bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0630_orf2
Length=155
Score E
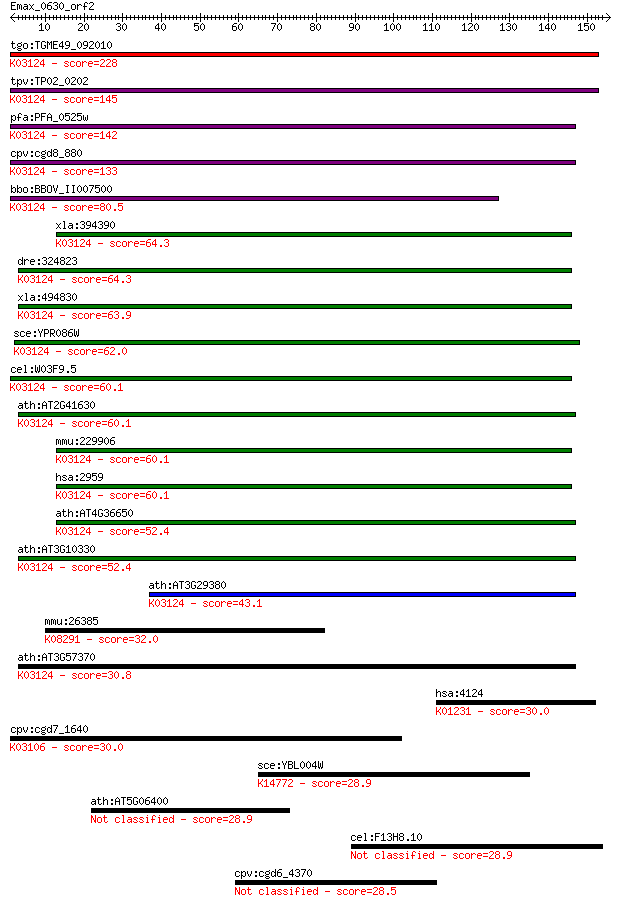
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_092010 transcription initiation factor IIB, putativ... 228 6e-60
tpv:TP02_0202 transcription initiation factor TFIIB; K03124 tr... 145 4e-35
pfa:PFA_0525w transcription initiation factor TFIIB, putative;... 142 5e-34
cpv:cgd8_880 transcription initiation factor TFIIB Sua7p; ZnR+... 133 2e-31
bbo:BBOV_II007500 18.m06622; transcription initiation factor T... 80.5 2e-15
xla:394390 gtf2b-a, XLTFIIB, gtf2b, tf2b, tfiib; general trans... 64.3 1e-10
dre:324823 gtf2b, wu:fc44c11, zgc:66258; general transcription... 64.3 1e-10
xla:494830 gtf2b-b, gtf2b, tf2b, tfiib; general transcription ... 63.9 2e-10
sce:YPR086W SUA7, SOH4; Sua7p; K03124 transcription initiation... 62.0 7e-10
cel:W03F9.5 ttb-1; TFII(two)B (general transcription factor) f... 60.1 3e-09
ath:AT2G41630 TFIIB; TFIIB (TRANSCRIPTION FACTOR II B); RNA po... 60.1 3e-09
mmu:229906 Gtf2b, MGC6859; general transcription factor IIB; K... 60.1 3e-09
hsa:2959 GTF2B, TF2B, TFIIB; general transcription factor IIB;... 60.1 3e-09
ath:AT4G36650 ATPBRP; ATPBRP (PLANT-SPECIFIC TFIIB-RELATED PRO... 52.4 5e-07
ath:AT3G10330 transcription initiation factor IIB-2 / general ... 52.4 6e-07
ath:AT3G29380 transcription factor IIB (TFIIB) family protein;... 43.1 4e-04
mmu:26385 Grk6, Gprk6; G protein-coupled receptor kinase 6 (EC... 32.0 0.79
ath:AT3G57370 transcription factor IIB (TFIIB) family protein;... 30.8 2.0
hsa:4124 MAN2A1, GOLIM7, MANA2, MANII; mannosidase, alpha, cla... 30.0 2.9
cpv:cgd7_1640 SRP54. signal recognition 54. GTpase. ; K03106 s... 30.0 3.0
sce:YBL004W UTP20; Component of the small-subunit (SSU) proces... 28.9 6.4
ath:AT5G06400 pentatricopeptide (PPR) repeat-containing protein 28.9 7.5
cel:F13H8.10 bpl-1; Biotin Protein Ligase family member (bpl-1) 28.9 7.5
cpv:cgd6_4370 SPBC24E9.10-like. RRM domain protein, no PWI domain 28.5 8.1
> tgo:TGME49_092010 transcription initiation factor IIB, putative
; K03124 transcription initiation factor TFIIB
Length=464
Score = 228 bits (581), Expect = 6e-60, Method: Compositional matrix adjust.
