bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0583_orf1
Length=146
Score E
Sequences producing significant alignments: (Bits) Value
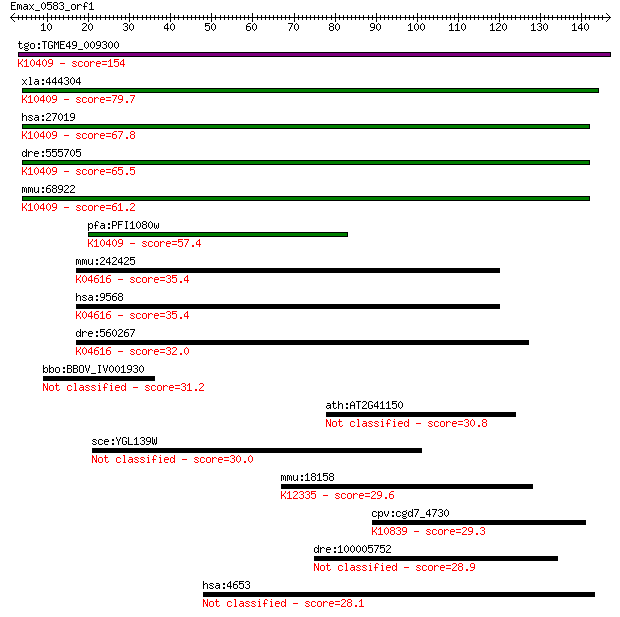
tgo:TGME49_009300 dynein intermediate chain, putative ; K10409... 154 1e-37
xla:444304 MGC80975 protein; K10409 dynein intermediate chain ... 79.7 3e-15
hsa:27019 DNAI1, CILD1, ICS, ICS1, MGC26204, PCD; dynein, axon... 67.8 1e-11
dre:555705 zgc:158666; K10409 dynein intermediate chain 1, axo... 65.5 6e-11
mmu:68922 Dnaic1, 1110066F04Rik, BB124644, Dnai1; dynein, axon... 61.2 1e-09
pfa:PFI1080w dynein intermediate chain 2, ciliary; K10409 dyne... 57.4 2e-08
mmu:242425 Gabbr2, GABABR2, Gb2, Gm425, Gpr51; gamma-aminobuty... 35.4 0.055
hsa:9568 GABBR2, FLJ36928, GABABR2, GPR51, GPRC3B, HG20, HRIHF... 35.4 0.055
dre:560267 si:dkey-190l1.2; K04616 gamma-aminobutyric acid (GA... 32.0 0.69
bbo:BBOV_IV001930 21.m02940; hypothetical protein 31.2 1.1
ath:AT2G41150 hypothetical protein 30.8 1.5
sce:YGL139W FLC3; Flc3p 30.0 2.6
mmu:18158 Nppb, AA408272, BNP; natriuretic peptide type B; K12... 29.6 3.6
cpv:cgd7_4730 RAD23p, UB+UBA domains protein ; K10839 UV excis... 29.3 4.7
dre:100005752 MGC162803, cb117, cgn, im:7137721, sb:cb117, sb:... 28.9 5.5
hsa:4653 MYOC, GLC1A, GPOA, JOAG, JOAG1, TIGR, myocilin; myoci... 28.1 9.9
> tgo:TGME49_009300 dynein intermediate chain, putative ; K10409
dynein intermediate chain 1, axonemal
Length=619
Score = 154 bits (388), Expect = 1e-37, Method: Compositional matrix adjust.
