bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0598_orf1
Length=88
Score E
Sequences producing significant alignments: (Bits) Value
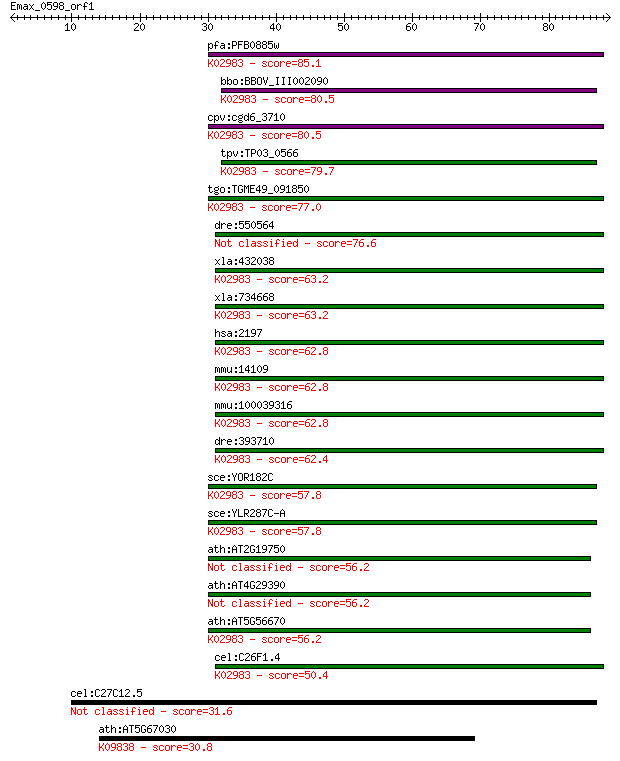
pfa:PFB0885w 40S ribosomal protein S30, putative; K02983 small... 85.1 5e-17
bbo:BBOV_III002090 17.m07202; 40S ribosomal protein S30; K0298... 80.5 1e-15
cpv:cgd6_3710 40S ribosomal protein S30 ; K02983 small subunit... 80.5 1e-15
tpv:TP03_0566 40S ribosomal protein S30; K02983 small subunit ... 79.7 2e-15
tgo:TGME49_091850 40S ribosomal protein S30, putative ; K02983... 77.0 1e-14
dre:550564 faub, im:7142593, zgc:110695; Finkel-Biskis-Reilly ... 76.6 2e-14
xla:432038 hypothetical protein MGC80045; K02983 small subunit... 63.2 2e-10
xla:734668 hypothetical protein MGC116435; K02983 small subuni... 63.2 2e-10
hsa:2197 FAU, FAU1, FLJ22986, Fub1, Fubi, MNSFbeta, RPS30, asr... 62.8 2e-10
mmu:14109 Fau, MNSFbeta; Finkel-Biskis-Reilly murine sarcoma v... 62.8 2e-10
mmu:100039316 Gm9843; predicted gene 9843; K02983 small subuni... 62.8 2e-10
dre:393710 faua, MGC73171, fau, zgc:73171; Finkel-Biskis-Reill... 62.4 4e-10
sce:YOR182C RPS30B; Rps30bp; K02983 small subunit ribosomal pr... 57.8 9e-09
sce:YLR287C-A RPS30A; Rps30ap; K02983 small subunit ribosomal ... 57.8 9e-09
ath:AT2G19750 40S ribosomal protein S30 (RPS30A) 56.2 2e-08
ath:AT4G29390 40S ribosomal protein S30 (RPS30B) 56.2 2e-08
ath:AT5G56670 40S ribosomal protein S30 (RPS30C); K02983 small... 56.2 2e-08
cel:C26F1.4 rps-30; Ribosomal Protein, Small subunit family me... 50.4 1e-06
cel:C27C12.5 tag-324; Temporarily Assigned Gene name family me... 31.6 0.68
ath:AT5G67030 ABA1; ABA1 (ABA DEFICIENT 1); zeaxanthin epoxida... 30.8 1.1
> pfa:PFB0885w 40S ribosomal protein S30, putative; K02983 small
subunit ribosomal protein S30e
Length=58
Score = 85.1 bits (209), Expect = 5e-17, Method: Compositional matrix adjust.
