bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0569_orf1
Length=127
Score E
Sequences producing significant alignments: (Bits) Value
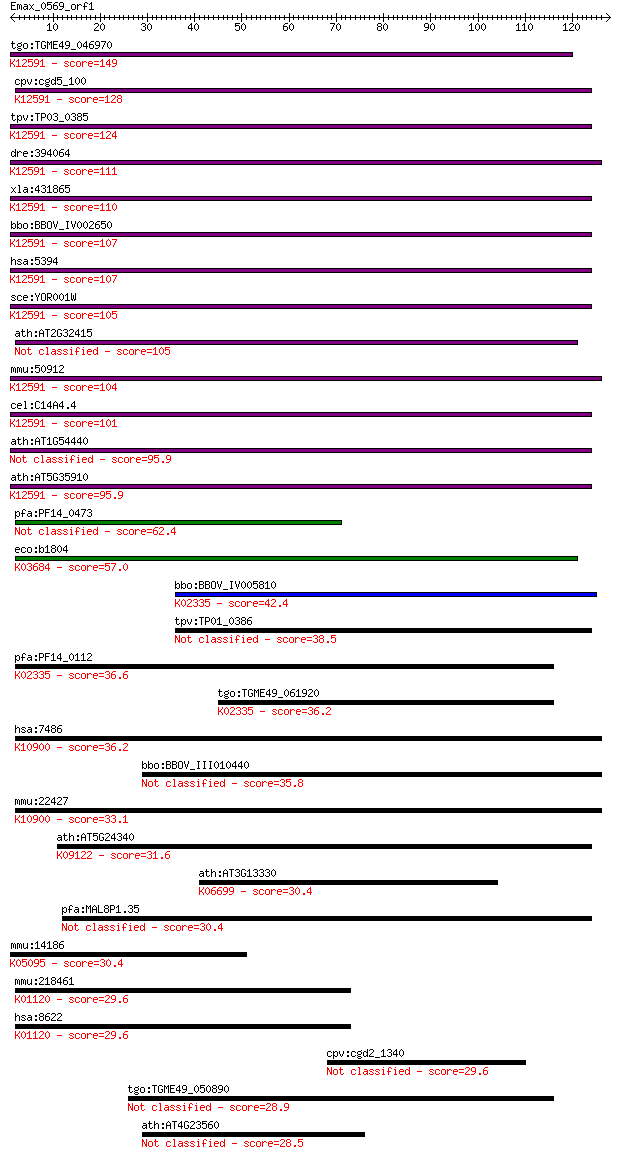
tgo:TGME49_046970 exosome component 10, putative ; K12591 exos... 149 2e-36
cpv:cgd5_100 RRPp/PMC2 like exosome 3'-5' exoribonuclease subu... 128 4e-30
tpv:TP03_0385 hypothetical protein; K12591 exosome complex exo... 124 1e-28
dre:394064 MGC55695, Pmscl2, exosc10; zgc:55695; K12591 exosom... 111 5e-25
xla:431865 exosc10, MGC83774; exosome component 10; K12591 exo... 110 1e-24
bbo:BBOV_IV002650 21.m02990; exosome component 10; K12591 exos... 107 9e-24
hsa:5394 EXOSC10, PM-Scl, PM/Scl-100, PMSCL, PMSCL2, RRP6, Rrp... 107 1e-23
sce:YOR001W RRP6; Nuclear exosome exonuclease component; has 3... 105 3e-23
ath:AT2G32415 3'-5' exonuclease/ nucleic acid binding 105 3e-23
mmu:50912 Exosc10, PM-Scl, PM/Scl-100, Pmscl2, RRP6, p2, p3, p... 104 7e-23
cel:C14A4.4 crn-3; Cell-death-Related Nuclease family member (... 101 7e-22
ath:AT1G54440 3'-5' exonuclease/ nucleic acid binding 95.9 2e-20
ath:AT5G35910 3'-5' exonuclease domain-containing protein / he... 95.9 3e-20
pfa:PF14_0473 Rrp6 homologue, putative 62.4 3e-10
eco:b1804 rnd, ECK1802, JW1793; ribonuclease D (EC:3.1.26.3); ... 57.0 1e-08
bbo:BBOV_IV005810 23.m05916; DNA polymerase I (EC:2.7.7.7); K0... 42.4 4e-04
tpv:TP01_0386 hypothetical protein 38.5 0.005
pfa:PF14_0112 POM1; Pfprex; K02335 DNA polymerase I [EC:2.7.7.7] 36.6 0.021
tgo:TGME49_061920 DNA polymerase I, putative (EC:2.7.7.7); K02... 36.2 0.025
hsa:7486 WRN, DKFZp686C2056, RECQ3, RECQL2, RECQL3; Werner syn... 36.2 0.025
bbo:BBOV_III010440 17.m07901; 3'-5' exonuclease domain contain... 35.8 0.036
mmu:22427 Wrn, AI846146; Werner syndrome homolog (human) (EC:3... 33.1 0.20
ath:AT5G24340 3'-5' exonuclease domain-containing protein; K09... 31.6 0.64
ath:AT3G13330 binding; K06699 proteasome activator subunit 4 30.4
pfa:MAL8P1.35 exonuclease, putative 30.4 1.4