Identities = 76/145 (52%), Positives = 105/145 (72%), Gaps = 1/145 (0%)
Query 3 EDQVQLAPEELRQRMPPKILYPQNPRAPQNITRFSFKENQFKKEGAVEQTVFHLFIDSTL 62
+DQ+QL+PEELR+RMPP+ILYPQNPRAP+N+ +SFKE FK++ ++QTVFHL +D L
Sbjct 55 KDQLQLSPEELRKRMPPRILYPQNPRAPKNLALYSFKEKVFKRDEQIDQTVFHLSVDGAL 114
Query 63 LLKDSPEAAEQEELLRLRDEAEERRRELEAVEVSIEDD-GATQEEEGRVMRNQFNNVDRA 121
L +S EA EQEE RLR+E + +++ E+V++ IEDD E+EGRV+RNQFN DRA
Sbjct 115 LHAESQEAIEQEEAWRLREEEDAKKKRKESVDIRIEDDLQQNTEDEGRVLRNQFNYGDRA 174
Query 122 TQIKSCPTVEQSCSTLPPPKKLCCG 146
+Q ++ E+ +T PPP G
Sbjct 175 SQTETKADRERGVATRPPPTVPVAG 199
> xla:444304 MGC80975 protein; K10409 dynein intermediate chain
1, axonemal
Length=702
Score = 79.7 bits (195), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 54/156 (34%), Positives = 80/156 (51%), Gaps = 17/156 (10%)
Query 4 DQVQLAPEELRQRMPPKILYPQNPRAPQNITRFSFKENQFKKEGAVEQTVFHLFIDSTLL 63
DQ++L EL + +IL NP APQNI R+SFKE +K VEQ H +D +L
Sbjct 57 DQLELTEAELNEEFT-RILTANNPHAPQNIVRYSFKERAYKPVSIVEQLAIHFSLDGNML 115
Query 64 LKDSPEAAEQEELLRLRD----------------EAEERRRELEAVEVSIEDDGATQEEE 107
KDS EA Q+ + + + EAEE A E + + +T+ +
Sbjct 116 HKDSDEARRQKLRMGVLEGSTQTDTTAPETGDNMEAEEHPETPVAGEEGADREDSTKSRK 175
Query 108 GRVMRNQFNNVDRATQIKSCPTVEQSCSTLPPPKKL 143
R + NQFN +RA+Q + P E++C T PPP+++
Sbjct 176 ERKLTNQFNYSERASQSLNNPLRERACQTEPPPREV 211
> hsa:27019 DNAI1, CILD1, ICS, ICS1, MGC26204, PCD; dynein, axonemal,
intermediate chain 1; K10409 dynein intermediate chain
1, axonemal
Length=699
Score = 67.8 bits (164), Expect = 1e-11, Method: Composition-based stats.
Identities = 51/167 (30%), Positives = 76/167 (45%), Gaps = 30/167 (17%)
Query 4 DQVQLAPEELRQRMPPKILYPQNPRAPQNITRFSFKENQFKKEGAVEQTVFHLFIDSTLL 63
DQ++L EL++ +IL NP APQNI R+SFKE +K G V Q H L+
Sbjct 53 DQLELTDAELKEEFT-RILTANNPHAPQNIVRYSFKEGTYKPIGFVNQLAVHYTQVGNLI 111
Query 64 LKDSPEAAEQ---EELLRLRDEAEERRREL--------------------------EAVE 94
KDS E Q +EL+ E+ + E A +
Sbjct 112 PKDSDEGRRQHYRDELVAGSQESVKVISETGNLEEDEEPKELETEPGSQTDVPAAGAAEK 171
Query 95 VSIEDDGATQEEEGRVMRNQFNNVDRATQIKSCPTVEQSCSTLPPPK 141
V+ E+ ++ + R + NQFN +RA+Q + P ++ C T PPP+
Sbjct 172 VTEEELMTPKQPKERKLTNQFNFSERASQTYNNPVRDRECQTEPPPR 218
> dre:555705 zgc:158666; K10409 dynein intermediate chain 1, axonemal
Length=660
Score = 65.5 bits (158), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 48/147 (32%), Positives = 69/147 (46%), Gaps = 27/147 (18%)
Query 4 DQVQLAPEELRQRMPPKILYPQNPRAPQNITRFSFKENQFKKEGAVEQTVFHLFIDSTLL 63
DQ+ L EEL++ + ++L NP APQN+ R+SFKE +K V+Q H ++ L+
Sbjct 62 DQLDLTEEELKEEIT-RVLTANNPNAPQNVVRYSFKEGCYKTIVFVDQMAIHFELEGNLV 120
Query 64 LKDSPEAAEQEELLRLRDEAEERRRELEAVEVSIEDDGATQEEEGRVMR---------NQ 114
K+S E R L A ED G + E ++ NQ
Sbjct 121 HKESDEG-----------------RRLMAKHTEREDCGGDERLENVPIKPGKREQKVTNQ 163
Query 115 FNNVDRATQIKSCPTVEQSCSTLPPPK 141
FN +DRA+Q + E SC T PPP+
Sbjct 164 FNFIDRASQTLNNLPREISCQTEPPPR 190
> mmu:68922 Dnaic1, 1110066F04Rik, BB124644, Dnai1; dynein, axonemal,
intermediate chain 1; K10409 dynein intermediate chain
1, axonemal
Length=701
Score = 61.2 bits (147), Expect = 1e-09, Method: Composition-based stats.