Identities = 42/58 (72%), Positives = 50/58 (86%), Gaps = 0/58 (0%)
Query 30 MGKVHGSLARAGKVKNQTPKVQKQQKRKPVTGRAKKRQLYNRRFTTTLNRRRGPNAQS 87
MGKVHGSLARAGKVKNQTPKV K K+K +TGRAKKRQLYNRRF+ R++GPN+++
Sbjct 1 MGKVHGSLARAGKVKNQTPKVPKLDKKKRLTGRAKKRQLYNRRFSDNGGRKKGPNSKA 58
> bbo:BBOV_III002090 17.m07202; 40S ribosomal protein S30; K02983
small subunit ribosomal protein S30e
Length=60
Score = 80.5 bits (197), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 42/56 (75%), Positives = 47/56 (83%), Gaps = 1/56 (1%)
Query 32 KVHGSLARAGKVKNQTPKVQKQQKRKPVTGRAKKRQLYNRRFTTTLNRR-RGPNAQ 86
KVHGSLARAGKV+N TPKV KQ+K+KP TGRAKKRQ YNRRF+ +L R RGPN Q
Sbjct 4 KVHGSLARAGKVRNHTPKVAKQEKKKPPTGRAKKRQQYNRRFSNSLGMRPRGPNGQ 59
> cpv:cgd6_3710 40S ribosomal protein S30 ; K02983 small subunit
ribosomal protein S30e
Length=59
Score = 80.5 bits (197), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 40/58 (68%), Positives = 47/58 (81%), Gaps = 0/58 (0%)
Query 30 MGKVHGSLARAGKVKNQTPKVQKQQKRKPVTGRAKKRQLYNRRFTTTLNRRRGPNAQS 87
M K HGSLARAGKVKNQTP+V+KQ+K+K +TGRAKKRQ YNRRF + + RR N QS
Sbjct 2 MAKAHGSLARAGKVKNQTPRVEKQEKKKALTGRAKKRQQYNRRFVSVVAGRRKCNPQS 59
> tpv:TP03_0566 40S ribosomal protein S30; K02983 small subunit
ribosomal protein S30e
Length=60
Score = 79.7 bits (195), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 40/56 (71%), Positives = 48/56 (85%), Gaps = 1/56 (1%)
Query 32 KVHGSLARAGKVKNQTPKVQKQQKRKPVTGRAKKRQLYNRRFTTTL-NRRRGPNAQ 86
KVHGSLARAGKV+N TPKV KQ+K+KP+TGRAKKRQ YNRRF + + +RRG N+Q
Sbjct 4 KVHGSLARAGKVRNHTPKVAKQEKKKPLTGRAKKRQTYNRRFASNVAGKRRGQNSQ 59
> tgo:TGME49_091850 40S ribosomal protein S30, putative ; K02983
small subunit ribosomal protein S30e
Length=59
Score = 77.0 bits (188), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 46/58 (79%), Positives = 54/58 (93%), Gaps = 0/58 (0%)
Query 30 MGKVHGSLARAGKVKNQTPKVQKQQKRKPVTGRAKKRQLYNRRFTTTLNRRRGPNAQS 87
MGKVHGSLARAGKVKNQTPKV K++K+KP TGRAKKRQ +NRRFTT++ R+RGPNAQ+
Sbjct 1 MGKVHGSLARAGKVKNQTPKVAKKEKKKPPTGRAKKRQQFNRRFTTSVGRKRGPNAQT 58
> dre:550564 faub, im:7142593, zgc:110695; Finkel-Biskis-Reilly
murine sarcoma virus (FBR-MuSV) ubiquitously expressed b
Length=133
Score = 76.6 bits (187), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 40/60 (66%), Positives = 46/60 (76%), Gaps = 3/60 (5%)
Query 31 GKVHGSLARAGKVKNQTPKVQKQQKRKPVTGRAKKRQLYNRRFTT---TLNRRRGPNAQS 87
GKVHGSLARAGKV+ QTPKV KQ+KRK TGRAK+R YNRRF T +++GPNA S
Sbjct 74 GKVHGSLARAGKVRGQTPKVDKQEKRKKRTGRAKRRIQYNRRFVNVVPTFGKKKGPNANS 133
> xla:432038 hypothetical protein MGC80045; K02983 small subunit
ribosomal protein S30e
Length=133
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 39/60 (65%), Positives = 46/60 (76%), Gaps = 3/60 (5%)
Query 31 GKVHGSLARAGKVKNQTPKVQKQQKRKPVTGRAKKRQLYNRRFTT---TLNRRRGPNAQS 87
GKVHGSLARAGKV+ QTPKV KQ+K+K TGRAK+R YNRRF T +++GPNA S
Sbjct 74 GKVHGSLARAGKVRGQTPKVAKQEKKKKKTGRAKRRMQYNRRFVNVVPTFGKKKGPNANS 133
> xla:734668 hypothetical protein MGC116435; K02983 small subunit
ribosomal protein S30e
