bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0587_orf2
Length=147
Score E
Sequences producing significant alignments: (Bits) Value
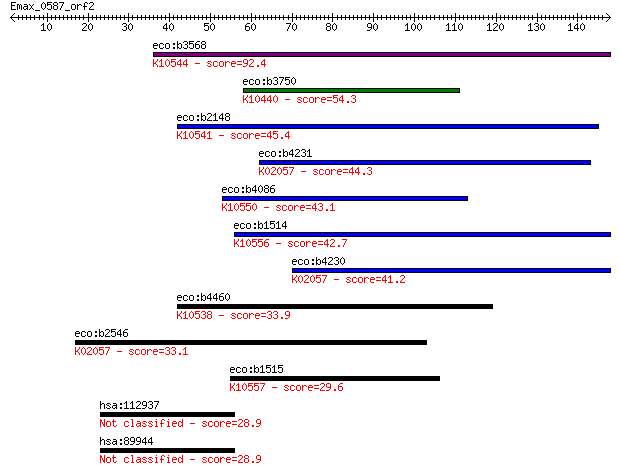
eco:b3568 xylH, ECK3557, JW3540; D-xylose ABC transporter perm... 92.4 5e-19
eco:b3750 rbsC, ECK3744, JW3729, rbsP, rbsT; D-ribose transpor... 54.3 1e-07
eco:b2148 mglC, ECK2141, JW2135, mglP; methyl-galactoside tran... 45.4 7e-05
eco:b4231 yjfF, ECK4226, JW5754; predicted sugar transporter s... 44.3 1e-04
eco:b4086 alsC, ECK4079, JW4047, yjcV; D-allose transporter su... 43.1 3e-04
eco:b1514 lsrC, ECK1507, JW1507, ydeY; Autoinducer 2 import sy... 42.7 5e-04
eco:b4230 ytfT, ECK4225, JW5753; predicted sugar transporter s... 41.2 0.001
eco:b4460 araH, ECK1897, JW1887; fused L-arabinose transporter... 33.9 0.18
eco:b2546 yphD, ECK2543, JW2530; predicted sugar transporter s... 33.1 0.31
eco:b1515 lsrD, ECK1508, JW1508, ydeZ; autoinducer 2 import sy... 29.6 3.4
hsa:112937 GLB1L3, FLJ90231; galactosidase, beta 1-like 3 28.9
hsa:89944 GLB1L2, MST114, MSTP114; galactosidase, beta 1-like ... 28.9 6.1
> eco:b3568 xylH, ECK3557, JW3540; D-xylose ABC transporter permease
subunit; K10544 D-xylose transport system permease protein
Length=393
Score = 92.4 bits (228), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 54/113 (47%), Positives = 77/113 (68%), Gaps = 1/113 (0%)
Query 36 NIDIRAYTMIGALVAIWIFFGLLND-TFLSARNLSNLFTQMSVTAILAIGMVLVIVAGHI 94
+++++ + MI A++AI +FF D +LSARN+SNL Q ++T ILA+GMV VI++ I
Sbjct 24 SLNLQVFVMIAAIIAIMLFFTWTTDGAYLSARNVSNLLRQTAITGILAVGMVFVIISAEI 83
Query 95 DLSVGSIVGLTGGIAAILSNWMGMPTSVVIAGTLAAGALIGMIQGWLVAYHAI 147
DLSVGS++GL GG+AAI W+G P + I TL G L+G GW VAY +
Sbjct 84 DLSVGSMMGLLGGVAAICDVWLGWPLPLTIIVTLVLGLLLGAWNGWWVAYRKV 136
> eco:b3750 rbsC, ECK3744, JW3729, rbsP, rbsT; D-ribose transporter
subunit; K10440 ribose transport system permease protein
Length=321
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 28/53 (52%), Positives = 37/53 (69%), Gaps = 0/53 (0%)
Query 58 LNDTFLSARNLSNLFTQMSVTAILAIGMVLVIVAGHIDLSVGSIVGLTGGIAA 110
L+ F + NL N+ Q SV AI+A+GM LVI+ IDLSVGS++ LTG +AA
Sbjct 36 LSPNFFTINNLFNILQQTSVNAIMAVGMTLVILTSGIDLSVGSLLALTGAVAA 88
> eco:b2148 mglC, ECK2141, JW2135, mglP; methyl-galactoside transporter
subunit; K10541 methyl-galactoside transport system
permease protein
Length=336
Score = 45.4 bits (106), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 47/114 (41%), Positives = 63/114 (55%), Gaps = 14/114 (12%)
Query 42 YTMIGALVAIWIFFGLLNDTFLSARNLSNLFTQMSVTAILAIGMVLVIVAGHIDLSVGSI 101