Identities = 113/159 (71%), Positives = 131/159 (82%), Gaps = 7/159 (4%)
Query 1 RLIAEAFCLRDYILERAKEIAKELLDAG-QLRSRSTCVCMLAITYLACREGGATRTVKEL 59
RLI EAF LRD +LERAKEI K+L+ G QLR+RS MLAITYLACRE G TRTVKEL
Sbjct 240 RLIGEAFALRDNVLERAKEITKDLMQDGVQLRTRSNTTTMLAITYLACREAGVTRTVKEL 299
Query 60 IVYDRSIKEKELGKQINKIKRLLPNRGGGNNQENVQQLLPRYCSKLQLSMHVSDVAEYVA 119
+VYDR+I EKELGK IN+IK+LLP RGG N+ E+ QLLPRYCS+LQLSMHV+DVAE+VA
Sbjct 300 VVYDRAISEKELGKAINRIKKLLPQRGGVNSAESATQLLPRYCSRLQLSMHVADVAEHVA 359
Query 120 KRAAQVFVSSH------RPNSVAAAAIWLVVQLLSSSSS 152
KRA QV +SSH RPNSVAAAAIWLVV+LL+SS++
Sbjct 360 KRATQVIISSHRHVLARRPNSVAAAAIWLVVKLLNSSTN 398
> tpv:TP02_0202 transcription initiation factor TFIIB; K03124
transcription initiation factor TFIIB
Length=342
Score = 145 bits (367), Expect = 4e-35, Method: Compositional matrix adjust.
Identities = 70/152 (46%), Positives = 104/152 (68%), Gaps = 1/152 (0%)
Query 1 RLIAEAFCLRDYILERAKEIAKELLDAGQLRSRSTCVCMLAITYLACREGGATRTVKELI 60
R IA++ +RD ++ERAKEI KEL G LRSR+ + LA+ YLACRE +R+++EL+
Sbjct 119 RTIADSLNIRDQVVERAKEILKELNQMGHLRSRANSLNTLAVLYLACRESSVSRSLRELV 178
Query 61 VYDRSIKEKELGKQINKIKRLLPNRGGGNNQENVQQLLPRYCSKLQLSMHVSDVAEYVAK 120
+YDRS+ KELG+ IN++K++LPNRG E+V QL+PR+CS+L LS + E VA+
Sbjct 179 IYDRSLTVKELGRAINRLKKVLPNRGNAPT-EDVSQLMPRFCSRLNLSNEIMTTCEAVAQ 237
Query 121 RAAQVFVSSHRPNSVAAAAIWLVVQLLSSSSS 152
++ + S+HR S+A I+LV +L + S
Sbjct 238 KSFVLLTSAHRTTSLAGGIIYLVTRLFFADDS 269
> pfa:PFA_0525w transcription initiation factor TFIIB, putative;
K03124 transcription initiation factor TFIIB
Length=367
Score = 142 bits (358), Expect = 5e-34, Method: Compositional matrix adjust.