Length=133
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 39/60 (65%), Positives = 46/60 (76%), Gaps = 3/60 (5%)
Query 31 GKVHGSLARAGKVKNQTPKVQKQQKRKPVTGRAKKRQLYNRRFTT---TLNRRRGPNAQS 87
GKVHGSLARAGKV+ QTPKV KQ+K+K TGRAK+R YNRRF T +++GPNA S
Sbjct 74 GKVHGSLARAGKVRGQTPKVAKQEKKKKKTGRAKRRMQYNRRFVNVVPTFGKKKGPNANS 133
> hsa:2197 FAU, FAU1, FLJ22986, Fub1, Fubi, MNSFbeta, RPS30, asr1;
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously
expressed; K02983 small subunit ribosomal protein
S30e
Length=133
Score = 62.8 bits (151), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 39/60 (65%), Positives = 46/60 (76%), Gaps = 3/60 (5%)
Query 31 GKVHGSLARAGKVKNQTPKVQKQQKRKPVTGRAKKRQLYNRRFTT---TLNRRRGPNAQS 87
GKVHGSLARAGKV+ QTPKV KQ+K+K TGRAK+R YNRRF T +++GPNA S
Sbjct 74 GKVHGSLARAGKVRGQTPKVAKQEKKKKKTGRAKRRMQYNRRFVNVVPTFGKKKGPNANS 133
> mmu:14109 Fau, MNSFbeta; Finkel-Biskis-Reilly murine sarcoma
virus (FBR-MuSV) ubiquitously expressed (fox derived); K02983
small subunit ribosomal protein S30e
Length=133
Score = 62.8 bits (151), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 39/60 (65%), Positives = 46/60 (76%), Gaps = 3/60 (5%)
Query 31 GKVHGSLARAGKVKNQTPKVQKQQKRKPVTGRAKKRQLYNRRFTT---TLNRRRGPNAQS 87
GKVHGSLARAGKV+ QTPKV KQ+K+K TGRAK+R YNRRF T +++GPNA S
Sbjct 74 GKVHGSLARAGKVRGQTPKVAKQEKKKKKTGRAKRRMQYNRRFVNVVPTFGKKKGPNANS 133
> mmu:100039316 Gm9843; predicted gene 9843; K02983 small subunit
ribosomal protein S30e
Length=133
Score = 62.8 bits (151), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 39/60 (65%), Positives = 46/60 (76%), Gaps = 3/60 (5%)
Query 31 GKVHGSLARAGKVKNQTPKVQKQQKRKPVTGRAKKRQLYNRRFTT---TLNRRRGPNAQS 87
GKVHGSLARAGKV+ QTPKV KQ+K+K TGRAK+R YNRRF T +++GPNA S
Sbjct 74 GKVHGSLARAGKVRGQTPKVAKQEKKKKKTGRAKRRMQYNRRFVNVVPTFGKKKGPNANS 133
> dre:393710 faua, MGC73171, fau, zgc:73171; Finkel-Biskis-Reilly
murine sarcoma virus (FBR-MuSV) ubiquitously expressed a;
K02983 small subunit ribosomal protein S30e
Length=133
Score = 62.4 bits (150), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 39/60 (65%), Positives = 46/60 (76%), Gaps = 3/60 (5%)
Query 31 GKVHGSLARAGKVKNQTPKVQKQQKRKPVTGRAKKRQLYNRRFTT---TLNRRRGPNAQS 87
GKVHGSLARAGKV+ QTPKV KQ+K+K TGRAK+R YNRRF T +++GPNA S
Sbjct 74 GKVHGSLARAGKVRGQTPKVDKQEKKKKKTGRAKRRIQYNRRFVNVVPTFGKKKGPNANS 133
> sce:YOR182C RPS30B; Rps30bp; K02983 small subunit ribosomal
protein S30e
Length=63
Score = 57.8 bits (138), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 38/63 (60%), Positives = 44/63 (69%), Gaps = 6/63 (9%)
Query 30 MGKVHGSLARAGKVKNQTPKVQKQQKRKPVTGRAKKRQLYNRRF--TTTLNRRR----GP 83
M KVHGSLARAGKVK+QTPKV+K +K K GRA KR LY RRF T +N +R GP
Sbjct 1 MAKVHGSLARAGKVKSQTPKVEKTEKPKKPKGRAYKRLLYTRRFVNVTLVNGKRRMNPGP 60
Query 84 NAQ 86
+ Q
Sbjct 61 SVQ 63
> sce:YLR287C-A RPS30A; Rps30ap; K02983 small subunit ribosomal
protein S30e
Length=63
Score = 57.8 bits (138), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 38/63 (60%), Positives = 44/63 (69%), Gaps = 6/63 (9%)