Y ++ L+AI IF + TFLS NLSN+ TQ SV I+A+G+ +IV DLS G
Sbjct 19 YVVLLVLLAIIIF---QDPTFLSLLNLSNILTQSSVRIIIALGVAGLIVTQGTDLSAGRQ 75
Query 102 VGLTGGIAAILSNWMG-----------MPTSVVIAGTLAAGALIGMIQGWLVAY 144
VGL +AA L M MP ++VI A GA+IG+I G ++AY
Sbjct 76 VGLAAVVAATLLQSMDNANKVFPEMATMPIALVILIVCAIGAVIGLINGLIIAY 129
> eco:b4231 yjfF, ECK4226, JW5754; predicted sugar transporter
subunit: membrane component of ABC superfamily; K02057 simple
sugar transport system permease protein
Length=331
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 28/81 (34%), Positives = 41/81 (50%), Gaps = 0/81 (0%)
Query 62 FLSARNLSNLFTQMSVTAILAIGMVLVIVAGHIDLSVGSIVGLTGGIAAILSNWMGMPTS 121
F S R + N+ T + I+A+GM VI++G IDLSVGS++ TG A + G+
Sbjct 29 FASTRVICNILTDNAFLGIIAVGMTFVILSGGIDLSVGSVIAFTGVFLAKVIGDFGLSPL 88
Query 122 VVIAGTLAAGALIGMIQGWLV 142
+ L G G G L+
Sbjct 89 LAFPLVLVMGCAFGAFMGLLI 109
> eco:b4086 alsC, ECK4079, JW4047, yjcV; D-allose transporter
subunit; K10550 D-allose transport system permease protein
Length=326
Score = 43.1 bits (100), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 24/60 (40%), Positives = 37/60 (61%), Gaps = 0/60 (0%)
Query 53 IFFGLLNDTFLSARNLSNLFTQMSVTAILAIGMVLVIVAGHIDLSVGSIVGLTGGIAAIL 112
IF L + FL+ N++ +F Q SVT ++ +G I+ IDLSVG+I+ L+G + A L
Sbjct 38 IFGSLSPEYFLTTNNITQIFVQSSVTVLIGMGEFFAILVAGIDLSVGAILALSGMVTAKL 97
> eco:b1514 lsrC, ECK1507, JW1507, ydeY; Autoinducer 2 import
system permease protein; K10556 AI-2 transport system permease
protein
Length=342
Score = 42.7 bits (99), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 32/92 (34%), Positives = 50/92 (54%), Gaps = 1/92 (1%)
Query 56 GLLNDTFLSARNLSNLFTQMSVTAILAIGMVLVIVAGHIDLSVGSIVGLTGGIAAILSNW 115
G L+ +LS + L+ +++ + +LA+G LV++ +ID+SVGSI G+ + +L N
Sbjct 25 GFLDRQYLSVQTLTMVYSSAQILILLAMGATLVMLTRNIDVSVGSITGMCAVLLGMLLN- 83
Query 116 MGMPTSVVIAGTLAAGALIGMIQGWLVAYHAI 147
G V TL G L G G LVA+ I
Sbjct 84 AGYSLPVACVATLLLGLLAGFFNGVLVAWLKI 115
> eco:b4230 ytfT, ECK4225, JW5753; predicted sugar transporter
subunit: membrane component of ABC superfamily; K02057 simple
sugar transport system permease protein
Length=341
Score = 41.2 bits (95), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 29/78 (37%), Positives = 43/78 (55%), Gaps = 1/78 (1%)
Query 70 NLFTQMSVTAILAIGMVLVIVAGHIDLSVGSIVGLTGGIAAILSNWMGMPTSVVIAGTLA 129
++ + + A+LAIGM LVI G IDLSVG+++ + G A ++ G +V+ L
Sbjct 58 DILNRAAPVALLAIGMTLVIATGGIDLSVGAVMAIAGATTAAMTV-AGFSLPIVLLSALG 116
Query 130 AGALIGMIQGWLVAYHAI 147
G L G+ G LVA I