mmu:14186 Fgfr4, Fgfr-4; fibroblast growth factor receptor 4 (... 30.4 1.6
mmu:218461 Pde8b, B230331L10Rik, C030047E14Rik; phosphodiester... 29.6 2.2
hsa:8622 PDE8B, ADSD, FLJ11212, PPNAD3; phosphodiesterase 8B (... 29.6 2.2
cpv:cgd2_1340 possible phosphatidylinositol 3- and 4-kinase fa... 29.6 2.3
tgo:TGME49_050890 3'-5' exonuclease domain-containing protein 28.9 4.3
ath:AT4G23560 AtGH9B15 (Arabidopsis thaliana glycosyl hydrolas... 28.5 5.0
> tgo:TGME49_046970 exosome component 10, putative ; K12591 exosome
complex exonuclease RRP6 [EC:3.1.13.-]
Length=1353
Score = 149 bits (376), Expect = 2e-36, Method: Composition-based stats.
Identities = 69/119 (57%), Positives = 82/119 (68%), Gaps = 0/119 (0%)
Query 1 CLIQLGTAEYVYVIDALKLFDHLHILNAITANPTIVKVFHNGDNDIAWLQRDFNVFVVNA 60
CL+QL T E Y+ID LFDHLH+LN ITANP I+K+FH D+DI WLQRDF+V+VVN
Sbjct 502 CLLQLSTREKDYIIDPFALFDHLHVLNTITANPKILKIFHGADSDIIWLQRDFSVYVVNM 561
Query 61 FDTGKAAKYLGVPGGTSLRNILQREFKVIKNERMSTCDWSRRPLTWDMRKYAVKDVGYL 119
FDT AA+ L VPGG SL N+LQ V N++ DW RRPLT +M YA D YL
Sbjct 562 FDTCVAARALAVPGGASLANLLQTYCHVEANKQYQLADWRRRPLTPEMETYARSDTHYL 620
> cpv:cgd5_100 RRPp/PMC2 like exosome 3'-5' exoribonuclease subunit
with an RNAseD domain and an HRDc domain ; K12591 exosome
complex exonuclease RRP6 [EC:3.1.13.-]
Length=957
Score = 128 bits (321), Expect = 4e-30, Method: Composition-based stats.
Identities = 59/122 (48%), Positives = 81/122 (66%), Gaps = 0/122 (0%)
Query 2 LIQLGTAEYVYVIDALKLFDHLHILNAITANPTIVKVFHNGDNDIAWLQRDFNVFVVNAF 61
LIQL T + Y+ID LF+ + +LN +TANP I+KV H D DI WLQRDF+V++VN F
Sbjct 322 LIQLSTRTHDYIIDPFNLFNEIQMLNELTANPKILKVLHGSDYDIIWLQRDFSVYIVNMF 381
Query 62 DTGKAAKYLGVPGGTSLRNILQREFKVIKNERMSTCDWSRRPLTWDMRKYAVKDVGYLLC 121
DTG+AA+ L PGG SL+N+L + ++R DW RPL+ ++ +YA D YLL
Sbjct 382 DTGQAARILNTPGGYSLKNLLSIYCSLDIDKRFQLADWRERPLSNELIEYARGDTHYLLY 441
Query 122 LY 123
+Y
Sbjct 442 IY 443
> tpv:TP03_0385 hypothetical protein; K12591 exosome complex exonuclease
RRP6 [EC:3.1.13.-]
Length=996
Score = 124 bits (310), Expect = 1e-28, Method: Composition-based stats.