Identities = 52/179 (29%), Positives = 72/179 (40%), Gaps = 46/179 (25%)
Query 4 DQVQLAPEELRQRMPPKILYPQNPRAPQNITRFSFKENQFKKEGAVEQTVFHLFIDSTLL 63
DQ++L EL++ +IL NP APQNI R+SFKE +K G V Q H L+
Sbjct 46 DQLELTDAELKEEFT-RILTANNPHAPQNIVRYSFKEGTYKLIGFVNQMAVHFSQVGNLI 104
Query 64 LKDSPEAAEQEELLRLRDEAEERRRE---------------------------------- 89
KDS E Q RDE +E
Sbjct 105 PKDSDEGRRQH----YRDEMVAGSQESIKVVTSEAENLEEEEEPKEGEGEAEAEAEAGSQ 160
Query 90 ------LEAVEVSIEDD-GATQEEEGRVMRNQFNNVDRATQIKSCPTVEQSCSTLPPPK 141
E E IE++ A + + R + NQFN +RA+Q + P ++ C PPP+
Sbjct 161 TDIPAAAETTEKVIEEELMAPVQPKERKLTNQFNFSERASQTFNNPLRDRECQMEPPPR 219
> pfa:PFI1080w dynein intermediate chain 2, ciliary; K10409 dynein
intermediate chain 1, axonemal
Length=820
Score = 57.4 bits (137), Expect = 2e-08, Method: Composition-based stats.
Identities = 27/63 (42%), Positives = 39/63 (61%), Gaps = 0/63 (0%)
Query 20 KILYPQNPRAPQNITRFSFKENQFKKEGAVEQTVFHLFIDSTLLLKDSPEAAEQEELLRL 79
+ L+P+NPRA N+ +SF QF ++ TVFHL I S L+ KDS E +Q EL++
Sbjct 47 RFLFPKNPRAFTNLVEYSFSNEQFVTIENIDHTVFHLDIKSCLVRKDSDEGKKQMELIKK 106
Query 80 RDE 82
+ E
Sbjct 107 KIE 109
> mmu:242425 Gabbr2, GABABR2, Gb2, Gm425, Gpr51; gamma-aminobutyric
acid (GABA) B receptor, 2; K04616 gamma-aminobutyric acid
(GABA) B receptor 2
Length=940
Score = 35.4 bits (80), Expect = 0.055, Method: Composition-based stats.