Identities = 70/146 (47%), Positives = 102/146 (69%), Gaps = 1/146 (0%)
Query 1 RLIAEAFCLRDYILERAKEIAKELLDAGQLRSRSTCVCMLAITYLACREGGATRTVKELI 60
+LI + F LR ++ERAKEI KEL D QL++R + MLA+ YLACRE G +++KELI
Sbjct 153 KLICDTFFLRSNVIERAKEITKELQDMEQLKNRINNLNMLAVVYLACREAGHIKSIKELI 212
Query 61 VYDRSIKEKELGKQINKIKRLLPNRGGGNNQENVQQLLPRYCSKLQLSMHVSDVAEYVAK 120
+DRS KEK+LGK INK+K++LP+R N EN+ L+ ++LQLS+ + + EYV K
Sbjct 213 TFDRSYKEKDLGKTINKLKKVLPSRAFVYN-ENISHLIYSLSNRLQLSIDLIEAIEYVVK 271
Query 121 RAAQVFVSSHRPNSVAAAAIWLVVQL 146
+A+ + +SHR NS+ +I L+V+L
Sbjct 272 KASTLITTSHRLNSLCGGSIHLIVEL 297
> cpv:cgd8_880 transcription initiation factor TFIIB Sua7p; ZnR+2cyclins
; K03124 transcription initiation factor TFIIB
Length=456
Score = 133 bits (335), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 73/146 (50%), Positives = 94/146 (64%), Gaps = 3/146 (2%)
Query 1 RLIAEAFCLRDYILERAKEIAKELLDAGQLRSRSTCVCMLAITYLACREGGATRTVKELI 60
R IA +F L D I+ER KEI KE + L+S V LAI YLACRE G +RTVKEL+
Sbjct 255 RQIANSFSLHDNIIERCKEIVKEQSNLNILKSGKKHV--LAIVYLACREEGVSRTVKELL 312
Query 61 VYDRSIKEKELGKQINKIKRLLPNRGGGNNQENVQQLLPRYCSKLQLSMHVSDVAEYVAK 120
+DR+I E+EL + INK+K+ LP R G + +L+PR+C LQLS + +AEYV +
Sbjct 313 SFDRTISERELVRSINKLKKDLPRR-GPTLSSSAAELMPRFCHYLQLSHEIVGIAEYVCR 371
Query 121 RAAQVFVSSHRPNSVAAAAIWLVVQL 146
A Q SHRPNS+AA AI+ V L
Sbjct 372 TAEQYINKSHRPNSLAAGAIYFVCNL 397
> bbo:BBOV_II007500 18.m06622; transcription initiation factor
TFIIB; K03124 transcription initiation factor TFIIB
Length=253
Score = 80.5 bits (197), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 44/127 (34%), Positives = 73/127 (57%), Gaps = 6/127 (4%)
Query 1 RLIAEAFCLRDYILERAKEIAKELLDAGQLRSRSTCVCMLAITYLACREGGATRTVKELI 60
R + + L D ++ER+KE+ KEL +AG L+ R + +L+I Y+A RE G R++ EL
Sbjct 121 RQVDDTMNLSDLVIERSKEMLKELDNAGHLKGRCNMLNVLSIVYMASREVGVCRSLDELT 180
Query 61 VYDRSIKEKELGKQINKIKRLLPNRGGGNNQENVQQLLPRYCSKLQLSMHVSDVAEY-VA 119
+Y+ I +++L + I ++K+LLP RG E+ Q++P Q S D+ Y +A
Sbjct 181 IYEAKISQRDLSRAIGRMKKLLPQRGNA-TVEDSAQIIPSE----QSSNATKDITPYDIA 235
Query 120 KRAAQVF 126
R +F
Sbjct 236 SRRYHIF 242
> xla:394390 gtf2b-a, XLTFIIB, gtf2b, tf2b, tfiib; general transcription
factor 2B; K03124 transcription initiation factor
TFIIB
Length=316
Score = 64.3 bits (155), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 44/139 (31%), Positives = 70/139 (50%), Gaps = 15/139 (10%)
Query 13 ILERAKEIAKELLDAGQLRSRSTCVCMLAITYLACREGGATRTVKELIVYDRSIKEKELG 72
I++R + K++ + L+ RS A Y+ACR+ G RT KE+ R I +KE+G