Identities = 58/123 (47%), Positives = 79/123 (64%), Gaps = 1/123 (0%)
Query 1 CLIQLGTAEYVYVIDALKLFDHLHILNAITANPTIVKVFHNGDNDIAWLQRDFNVFVVNA 60
CL+QL T E Y+ID K+F ++ LN +T +P I+K+ H ND+ WLQRDFN+FVVN
Sbjct 511 CLVQLSTPEENYIIDPFKIFGKMNKLNRLTTDPKILKIMHGASNDVVWLQRDFNIFVVNL 570
Query 61 FDTGKAAKYLGVPGGTSLRNILQREFKVIKNERMSTCDWSRRPLTWDMRKYAVKDVGYLL 120
FDT +AAK L + SL ++Q+ F + N+R DWS+RPL +M YA D YL+
Sbjct 571 FDTREAAKVLNL-AEQSLAKLIQKYFNIKLNKRFQLSDWSKRPLDAEMLDYACCDSHYLI 629
Query 121 CLY 123
LY
Sbjct 630 PLY 632
> dre:394064 MGC55695, Pmscl2, exosc10; zgc:55695; K12591 exosome
complex exonuclease RRP6 [EC:3.1.13.-]
Length=899
Score = 111 bits (278), Expect = 5e-25, Method: Composition-based stats.
Identities = 57/125 (45%), Positives = 78/125 (62%), Gaps = 1/125 (0%)
Query 1 CLIQLGTAEYVYVIDALKLFDHLHILNAITANPTIVKVFHNGDNDIAWLQRDFNVFVVNA 60
CL+Q+ T E ++ID L+L ++ILN +P IVKVFH D+DI WLQ+DF ++VVN
Sbjct 325 CLMQISTREEDFIIDTLELRSEMYILNETFTDPAIVKVFHGADSDIEWLQKDFGLYVVNM 384
Query 61 FDTGKAAKYLGVPGGTSLRNILQREFKVIKNERMSTCDWSRRPLTWDMRKYAVKDVGYLL 120
FDT AA+ L + G SL ++L+ V ++R DW RPL +M KYA D YLL
Sbjct 385 FDTHHAARCLNL-GRNSLDHLLKVYCDVSSDKRYQLADWRIRPLPDEMLKYAQADTHYLL 443
Query 121 CLYYR 125
+Y R
Sbjct 444 YVYDR 448
> xla:431865 exosc10, MGC83774; exosome component 10; K12591 exosome
complex exonuclease RRP6 [EC:3.1.13.-]
Length=883
Score = 110 bits (275), Expect = 1e-24, Method: Composition-based stats.
Identities = 55/123 (44%), Positives = 79/123 (64%), Gaps = 1/123 (0%)
Query 1 CLIQLGTAEYVYVIDALKLFDHLHILNAITANPTIVKVFHNGDNDIAWLQRDFNVFVVNA 60
CL+Q+ T Y+ID L+L +L+ILN NP+I+KVFH D+DI WLQ+DF +++VN
Sbjct 323 CLMQISTRTEDYIIDVLELRSNLYILNESFTNPSIIKVFHGADSDIEWLQKDFGLYIVNM 382
Query 61 FDTGKAAKYLGVPGGTSLRNILQREFKVIKNERMSTCDWSRRPLTWDMRKYAVKDVGYLL 120
FDT +AA+ L + G SL ++L+ V ++R DW RPL +M +YA D YLL
Sbjct 383 FDTHQAARILNL-GRHSLDHLLRLYCNVESDKRYQLADWRIRPLPEEMLEYARVDTHYLL 441
Query 121 CLY 123
+Y
Sbjct 442 YIY 444
> bbo:BBOV_IV002650 21.m02990; exosome component 10; K12591 exosome
complex exonuclease RRP6 [EC:3.1.13.-]
Length=879
Score = 107 bits (267), Expect = 9e-24, Method: Composition-based stats.
Identities = 53/123 (43%), Positives = 74/123 (60%), Gaps = 0/123 (0%)
Query 1 CLIQLGTAEYVYVIDALKLFDHLHILNAITANPTIVKVFHNGDNDIAWLQRDFNVFVVNA 60
CL+Q+ A+ +VID +FD + LN +T +P I+KV H ++DI WLQRDF V+VVN
Sbjct 355 CLVQITGADDDWVIDPFSIFDEMWRLNDVTTDPRILKVMHGAESDILWLQRDFGVYVVNL 414
Query 61 FDTGKAAKYLGVPGGTSLRNILQREFKVIKNERMSTCDWSRRPLTWDMRKYAVKDVGYLL 120
FDT KAA L + G SL ++++ + ++ DW RP+ DM YA D YLL
Sbjct 415 FDTLKAADVLCLSCGHSLSSLVRHFLGIHLDKSYQLADWRIRPIPRDMLTYATADTHYLL 474
Query 121 CLY 123
LY
Sbjct 475 DLY 477
> hsa:5394 EXOSC10, PM-Scl, PM/Scl-100, PMSCL, PMSCL2, RRP6, Rrp6p,
p2, p3, p4; exosome component 10; K12591 exosome complex
exonuclease RRP6 [EC:3.1.13.-]
Length=885
Score = 107 bits (266), Expect = 1e-23, Method: Composition-based stats.