Sbjct 117 TGILAGLWNGILVAILKI 134
> eco:b4460 araH, ECK1897, JW1887; fused L-arabinose transporter
subunits of ABC superfamily: membrane components; K10538
L-arabinose transport system permease protein
Length=328
Score = 33.9 bits (76), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 21/82 (25%), Positives = 41/82 (50%), Gaps = 5/82 (6%)
Query 42 YTMIGALVAIWIFFGLLNDTFLSARNLSNLFTQMSVTAILAIGMVLVIVAGHIDLSVGSI 101
Y M+ ++I + F + N+ L +S++ ++A GM+ + +G DLSV S+
Sbjct 25 YGMLVVFAVLFIACAIFVPNFATFINMKGLGLAISMSGMVACGMLFCLASGDFDLSVASV 84
Query 102 VGLTGGIAAILSN-----WMGM 118
+ G A++ N W+G+
Sbjct 85 IACAGVTTAVVINLTESLWIGV 106
> eco:b2546 yphD, ECK2543, JW2530; predicted sugar transporter
subunit: membrane component of ABC superfamily; K02057 simple
sugar transport system permease protein
Length=332
Score = 33.1 bits (74), Expect = 0.31, Method: Compositional matrix adjust.
Identities = 28/89 (31%), Positives = 40/89 (44%), Gaps = 18/89 (20%)
Query 17 VEIKAVVNEHQREKKNIFGNIDIRAYTMIGALVAI---WIFFGLLNDTFLSARNLSNLFT 73
V +K V+ H E IG LV I ++ F L F+S N N+
Sbjct 14 VSLKQFVSRHINE---------------IGLLVVIAILYLVFSLNAPGFISLNNQMNVLR 58
Query 74 QMSVTAILAIGMVLVIVAGHIDLSVGSIV 102
+ I A M L+I++G ID+SVG +V
Sbjct 59 DAATIGIAAWAMTLIIISGEIDVSVGPMV 87
> eco:b1515 lsrD, ECK1508, JW1508, ydeZ; autoinducer 2 import
system permease protein; K10557 AI-2 transport system permease
protein
Length=330
Score = 29.6 bits (65), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 18/51 (35%), Positives = 27/51 (52%), Gaps = 0/51 (0%)
Query 55 FGLLNDTFLSARNLSNLFTQMSVTAILAIGMVLVIVAGHIDLSVGSIVGLT 105
FG +N L L + I+A+ + +VIV+G ID+S GS +GL
Sbjct 22 FGAINPRMLDLNMLLFSTSDFICIGIVALPLTMVIVSGGIDISFGSTIGLC 72
> hsa:112937 GLB1L3, FLJ90231; galactosidase, beta 1-like 3
Length=653
Score = 28.9 bits (63), Expect = 5.4, Method: Composition-based stats.
Identities = 9/33 (27%), Positives = 21/33 (63%), Gaps = 0/33 (0%)
Query 23 VNEHQREKKNIFGNIDIRAYTMIGALVAIWIFF 55
++E +R K + GN+D+ A+ ++ A + +W+
Sbjct 127 LHEPERGKFDFSGNLDLEAFVLMAAEIGLWVIL 159
> hsa:89944 GLB1L2, MST114, MSTP114; galactosidase, beta 1-like
2 (EC:3.2.1.23)
Length=636
Score = 28.9 bits (63), Expect = 6.1, Method: Composition-based stats.
Identities = 9/33 (27%), Positives = 21/33 (63%), Gaps = 0/33 (0%)
Query 23 VNEHQREKKNIFGNIDIRAYTMIGALVAIWIFF 55
++E +R K + GN+D+ A+ ++ A + +W+
Sbjct 101 LHEPERGKFDFSGNLDLEAFVLMAAEIGLWVIL 133
Lambda K H
0.325 0.140 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2938175820
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40