Sbjct 134 IIDRTNNLFKQVYEQKSLKGRSNDAIASACLYIACRQEGVPRTFKEICAVSR-ISKKEIG 192
Query 73 KQINKIKRLLPNRGGGNNQENVQ-----QLLPRYCSKLQLSMHVSDVAEYVAKRAAQV-F 126
+ I + L + NV + R+CS L L+ V A ++A++A ++
Sbjct 193 RCFKLILKAL--------ETNVDLITTGDFMSRFCSNLGLTKQVQMAATHIARKAVELDL 244
Query 127 VSSHRPNSVAAAAIWLVVQ 145
V P SVAAAAI++ Q
Sbjct 245 VPGRSPISVAAAAIYMASQ 263
> dre:324823 gtf2b, wu:fc44c11, zgc:66258; general transcription
factor IIB; K03124 transcription initiation factor TFIIB
Length=316
Score = 64.3 bits (155), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 45/144 (31%), Positives = 71/144 (49%), Gaps = 5/144 (3%)
Query 3 IAEAFCLRDYILERAKEIAKELLDAGQLRSRSTCVCMLAITYLACREGGATRTVKELIVY 62
+A+ L I++R + K++ + L+ RS A Y+ACR+ G RT KE+
Sbjct 124 MADRINLPRNIIDRTNNLFKQVYEQKSLKGRSNDAIASACLYIACRQEGVPRTFKEICAV 183
Query 63 DRSIKEKELGKQINKIKRLLPNRGGGNNQENVQQLLPRYCSKLQLSMHVSDVAEYVAKRA 122
R I +KE+G+ I + L + + R+CS L L V A Y+A++A
Sbjct 184 SR-ISKKEIGRCFKLILKALET---SVDLITTGDFMSRFCSNLGLPKQVQMAATYIARKA 239
Query 123 AQV-FVSSHRPNSVAAAAIWLVVQ 145
++ V P SVAAAAI++ Q
Sbjct 240 VELDLVPGRSPISVAAAAIYMASQ 263
> xla:494830 gtf2b-b, gtf2b, tf2b, tfiib; general transcription
factor 2B; K03124 transcription initiation factor TFIIB
Length=316
Score = 63.9 bits (154), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 46/149 (30%), Positives = 74/149 (49%), Gaps = 15/149 (10%)
Query 3 IAEAFCLRDYILERAKEIAKELLDAGQLRSRSTCVCMLAITYLACREGGATRTVKELIVY 62
+A+ L I++R + K++ + L+ RS A Y+ACR+ G RT KE+
Sbjct 124 MADRINLPRNIVDRTNNLFKQVYEQKSLKGRSNDAIASACLYIACRQEGVPRTFKEICAV 183
Query 63 DRSIKEKELGKQINKIKRLLPNRGGGNNQENVQ-----QLLPRYCSKLQLSMHVSDVAEY 117
R I +KE+G+ I + L + NV + R+CS L L+ V A +
Sbjct 184 SR-ISKKEIGRCFKLILKAL--------ETNVDLITTGDFMSRFCSNLGLTKQVQMAATH 234
Query 118 VAKRAAQV-FVSSHRPNSVAAAAIWLVVQ 145
+A++A ++ V P SVAAAAI++ Q
Sbjct 235 IARKAVELDLVPGRSPISVAAAAIYMASQ 263
> sce:YPR086W SUA7, SOH4; Sua7p; K03124 transcription initiation
factor TFIIB
Length=345
Score = 62.0 bits (149), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 46/158 (29%), Positives = 77/158 (48%), Gaps = 15/158 (9%)
Query 2 LIAEAFCLRDYILERAKEIAKELLDAGQLRSRSTCVCMLAITYLACREGGATRTVKEL-- 59
++ +A L + + AKE K D L+ +S M A + CR RT KE+
Sbjct 135 MLCDAAELPKIVKDCAKEAYKLCHDEKTLKGKSMESIMAASILIGCRRAEVARTFKEIQS 194
Query 60 IVYDRSIKEKELGKQINKIKRLLPNRG-------GGNNQENVQQL--LPRYCSKLQLSMH 110
+++ +K KE GK +N +K +L + +N Q L +PR+CS L L M
