-4-phosphate
5-kinase-like protein 1 [EC:2.7.1.68]
Length=395
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 29/101 (28%), Positives = 49/101 (48%), Gaps = 11/101 (10%)
Query 73 VFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGSLFYYTTDGRFMI 132
F RLR G+ Y ++GP G+ L + S KS + F+ T D RF +
Sbjct 106 AFARLRKSIGLTEEDYQATLGP----GDPYL-------QFFSTSKSKASFFLTHDQRFFV 154
Query 133 KTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALRL 173
KT + + + L Y +H+ +++L RL G+++LR+
Sbjct 155 KTQRRHEVHVLLAHLPRYVEHLQQYPHSLLARLLGVYSLRV 195
> hsa:138429 PIP5KL1, MGC46424, PIPKH, bA203J24.5; phosphatidylinositol-4-phosphate
5-kinase-like 1 (EC:2.7.1.68); K13712
phosphatidylinositol-4-phosphate 5-kinase-like protein 1 [EC:2.7.1.68]
Length=394
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 28/101 (27%), Positives = 48/101 (47%), Gaps = 11/101 (10%)
Query 73 VFRRLRALFGIDSLCYIRSVGPDQLLGNLILGNLSSLSELVSEGKSGSLFYYTTDGRFMI 132
F LR G+ Y ++GP G L + +S KS + F+ + D RF +
Sbjct 104 AFAWLRRSLGLAEEDYQAALGP----GGPYL-------QFLSTSKSKASFFLSHDQRFFL 152
Query 133 KTVSKETAFFMRSILFDYYKHVSTCTNTMLTRLCGLHALRL 173
KT + + + L Y +H+ +++L RL G+H+LR+
Sbjct 153 KTQGRREVQALLAHLPRYVQHLQRHPHSLLARLLGVHSLRV 193
Lambda K H
0.327 0.140 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4406352944
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40