Identities = 28/103 (27%), Positives = 47/103 (45%), Gaps = 8/103 (7%)
Query 17 MPPKILYPQNPRAPQNITRFSFKENQFKKEGAVEQTVFHLFIDSTLLLKDSPEAAEQEEL 76
+P I NP A RF F +NQ K++ +V + ST L+ Q E
Sbjct 740 VPKLITLRTNPDAATQNRRFQFTQNQKKEDSKTSTSVTSVNQASTSRLE-----GLQSEN 794
Query 77 LRLRDEAEERRRELEAVEVSIEDDGATQEEEGRVMRNQFNNVD 119
RLR + E ++LE V + ++D T E+ + +N + ++
Sbjct 795 HRLRMKITELDKDLEEVTMQLQD---TPEKTTYIKQNHYQELN 834
> hsa:9568 GABBR2, FLJ36928, GABABR2, GPR51, GPRC3B, HG20, HRIHFB2099;
gamma-aminobutyric acid (GABA) B receptor, 2; K04616
gamma-aminobutyric acid (GABA) B receptor 2
Length=941
Score = 35.4 bits (80), Expect = 0.055, Method: Composition-based stats.
Identities = 28/103 (27%), Positives = 47/103 (45%), Gaps = 8/103 (7%)
Query 17 MPPKILYPQNPRAPQNITRFSFKENQFKKEGAVEQTVFHLFIDSTLLLKDSPEAAEQEEL 76
+P I NP A RF F +NQ K++ +V + ST L+ Q E
Sbjct 741 VPKLITLRTNPDAATQNRRFQFTQNQKKEDSKTSTSVTSVNQASTSRLE-----GLQSEN 795
Query 77 LRLRDEAEERRRELEAVEVSIEDDGATQEEEGRVMRNQFNNVD 119
RLR + E ++LE V + ++D T E+ + +N + ++
Sbjct 796 HRLRMKITELDKDLEEVTMQLQD---TPEKTTYIKQNHYQELN 835
> dre:560267 si:dkey-190l1.2; K04616 gamma-aminobutyric acid (GABA)
B receptor 2
Length=935
Score = 32.0 bits (71), Expect = 0.69, Method: Composition-based stats.
Identities = 26/110 (23%), Positives = 48/110 (43%), Gaps = 8/110 (7%)
Query 17 MPPKILYPQNPRAPQNITRFSFKENQFKKEGAVEQTVFHLFIDSTLLLKDSPEAAEQEEL 76
+P + NP A R F +NQ K++ +V + +T L Q +
Sbjct 736 VPKFVTMRTNPDAATQNRRLKFTQNQKKEDSKTSTSVTSVNQANTSRLD-----GLQSDN 790
Query 77 LRLRDEAEERRRELEAVEVSIEDDGATQEEEGRVMRNQFNNVDRATQIKS 126
RLR + E +ELE V + ++D T E+ + +N + + + I++
Sbjct 791 HRLRMKITELDKELEEVTMQLQD---TPEKTPYIKQNHYQDANNILSIRN 837
> bbo:BBOV_IV001930 21.m02940; hypothetical protein
Length=1006
Score = 31.2 bits (69), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 12/27 (44%), Positives = 17/27 (62%), Gaps = 0/27 (0%)
Query 9 APEELRQRMPPKILYPQNPRAPQNITR 35
A E LR+R+P I+ P+ PR P + R
Sbjct 816 AVERLRKRLPTGIIAPKKPRGPSRLAR 842
> ath:AT2G41150 hypothetical protein
Length=404
Score = 30.8 bits (68), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 19/49 (38%), Positives = 31/49 (63%), Gaps = 3/49 (6%)
Query 78 RLRDEAEERRREL---EAVEVSIEDDGATQEEEGRVMRNQFNNVDRATQ 123
RLRD AE+ + +L +A+ V D T+++ RV R+QF ++DR T+
Sbjct 245 RLRDAAEKIKAKLGDYDAIHVRRGDKLKTRKDRFRVERSQFPHLDRDTR 293
> sce:YGL139W FLC3; Flc3p
Length=802
Score = 30.0 bits (66), Expect = 2.6, Method: Composition-based stats.