Sbjct 195 LIH---VKTKEFGKTLNIMKNILRGKSEDGFLKIDTDNMSGAQNLTYIPRFCSHLGLPMQ 251
Query 111 VSDVAEYVAKRAAQVF-VSSHRPNSVAAAAIWLVVQLL 147
V+ AEY AK+ ++ ++ P ++A +I+L + L
Sbjct 252 VTTSAEYTAKKCKEIKEIAGKSPITIAVVSIYLNILLF 289
> cel:W03F9.5 ttb-1; TFII(two)B (general transcription factor)
family member (ttb-1); K03124 transcription initiation factor
TFIIB
Length=306
Score = 60.1 bits (144), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 46/151 (30%), Positives = 75/151 (49%), Gaps = 15/151 (9%)
Query 1 RLIAEAFCLRDYILERAKEIAKELLDAGQLRSRSTCVCMLAITYLACREGGATRTVKELI 60
R ++E L I + A I K++L++ LR ++ A Y+ACR+ G RT KE+
Sbjct 112 REMSERIHLPRNIQDSASRIFKDVLESKALRGKNNEAQAAACLYIACRKDGVPRTFKEIC 171
Query 61 VYDRSIKEKELGKQINKIKRLLPNRGGGNNQENVQQL-----LPRYCSKLQLSMHVSDVA 115
R + +KE+G+ I R L + N++Q+ + R+C L L + A
Sbjct 172 AVSR-VSKKEIGRCFKIIVRSL--------ETNLEQITSADFMSRFCGNLSLPNSIQAAA 222
Query 116 EYVAKRAAQV-FVSSHRPNSVAAAAIWLVVQ 145
+AK A + V+ P S+AAAAI++ Q
Sbjct 223 TRIAKCAVDMDLVAGRTPISIAAAAIYMASQ 253
> ath:AT2G41630 TFIIB; TFIIB (TRANSCRIPTION FACTOR II B); RNA
polymerase II transcription factor/ protein binding / transcription
regulator/ translation initiation factor/ zinc ion binding;
K03124 transcription initiation factor TFIIB
Length=312
Score = 60.1 bits (144), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 42/147 (28%), Positives = 70/147 (47%), Gaps = 4/147 (2%)
Query 3 IAEAFCLRDYILERAKEIAKELLDAGQLRSRSTCVCMLAITYLACREGGATRTVKELIVY 62
++E L I +RA E+ K L D R R+ A Y+ACR+ RT+KE+ V
Sbjct 115 MSERLGLVATIKDRANELYKRLEDQKSSRGRNQDALYAACLYIACRQEDKPRTIKEICVI 174
Query 63 DRSIKEKELGKQINKIKRLL---PNRGGGNNQENVQQLLPRYCSKLQLSMHVSDVAEYVA 119
+KE+G+ + I + L P + + + R+CS L +S H A+
Sbjct 175 ANGATKKEIGRAKDYIVKTLGLEPGQSVDLGTIHAGDFMRRFCSNLAMSNHAVKAAQEAV 234
Query 120 KRAAQVFVSSHRPNSVAAAAIWLVVQL 146
+++ + F P S+AA I+++ QL
Sbjct 235 QKSEE-FDIRRSPISIAAVVIYIITQL 260
> mmu:229906 Gtf2b, MGC6859; general transcription factor IIB;
K03124 transcription initiation factor TFIIB
Length=316
Score = 60.1 bits (144), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 41/134 (30%), Positives = 67/134 (50%), Gaps = 5/134 (3%)
Query 13 ILERAKEIAKELLDAGQLRSRSTCVCMLAITYLACREGGATRTVKELIVYDRSIKEKELG 72
I++R + K++ + L+ R+ A Y+ACR+ G RT KE+ R I +KE+G
Sbjct 134 IVDRTNNLFKQVYEQKSLKGRANDAIASACLYIACRQEGVPRTFKEICAVSR-ISKKEIG 192
Query 73 KQINKIKRLLPNRGGGNNQENVQQLLPRYCSKLQLSMHVSDVAEYVAKRAAQV-FVSSHR 131
+ I + L + + R+CS L L V A ++A++A ++ V
Sbjct 193 RCFKLILKALET---SVDLITTGDFMSRFCSNLCLPKQVQMAATHIARKAVELDLVPGRS 249