Identities = 54/123 (43%), Positives = 76/123 (61%), Gaps = 1/123 (0%)
Query 1 CLIQLGTAEYVYVIDALKLFDHLHILNAITANPTIVKVFHNGDNDIAWLQRDFNVFVVNA 60
CL+Q+ T ++ID L+L ++ILN +P IVKVFH D+DI WLQ+DF ++VVN
Sbjct 327 CLMQISTRTEDFIIDTLELRSDMYILNESLTDPAIVKVFHGADSDIEWLQKDFGLYVVNM 386
Query 61 FDTGKAAKYLGVPGGTSLRNILQREFKVIKNERMSTCDWSRRPLTWDMRKYAVKDVGYLL 120
FDT +AA+ L + G SL ++L+ V N++ DW RPL +M YA D YLL
Sbjct 387 FDTHQAARLLNL-GRHSLDHLLKLYCNVDSNKQYQLADWRIRPLPEEMLSYARDDTHYLL 445
Query 121 CLY 123
+Y
Sbjct 446 YIY 448
> sce:YOR001W RRP6; Nuclear exosome exonuclease component; has
3'-5' exonuclease activity; involved in RNA processing, maturation,
surveillance, degradation, tethering, and export; has
similarity to E. coli RNase D and to human PM-Sc1 100 (EXOSC10)
(EC:3.1.13.-); K12591 exosome complex exonuclease RRP6
[EC:3.1.13.-]
Length=733
Score = 105 bits (263), Expect = 3e-23, Method: Composition-based stats.
Identities = 54/123 (43%), Positives = 75/123 (60%), Gaps = 1/123 (0%)
Query 1 CLIQLGTAEYVYVIDALKLFDHLHILNAITANPTIVKVFHNGDNDIAWLQRDFNVFVVNA 60
CL+Q+ T E Y++D LKL ++LHILN + NP+IVKVFH DI WLQRD ++VV
Sbjct 252 CLMQISTRERDYLVDTLKLRENLHILNEVFTNPSIVKVFHGAFMDIIWLQRDLGLYVVGL 311
Query 61 FDTGKAAKYLGVPGGTSLRNILQREFKVIKNERMSTCDWSRRPLTWDMRKYAVKDVGYLL 120
FDT A+K +G+P SL +L+ +++ DW RPL+ M YA D +LL
Sbjct 312 FDTYHASKAIGLP-RHSLAYLLENFANFKTSKKYQLADWRIRPLSKPMTAYARADTHFLL 370
Query 121 CLY 123
+Y
Sbjct 371 NIY 373
> ath:AT2G32415 3'-5' exonuclease/ nucleic acid binding
Length=891
Score = 105 bits (263), Expect = 3e-23, Method: Composition-based stats.
Identities = 52/119 (43%), Positives = 74/119 (62%), Gaps = 1/119 (0%)
Query 2 LIQLGTAEYVYVIDALKLFDHLHILNAITANPTIVKVFHNGDNDIAWLQRDFNVFVVNAF 61
LIQ+ T E +++D + L D + IL + ++P I KVFH DND+ WLQRDF+++VVN F
Sbjct 159 LIQISTHEEDFLVDTIALHDVMSILRPVFSDPNICKVFHGADNDVIWLQRDFHIYVVNMF 218
Query 62 DTGKAAKYLGVPGGTSLRNILQREFKVIKNERMSTCDWSRRPLTWDMRKYAVKDVGYLL 120
DT KA + L P SL +L+ V N+ + DW +RPL+ +M +YA D YLL
Sbjct 219 DTAKACEVLSKP-QRSLAYLLETVCGVATNKLLQREDWRQRPLSEEMVRYARTDAHYLL 276
> mmu:50912 Exosc10, PM-Scl, PM/Scl-100, Pmscl2, RRP6, p2, p3,
p4; exosome component 10; K12591 exosome complex exonuclease
RRP6 [EC:3.1.13.-]
Length=887
Score = 104 bits (259), Expect = 7e-23, Method: Composition-based stats.
Identities = 53/125 (42%), Positives = 76/125 (60%), Gaps = 1/125 (0%)
Query 1 CLIQLGTAEYVYVIDALKLFDHLHILNAITANPTIVKVFHNGDNDIAWLQRDFNVFVVNA 60
CL+Q+ T +++D L+L ++ILN +P IVKVFH D+DI WLQ+DF ++VVN
Sbjct 327 CLMQISTRTEDFIVDTLELRSDMYILNESLTDPAIVKVFHGADSDIEWLQKDFGLYVVNM 386
Query 61 FDTGKAAKYLGVPGGTSLRNILQREFKVIKNERMSTCDWSRRPLTWDMRKYAVKDVGYLL 120
FDT +AA+ L + SL ++L+ V N++ DW RPL +M YA D YLL
Sbjct 387 FDTHQAARLLNL-ARHSLDHLLRLYCGVESNKQYQLADWRIRPLPEEMLSYARDDTHYLL 445
Query 121 CLYYR 125
+Y R
Sbjct 446 YIYDR 450
> cel:C14A4.4 crn-3; Cell-death-Related Nuclease family member
(crn-3); K12591 exosome complex exonuclease RRP6 [EC:3.1.13.-]
Length=876
Score = 101 bits (251), Expect = 7e-22, Method: Composition-based stats.