Query 30 MGKVHGSLARAGKVKNQTPKVQKQQKRKPVTGRAKKRQLYNRRF--TTTLNRRR----GP 83
M KVHGSLARAGKVK+QTPKV+K +K K GRA KR LY RRF T +N +R GP
Sbjct 1 MAKVHGSLARAGKVKSQTPKVEKTEKPKKPKGRAYKRLLYTRRFVNVTLVNGKRRMNPGP 60
Query 84 NAQ 86
+ Q
Sbjct 61 SVQ 63
> ath:AT2G19750 40S ribosomal protein S30 (RPS30A)
Length=62
Score = 56.2 bits (134), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 37/59 (62%), Positives = 44/59 (74%), Gaps = 3/59 (5%)
Query 30 MGKVHGSLARAGKVKNQTPKVQKQQKRKPVTGRAKKRQLYNRRFTTTL---NRRRGPNA 85
MGKVHGSLARAGKV+ QTPKV KQ K+K GRA KR +NRRF T + ++RGPN+
Sbjct 1 MGKVHGSLARAGKVRGQTPKVAKQDKKKKPRGRAHKRLQHNRRFVTAVVGFGKKRGPNS 59
> ath:AT4G29390 40S ribosomal protein S30 (RPS30B)
Length=62
Score = 56.2 bits (134), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 37/59 (62%), Positives = 44/59 (74%), Gaps = 3/59 (5%)
Query 30 MGKVHGSLARAGKVKNQTPKVQKQQKRKPVTGRAKKRQLYNRRFTTTL---NRRRGPNA 85
MGKVHGSLARAGKV+ QTPKV KQ K+K GRA KR +NRRF T + ++RGPN+
Sbjct 1 MGKVHGSLARAGKVRGQTPKVAKQDKKKKPRGRAHKRLQHNRRFVTAVVGFGKKRGPNS 59
> ath:AT5G56670 40S ribosomal protein S30 (RPS30C); K02983 small
subunit ribosomal protein S30e
Length=62
Score = 56.2 bits (134), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 37/59 (62%), Positives = 44/59 (74%), Gaps = 3/59 (5%)
Query 30 MGKVHGSLARAGKVKNQTPKVQKQQKRKPVTGRAKKRQLYNRRFTTTL---NRRRGPNA 85
MGKVHGSLARAGKV+ QTPKV KQ K+K GRA KR +NRRF T + ++RGPN+
Sbjct 1 MGKVHGSLARAGKVRGQTPKVAKQDKKKKPRGRAHKRLQHNRRFVTAVVGFGKKRGPNS 59
> cel:C26F1.4 rps-30; Ribosomal Protein, Small subunit family
member (rps-30); K02983 small subunit ribosomal protein S30e
Length=130
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 34/60 (56%), Positives = 42/60 (70%), Gaps = 3/60 (5%)
Query 31 GKVHGSLARAGKVKNQTPKVQKQQKRKPVTGRAKKRQLYNRRFTTTLN---RRRGPNAQS 87
GKVHGSLARAGKV+ QTPKV KQ K+K GRA +R Y RR+ + ++RGPN+ S
Sbjct 71 GKVHGSLARAGKVRAQTPKVDKQDKKKKKRGRAFRRVQYTRRYVNVASGPGKKRGPNSNS 130
> cel:C27C12.5 tag-324; Temporarily Assigned Gene name family
member (tag-324)
Length=480
Score = 31.6 bits (70), Expect = 0.68, Method: Composition-based stats.
Identities = 21/78 (26%), Positives = 34/78 (43%), Gaps = 1/78 (1%)
Query 10 VFGFFRQPANQYYLDSELSKM-GKVHGSLARAGKVKNQTPKVQKQQKRKPVTGRAKKRQL 68
+FG ++ +++ LS G +HGS + VQK+ +P+ A R L
Sbjct 383 IFGTLNSTRHKAFIERLLSNEDGSIHGSHEEIIITQKIEKTVQKETLEQPLEQVADTRPL 442
Query 69 YNRRFTTTLNRRRGPNAQ 86
RRF+ N + G Q
Sbjct 443 AERRFSLMPNNQLGVKVQ 460
> ath:AT5G67030 ABA1; ABA1 (ABA DEFICIENT 1); zeaxanthin epoxidase;
K09838 zeaxanthin epoxidase [EC:1.14.13.90]
Length=610
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 22/59 (37%), Positives = 32/59 (54%), Gaps = 4/59 (6%)
Query 14 FRQPANQYYLD--SELSKMGKVHGSLARAG--KVKNQTPKVQKQQKRKPVTGRAKKRQL 68
F + Q+YLD S K G V G +R VK T V+K++KR+ VT + KK ++
Sbjct 25 FSPVSKQFYLDLSSFSGKPGGVSGFRSRRALLGVKAATALVEKEEKREAVTEKKKKSRV 83
Lambda K H
0.321 0.134 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2021645584
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40