Query 132 PNSVAAAAIWLVVQ 145
P SVAAAAI++ Q
Sbjct 250 PISVAAAAIYMASQ 263
> hsa:2959 GTF2B, TF2B, TFIIB; general transcription factor IIB;
K03124 transcription initiation factor TFIIB
Length=316
Score = 60.1 bits (144), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 41/134 (30%), Positives = 67/134 (50%), Gaps = 5/134 (3%)
Query 13 ILERAKEIAKELLDAGQLRSRSTCVCMLAITYLACREGGATRTVKELIVYDRSIKEKELG 72
I++R + K++ + L+ R+ A Y+ACR+ G RT KE+ R I +KE+G
Sbjct 134 IVDRTNNLFKQVYEQKSLKGRANDAIASACLYIACRQEGVPRTFKEICAVSR-ISKKEIG 192
Query 73 KQINKIKRLLPNRGGGNNQENVQQLLPRYCSKLQLSMHVSDVAEYVAKRAAQV-FVSSHR 131
+ I + L + + R+CS L L V A ++A++A ++ V
Sbjct 193 RCFKLILKALET---SVDLITTGDFMSRFCSNLCLPKQVQMAATHIARKAVELDLVPGRS 249
Query 132 PNSVAAAAIWLVVQ 145
P SVAAAAI++ Q
Sbjct 250 PISVAAAAIYMASQ 263
> ath:AT4G36650 ATPBRP; ATPBRP (PLANT-SPECIFIC TFIIB-RELATED PROTEIN);
RNA polymerase II transcription factor/ rDNA binding;
K03124 transcription initiation factor TFIIB
Length=369
Score = 52.4 bits (124), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 39/135 (28%), Positives = 70/135 (51%), Gaps = 3/135 (2%)
Query 13 ILERAKEIAKELLDAGQLRSRSTCVCMLAITYLACREGGATRTVKELIVYDRSIKEKELG 72
I E A ++ ++ A LR+RS A A RE RT++E+ + ++++KE+G
Sbjct 150 ISEHAFQLFRDCCSATCLRNRSVEALATACLVQAIREAQEPRTLQEISIA-ANVQQKEIG 208
Query 73 KQINKIKRLLPNRGGGNNQENVQQLLPRYCSKLQLSMHVSDVAEYVAKRAA-QVFVSSHR 131
K I + L N ++ +PR+C+ LQL+ ++A ++ + + F +
Sbjct 209 KYIKILGEAL-QLSQPINSNSISVHMPRFCTLLQLNKSAQELATHIGEVVINKCFCTRRN 267
Query 132 PNSVAAAAIWLVVQL 146
P S++AAAI+L QL
Sbjct 268 PISISAAAIYLACQL 282
> ath:AT3G10330 transcription initiation factor IIB-2 / general
transcription factor TFIIB-2 (TFIIB2); K03124 transcription
initiation factor TFIIB
Length=312
Score = 52.4 bits (124), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 41/147 (27%), Positives = 68/147 (46%), Gaps = 4/147 (2%)
Query 3 IAEAFCLRDYILERAKEIAKELLDAGQLRSRSTCVCMLAITYLACREGGATRTVKELIVY 62
+A+ L I +RA EI K + D R R+ + A Y+ACR+ RTVKE+
Sbjct 115 MADRLGLVATIKDRANEIYKRVEDQKSSRGRNQDALLAACLYIACRQEDKPRTVKEICSV 174
Query 63 DRSIKEKELGKQINKIKRLLPNRGGG---NNQENVQQLLPRYCSKLQLSMHVSDVAEYVA 119
+KE+G+ I + L G + + R+CS L ++ A+
Sbjct 175 ANGATKKEIGRAKEYIVKQLGLETGQLVEMGTIHAGDFMRRFCSNLGMTNQTVKAAQESV 234
Query 120 KRAAQVFVSSHRPNSVAAAAIWLVVQL 146
+++ + F P S+AAA I+++ QL
Sbjct 235 QKSEE-FDIRRSPISIAAAVIYIITQL 260
> ath:AT3G29380 transcription factor IIB (TFIIB) family protein;