Identities = 54/123 (43%), Positives = 72/123 (58%), Gaps = 1/123 (0%)
Query 1 CLIQLGTAEYVYVIDALKLFDHLHILNAITANPTIVKVFHNGDNDIAWLQRDFNVFVVNA 60
CLIQ+ T + ++ID ++DH+ +LN ANP I+KVFH D+D+ WLQRD+ V VVN
Sbjct 317 CLIQISTRDEDFIIDPFPIWDHVGMLNEPFANPRILKVFHGSDSDVLWLQRDYGVHVVNL 376
Query 61 FDTGKAAKYLGVPGGTSLRNILQREFKVIKNERMSTCDWSRRPLTWDMRKYAVKDVGYLL 120
FDT A K L P SL + R V+ +++ DW RPL M YA +D YLL
Sbjct 377 FDTYVAMKKLKYP-KFSLAYLTLRFADVVLDKQYQLADWRARPLRNAMINYAREDTHYLL 435
Query 121 CLY 123
Y
Sbjct 436 YSY 438
> ath:AT1G54440 3'-5' exonuclease/ nucleic acid binding
Length=637
Score = 95.9 bits (237), Expect = 2e-20, Method: Composition-based stats.
Identities = 50/124 (40%), Positives = 71/124 (57%), Gaps = 2/124 (1%)
Query 1 CLIQLGTAEYVYVIDALKLFDHLH-ILNAITANPTIVKVFHNGDNDIAWLQRDFNVFVVN 59
CL+Q+ T Y++D KL+DH+ L + +P KV H D DI WLQRDF ++V N
Sbjct 154 CLMQISTRTEDYIVDIFKLWDHIGPYLRELFKDPKKKKVIHGADRDIIWLQRDFGIYVCN 213
Query 60 AFDTGKAAKYLGVPGGTSLRNILQREFKVIKNERMSTCDWSRRPLTWDMRKYAVKDVGYL 119
FDTG+A++ L + SL +L+ V N+ DW RPL M++YA +D YL
Sbjct 214 LFDTGQASRVLKLE-RNSLEFLLKHYCGVAANKEYQKADWRIRPLPDVMKRYAREDTHYL 272
Query 120 LCLY 123
L +Y
Sbjct 273 LYIY 276
> ath:AT5G35910 3'-5' exonuclease domain-containing protein /
helicase and RNase D C-terminal domain-containing protein /
HRDC domain-containing protein; K12591 exosome complex exonuclease
RRP6 [EC:3.1.13.-]
Length=838
Score = 95.9 bits (237), Expect = 3e-20, Method: Composition-based stats.
Identities = 51/124 (41%), Positives = 69/124 (55%), Gaps = 2/124 (1%)
Query 1 CLIQLGTAEYVYVIDALKLFDHLH-ILNAITANPTIVKVFHNGDNDIAWLQRDFNVFVVN 59
CL+Q+ T Y++D KL H+ L I +P KV H D DI WLQRDF ++V N
Sbjct 267 CLMQISTRTEDYIVDTFKLRIHIGPYLREIFKDPKKKKVMHGADRDIIWLQRDFGIYVCN 326
Query 60 AFDTGKAAKYLGVPGGTSLRNILQREFKVIKNERMSTCDWSRRPLTWDMRKYAVKDVGYL 119
FDTG+A++ L + SL +LQ V N+ DW RPL +M +YA +D YL
Sbjct 327 LFDTGQASRVLNLE-RNSLEFLLQHFCGVTANKEYQNADWRIRPLPEEMTRYAREDTHYL 385
Query 120 LCLY 123
L +Y
Sbjct 386 LYIY 389
> pfa:PF14_0473 Rrp6 homologue, putative
Length=1136
Score = 62.4 bits (150), Expect = 3e-10, Method: Composition-based stats.