Identities = 22/84 (26%), Positives = 39/84 (46%), Gaps = 4/84 (4%)
Query 21 ILYPQNPR---APQNITRFSFKENQFKKEGAVEQTVFHLFIDSTLLLKDSPEAAEQEEL- 76
+L+ +NP P R SF+ N K EG V ++V + + + KD + ++ E
Sbjct 582 MLFSKNPDLRFKPAKDDRTSFQRNTMKPEGTVNRSVANELLALGNVAKDHDDNSDYESND 641
Query 77 LRLRDEAEERRRELEAVEVSIEDD 100
+ DE ++ + E V+ DD
Sbjct 642 TGVNDELKQAQDETTPTTVTSSDD 665
> mmu:18158 Nppb, AA408272, BNP; natriuretic peptide type B; K12335
natriuretic peptide B
Length=121
Score = 29.6 bits (65), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 23/71 (32%), Positives = 39/71 (54%), Gaps = 16/71 (22%)
Query 67 SPEAAEQEELLRL-RDEAEER-RRELEAVEVSIEDDGATQEEEGRVMRNQFNNV------ 118
SPE + ++LL L R+++EE +R+L ++D G T+E RV+R+Q + +
Sbjct 35 SPEQFKMQKLLELIREKSEEMAQRQL------LKDQGLTKEHPKRVLRSQGSTLRVQQRP 88
Query 119 --DRATQIKSC 127
+ T I SC
Sbjct 89 QNSKVTHISSC 99
> cpv:cgd7_4730 RAD23p, UB+UBA domains protein ; K10839 UV excision
repair protein RAD23
Length=362
Score = 29.3 bits (64), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 21/52 (40%), Positives = 25/52 (48%), Gaps = 8/52 (15%)
Query 89 ELEAVEVSIEDDGATQEEEGRVMRNQFNNVDRATQIKSCPTVEQSCSTLPPP 140
ELE +I + G +E+ R MR FNN DRA VE S LP P
Sbjct 154 ELEKTITNIVNMGFEREQVTRAMRAAFNNPDRA--------VEYLTSGLPIP 197
> dre:100005752 MGC162803, cb117, cgn, im:7137721, sb:cb117, sb:cb117.;
si:dkey-204a24.2
Length=1161
Score = 28.9 bits (63), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 22/61 (36%), Positives = 35/61 (57%), Gaps = 3/61 (4%)
Query 75 ELLR--LRDEAEERRRELEAVEVSIEDDGATQEEEGRVMRNQFNNVDRATQIKSCPTVEQ 132
+LLR L D +R REL A++ +++D+ A+ ++E +R QF+ D S TV Q
Sbjct 503 QLLRQKLEDTLRQRERELTALKGALKDEVASHDKEMEALREQFSQ-DMDALRHSMETVSQ 561
Query 133 S 133
S
Sbjct 562 S 562
> hsa:4653 MYOC, GLC1A, GPOA, JOAG, JOAG1, TIGR, myocilin; myocilin,
trabecular meshwork inducible glucocorticoid response
Length=504
Score = 28.1 bits (61), Expect = 9.9, Method: Compositional matrix adjust.
Identities = 26/107 (24%), Positives = 48/107 (44%), Gaps = 12/107 (11%)
Query 48 AVEQTVFHLFIDSTLLLKDSPEAAEQE--ELLRLRDEAEERRRELEAVEVSIEDDGATQE 105
++E + L +D +++ E ++E L R RD+ E + RELE ++ D + E
Sbjct 94 SLESLLHQLTLDQAARPQETQEGLQRELGTLRRERDQLETQTRELETAYSNLLRDKSVLE 153
Query 106 EEGRVMRNQFNNVDRATQIKS----------CPTVEQSCSTLPPPKK 142
EE + +R + N+ R + S CP + +PP +
Sbjct 154 EEKKRLRQENENLARRLESSSQEVARLRRGQCPQTRDTARAVPPGSR 200
Lambda K H
0.313 0.130 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2872883024
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40