K03124 transcription initiation factor TFIIB
Length=336
Score = 43.1 bits (100), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 34/115 (29%), Positives = 60/115 (52%), Gaps = 8/115 (6%)
Query 37 VCMLAITYLACREGGATRTVKELIVYDRSIKEKELGKQINKIKRLLPNRGGGNNQE---- 92
+C +++ ACRE +RT+KE+ + +K++ K+ IKR+L + +
Sbjct 160 ICAASVS-TACRELQLSRTLKEIAEVANGVDKKDIRKESLVIKRVLESHQTSVSASQAII 218
Query 93 NVQQLLPRYCSKLQLSM-HVSDVAEYVAKRAAQVFVSSHRPNSVAAAAIWLVVQL 146
N +L+ R+CSKL +S + + E V K A+ F P SV AA I+++ +
Sbjct 219 NTGELVRRFCSKLDISQREIMAIPEAVEK--AENFDIRRNPKSVLAAIIFMISHI 271
> mmu:26385 Grk6, Gprk6; G protein-coupled receptor kinase 6 (EC:2.7.11.16);
K08291 G protein-coupled receptor kinase [EC:2.7.11.16]
Length=589
Score = 32.0 bits (71), Expect = 0.79, Method: Composition-based stats.
Identities = 25/75 (33%), Positives = 36/75 (48%), Gaps = 15/75 (20%)
Query 10 RDYILERAKEIAKELLDAGQLRSRSTCVCMLA---ITYLACREGGATRTVKELIVYDRSI 66
R+ + KE+A+E D ++RS C +L+ L CR GGA R +
Sbjct 394 REEVERLVKEVAEEYTDRFSSQARSLCSQLLSKDPAERLGCRGGGA-----------REV 442
Query 67 KEKELGKQINKIKRL 81
KE L K++N KRL
Sbjct 443 KEHPLFKKLN-FKRL 456
> ath:AT3G57370 transcription factor IIB (TFIIB) family protein;
K03124 transcription initiation factor TFIIB
Length=360
Score = 30.8 bits (68), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 32/144 (22%), Positives = 59/144 (40%), Gaps = 2/144 (1%)
Query 3 IAEAFCLRDYILERAKEIAKELLDAGQLRSRSTCVCMLAITYLACREGGATRTVKELIVY 62
I+ L I +A EI K + + + R+ V A Y+ACR+ TRT++E+ +
Sbjct 167 ISNGLKLPATIKGQANEIFKVVESYARGKERN--VLFAACIYIACRDNDMTRTMREISRF 224
Query 63 DRSIKEKELGKQINKIKRLLPNRGGGNNQENVQQLLPRYCSKLQLSMHVSDVAEYVAKRA 122
++ + + I L + R+CS +L + A A+
Sbjct 225 ANKASISDISETVGFIAEKLEINKNWYMSIETANFIKRFCSIFRLDKEAVEAALEAAESY 284
Query 123 AQVFVSSHRPNSVAAAAIWLVVQL 146
+ P SVAA ++++ +L
Sbjct 285 DYMTNGRRAPVSVAAGIVYVIARL 308
> hsa:4124 MAN2A1, GOLIM7, MANA2, MANII; mannosidase, alpha, class
2A, member 1 (EC:3.2.1.114); K01231 alpha-mannosidase II
[EC:3.2.1.114]
Length=1144
Score = 30.0 bits (66), Expect = 2.9, Method: Composition-based stats.
Identities = 13/41 (31%), Positives = 23/41 (56%), Gaps = 0/41 (0%)
Query 111 VSDVAEYVAKRAAQVFVSSHRPNSVAAAAIWLVVQLLSSSS 151
+S V+ YV+ QVF +S +P V +A+W +S ++
Sbjct 661 ISLVSVYVSSPTVQVFSASGKPVEVQVSAVWDTANTISETA 701
> cpv:cgd7_1640 SRP54. signal recognition 54. GTpase. ; K03106
signal recognition particle subunit SRP54
Length=515
Score = 30.0 bits (66), Expect = 3.0, Method: Composition-based stats.