Identities = 28/69 (40%), Positives = 44/69 (63%), Gaps = 0/69 (0%)
Query 2 LIQLGTAEYVYVIDALKLFDHLHILNAITANPTIVKVFHNGDNDIAWLQRDFNVFVVNAF 61
+I +GT Y+ID +F+ L+I+N IT +P I+K+ +N N I LQ+DF+++ VN F
Sbjct 582 IIMIGTNNMNYIIDVFNMFEDLYIINDITTDPNILKITYNAPNIINQLQKDFSIYFVNIF 641
Query 62 DTGKAAKYL 70
D + YL
Sbjct 642 DIAICSNYL 650
> eco:b1804 rnd, ECK1802, JW1793; ribonuclease D (EC:3.1.26.3);
K03684 ribonuclease D [EC:3.1.13.5]
Length=375
Score = 57.0 bits (136), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 39/119 (32%), Positives = 54/119 (45%), Gaps = 1/119 (0%)
Query 2 LIQLGTAEYVYVIDALKLFDHLHILNAITANPTIVKVFHNGDNDIAWLQRDFNVFVVNAF 61
LIQL E++ +ID L + D L AI +P+I K H G D+ F
Sbjct 43 LIQLFDGEHLALIDPLGITD-WSPLKAILRDPSITKFLHAGSEDLEVFLNVFGELPQPLI 101
Query 62 DTGKAAKYLGVPGGTSLRNILQREFKVIKNERMSTCDWSRRPLTWDMRKYAVKDVGYLL 120
DT A + G P ++++ V ++ S DW RPLT +YA DV YLL
Sbjct 102 DTQILAAFCGRPMSWGFASMVEEYSGVTLDKSESRTDWLARPLTERQCEYAAADVWYLL 160
> bbo:BBOV_IV005810 23.m05916; DNA polymerase I (EC:2.7.7.7);
K02335 DNA polymerase I [EC:2.7.7.7]
Length=1613
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 32/93 (34%), Positives = 45/93 (48%), Gaps = 5/93 (5%)
Query 36 VKVFHNGDNDIAWLQRD-FNVFVVNAFDTGKAAKYLGVPG---GTSLRNILQREFKVIKN 91
VKV HNG DI +L + FNV FDT AAK L L ++ +R ++ +
Sbjct 1115 VKVLHNGKFDINFLSHNGFNV-KGPIFDTMIAAKLLSATRFNWSCKLGHVAERYLNIVLD 1173
Query 92 ERMSTCDWSRRPLTWDMRKYAVKDVGYLLCLYY 124
+ DW+ PL + YA +D LL LY+
Sbjct 1174 KSQQFSDWTLDPLFEEQVIYASRDTAVLLPLYF 1206
> tpv:TP01_0386 hypothetical protein
Length=1786
Score = 38.5 bits (88), Expect = 0.005, Method: Composition-based stats.
Identities = 30/90 (33%), Positives = 43/90 (47%), Gaps = 2/90 (2%)
Query 36 VKVFHNGDNDIAWLQRDFNVFVVNAFDTGKAAKYLGVPGGTS--LRNILQREFKVIKNER 93
VKVFHNG DI +L+ F FDT A+K L S L ++ +R ++ ++
Sbjct 1291 VKVFHNGKFDINFLRVYGFEFEGPIFDTMVASKLLVASRYISCKLTHVSERYLNIVLDKT 1350
Query 94 MSTCDWSRRPLTWDMRKYAVKDVGYLLCLY 123
DWS L + Y+ +D LL LY
Sbjct 1351 QQYSDWSTLQLFEEQLLYSARDSFVLLPLY 1380
> pfa:PF14_0112 POM1; Pfprex; K02335 DNA polymerase I [EC:2.7.7.7]
Length=2016
Score = 36.6 bits (83), Expect = 0.021, Method: Composition-based stats.
Identities = 34/120 (28%), Positives = 54/120 (45%), Gaps = 7/120 (5%)
Query 2 LIQLGTAEY---VYVIDALKLFDHLHILNAITANPTIVKVFHNGDNDIAWLQRDFNVFVV 58
LIQ+ Y +Y + + D L L + N I+K+ NG D +L + N +
Sbjct 1485 LIQIAVENYPVIIYDMFNINKKDILDGLRKVLENKNIIKIIQNGKFDAKFLLHN-NFKIE 1543
Query 59 NAFDTGKAAKYLGVPG---GTSLRNILQREFKVIKNERMSTCDWSRRPLTWDMRKYAVKD 115
N FDT A+K L G L NI+++ VI +++ W+ L + YA +D
Sbjct 1544 NIFDTYIASKLLDKNKNMYGFKLNNIVEKYLNVILDKQQQNSVWNNSLLNNNQLFYAARD 1603
> tgo:TGME49_061920 DNA polymerase I, putative (EC:2.7.7.7); K02335
DNA polymerase I [EC:2.7.7.7]
Length=704
Score = 36.2 bits (82), Expect = 0.025, Method: Composition-based stats.
Identities = 27/75 (36%), Positives = 39/75 (52%), Gaps = 4/75 (5%)
Query 45 DIAWLQRDFNVFVVNA-FDTGKAAKYL--GV-PGGTSLRNILQREFKVIKNERMSTCDWS 100
++ LQ D VFV FDT AAK + GV G L +++R V+ ++RM DWS
Sbjct 180 EVGDLQSDGGVFVSGPLFDTLIAAKVVEAGVMRTGFKLLQVVERFLGVLMDKRMQASDWS 239
Query 101 RRPLTWDMRKYAVKD 115
L+ + YA +D
Sbjct 240 SPHLSQEQLLYAARD 254
> hsa:7486 WRN, DKFZp686C2056, RECQ3, RECQL2, RECQL3; Werner syndrome,
RecQ helicase-like (EC:3.6.4.12); K10900 werner syndrome
ATP-dependent helicase [EC:3.6.4.12]
Length=1432
Score = 36.2 bits (82), Expect = 0.025, Method: Composition-based stats.
Identities = 35/130 (26%), Positives = 63/130 (48%), Gaps = 9/130 (6%)
Query 2 LIQLGTAE---YVYVIDALKLFDHLHILNAITANPTIVKVFHNGDNDIAWLQRDFNVFVV 58
LIQL +E Y++ + ++ +F L + N + K + D L RDF++ +
Sbjct 99 LIQLCVSESKCYLFHVSSMSVFPQG--LKMLLENKAVKKAGVGIEGDQWKLLRDFDIKLK 156
Query 59 NAFD-TGKAAKYLGVPGGTSLRNILQREF--KVIKNERMSTCDWSRRPLTWDMRKYAVKD 115
N + T A K L SL ++++ +++K++ + +WS+ PLT D + YA D
Sbjct 157 NFVELTDVANKKLKCTETWSLNSLVKHLLGKQLLKDKSIRCSNWSKFPLTEDQKLYAATD 216
Query 116 VGYLLCLYYR 125
Y + YR
Sbjct 217 -AYAGFIIYR 225
> bbo:BBOV_III010440 17.m07901; 3'-5' exonuclease domain containing
protein
Length=230
Score = 35.8 bits (81), Expect = 0.036, Method: Compositional matrix adjust.
Identities = 22/97 (22%), Positives = 44/97 (45%), Gaps = 1/97 (1%)
Query 29 ITANPTIVKVFHNGDNDIAWLQRDFNVFVVNAFDTGKAAKYLGVPGGTSLRNILQREFKV 88
I ++P I+K+ H +D+ + R F V + D + L + SL++++QR +
Sbjct 112 ILSDPDILKISHGAPSDMRLMYRHFGVRSRSFVDLQSVCEELQL-RPCSLKSVVQRVLGL 170
Query 89 IKNERMSTCDWSRRPLTWDMRKYAVKDVGYLLCLYYR 125
+++ +W L+ KYA D L + +
Sbjct 171 RLSKKQQCSNWEAAELSQQQIKYAATDAWVTLAAFLK 207
> mmu:22427 Wrn, AI846146; Werner syndrome homolog (human) (EC:3.6.4.12);
K10900 werner syndrome ATP-dependent helicase [EC:3.6.4.12]
Length=1401
Score = 33.1 bits (74), Expect = 0.20, Method: Composition-based stats.
Identities = 34/130 (26%), Positives = 62/130 (47%), Gaps = 8/130 (6%)
Query 2 LIQLGTAE---YVYVIDALKLFDHLHILNAITANPTIVKVFHNGDNDIAWLQRDFNVFVV 58
+IQL +E Y++ I ++ +F L + N +I K + D L RDF+V +
Sbjct 93 VIQLCVSESKCYLFHISSMSVFPQG--LKMLLENKSIKKAGVGIEGDQWKLLRDFDVKLE 150
Query 59 NAFD-TGKAAKYLGVPGGTSLRNILQREF--KVIKNERMSTCDWSRRPLTWDMRKYAVKD 115
+ + T A + L SL +++ +++K++ + +WS PLT D + YA D
Sbjct 151 SFVELTDVANEKLKCAETWSLNGLVKHVLGKQLLKDKSIRCSNWSNFPLTEDQKLYAATD 210
Query 116 VGYLLCLYYR 125
L +Y +
Sbjct 211 AYAGLIIYQK 220
> ath:AT5G24340 3'-5' exonuclease domain-containing protein; K09122
hypothetical protein
Length=505
Score = 31.6 bits (70), Expect = 0.64, Method: Composition-based stats.