Identities = 29/105 (27%), Positives = 50/105 (47%), Gaps = 10/105 (9%)
Query 1 RLIAEAFCLRDYILERAKEIAKELLDAGQLRSRSTCVCMLAITYLACR--EGGATRTVKE 58
RL+ F LRD E + +L+ G + + + + LA + G R K
Sbjct 337 RLVQGIFTLRDMY-----EQFQNMLNMGSPSALLSMIPGMGPNILAKEDEQAGIERLKKF 391
Query 59 LIVYDRSIKEKELG--KQINKIKRLLPNRGGGNNQENVQQLLPRY 101
+++ D S+ E EL K IN + +RG G + ++VQ+LL ++
Sbjct 392 MVIMD-SMTESELDNEKTINASRVERISRGSGTSNQDVQELLSQH 435
> sce:YBL004W UTP20; Component of the small-subunit (SSU) processome,
which is involved in the biogenesis of the 18S rRNA;
K14772 U3 small nucleolar RNA-associated protein 20
Length=2493
Score = 28.9 bits (63), Expect = 6.4, Method: Composition-based stats.
Identities = 22/70 (31%), Positives = 33/70 (47%), Gaps = 2/70 (2%)
Query 65 SIKEKELGKQINKIKRLLPNRGGGNNQENVQQLLPRYCSKLQLSMHVSDVAEYVAKRAAQ 124
+I E G ++ +K LL R NQ + +LL RY L L+ + +E + K Q
Sbjct 1760 NISLTEFGTLLSPVKALLMVRINLRNQNKLSELLRRYL--LGLNHNSDSESESILKFCHQ 1817
Query 125 VFVSSHRPNS 134
+F S NS
Sbjct 1818 LFQESEMSNS 1827
> ath:AT5G06400 pentatricopeptide (PPR) repeat-containing protein
Length=1030
Score = 28.9 bits (63), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 21/60 (35%), Positives = 31/60 (51%), Gaps = 12/60 (20%)
Query 22 KELLDAGQLRSRSTCVCMLAITYLACREGG-----ATRTVKELI----VYDRSIKEKELG 72
KE+ D G + S ST C++ + C + G ATRT +E+I V DR + + LG
Sbjct 737 KEMKDMGLIPSSSTFKCLITVL---CEKKGRNVEEATRTFREMIRSGFVPDRELVQDYLG 793
> cel:F13H8.10 bpl-1; Biotin Protein Ligase family member (bpl-1)
Length=1215
Score = 28.9 bits (63), Expect = 7.5, Method: Composition-based stats.
Identities = 20/69 (28%), Positives = 33/69 (47%), Gaps = 4/69 (5%)
Query 89 NNQENVQQLLPRYCSKLQLSMHVSDVAEYVAKRAAQVFVSSHRPNSVAAAAIWL----VV 144
NN++N Q+ + ++ VS EY R+ + F S+ P+ VA + +
Sbjct 586 NNRQNSQRRFSAFSTESDRGRSVSPSFEYYQNRSLRGFTSAANPHRVATRNSTVRQVPLY 645
Query 145 QLLSSSSSS 153
Q L+S SSS
Sbjct 646 QFLTSRSSS 654
> cpv:cgd6_4370 SPBC24E9.10-like. RRM domain protein, no PWI domain.
Length=556
Score = 28.5 bits (62), Expect = 8.1, Method: Composition-based stats.
Identities = 20/53 (37%), Positives = 29/53 (54%), Gaps = 2/53 (3%)
Query 59 LIVYDRSIKEKELGK-QINKIKRLLPNRGGGNNQENVQQLLPRYCSKLQLSMH 110
L+ Y I +KE+ K Q N+I LL +RGG E + L SKL++S+
Sbjct 135 LVKYQHGI-DKEIAKWQSNRINELLKHRGGDCTIETILNELSTSDSKLRISIQ 186
Lambda K H
0.319 0.131 0.364
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3386671600
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40