Identities = 31/137 (22%), Positives = 59/137 (43%), Gaps = 30/137 (21%)
Query 11 VYVID--ALKLFDHLHILNAITANPTIVKVFHNGDNDIAWLQRDFNVFVVNAFDTG--KA 66
V++ID ++ L +LN + +P ++K+ D+ +L + F + + G +
Sbjct 68 VFLIDLSSIHLPSVWELLNDMFVSPDVLKLGFRFKQDLVYLS---STFTQHGCEGGFQEV 124
Query 67 AKYLGVPGGTSLRNILQRE--------------------FKVIKNERMSTCDWSRRPLTW 106
+YL + TS+ N LQ + + ++ + DWS RPLT
Sbjct 125 KQYLDI---TSIYNYLQHKRFGRKAPKDIKSLAAICKEMLDISLSKELQCSDWSYRPLTE 181
Query 107 DMRKYAVKDVGYLLCLY 123
+ + YA D LL ++
Sbjct 182 EQKLYAATDAHCLLQIF 198
> ath:AT3G13330 binding; K06699 proteasome activator subunit 4
Length=1781
Score = 30.4 bits (67), Expect = 1.4, Method: Composition-based stats.
Identities = 17/70 (24%), Positives = 34/70 (48%), Gaps = 8/70 (11%)
Query 41 NGD--NDIAWLQRDFNVFVVNAFDTGKAAKYLGVPGG-----TSLRNILQREFKVIKNER 93
NGD +D+ W++ F+ F++++F +G+A+ L V G SL+ ++ ++
Sbjct 1513 NGDSLDDVKWMETLFH-FIISSFKSGRASYLLDVIAGFLYPVMSLQETSHKDLSILAKAA 1571
Query 94 MSTCDWSRRP 103
W P
Sbjct 1572 FELLKWRVFP 1581
> pfa:MAL8P1.35 exonuclease, putative
Length=406
Score = 30.4 bits (67), Expect = 1.4, Method: Composition-based stats.
Identities = 32/128 (25%), Positives = 51/128 (39%), Gaps = 19/128 (14%)
Query 12 YVIDALKLFDHLHILNAITANPTIVKVFHNGDNDIAWLQRDFNVFVVNAFDTGKAAKYLG 71
Y+ D LK + I N +K+ H+ D + L + + N +DT +A +L
Sbjct 140 YIFDLLKT-SVIKSAQKIIENKKTLKLIHDCREDSSALYNQLGMKLENVYDTSRA--HLL 196
Query 72 VPGGTSLRNILQREFKVIKNERMSTCD----------------WSRRPLTWDMRKYAVKD 115
+ +I Q F + N+ + D W RPL+ YA+K+
Sbjct 197 LMEKQKKNDIYQVSFAQLINDYLGINDASLSFIKKEMYKNEKIWETRPLSNISIIYALKN 256
Query 116 VGYLLCLY 123
V YL LY
Sbjct 257 VKYLYKLY 264
> mmu:14186 Fgfr4, Fgfr-4; fibroblast growth factor receptor 4
(EC:2.7.10.1); K05095 fibroblast growth factor receptor 4 [EC:2.7.10.1]
Length=799
Score = 30.4 bits (67), Expect = 1.6, Method: Composition-based stats.
Identities = 21/62 (33%), Positives = 31/62 (50%), Gaps = 12/62 (19%)
Query 1 CLIQ--LGTAEYVYVIDALKLFDHLHILNA-ITANPTIV---------KVFHNGDNDIAW 48
CL++ LG+ Y Y++D L+ H IL A + AN T V KV+ + I W
Sbjct 221 CLVENSLGSIRYSYLLDVLERSPHRPILQAGLPANTTAVVGSDVELLCKVYSDAQPHIQW 280
Query 49 LQ 50
L+
Sbjct 281 LK 282
> mmu:218461 Pde8b, B230331L10Rik, C030047E14Rik; phosphodiesterase
8B (EC:3.1.4.17); K01120 3',5'-cyclic-nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=788
Score = 29.6 bits (65), Expect = 2.2, Method: Composition-based stats.
Identities = 23/74 (31%), Positives = 36/74 (48%), Gaps = 11/74 (14%)
Query 2 LIQLGTAEYVYVIDALKLFDHLHILNAITAN---PTIVKVFHNGDNDIAWLQRDFNVFVV 58
L +L EYV+ + H H+ IT N P+I ++ DN+ +W DFN+F +
Sbjct 416 LRRLSGNEYVFTKNVHH--SHSHLSMPITINDVPPSIAQLL---DNEESW---DFNIFEL 467
Query 59 NAFDTGKAAKYLGV 72
A + YLG+
Sbjct 468 EAVTHKRPLVYLGL 481
> hsa:8622 PDE8B, ADSD, FLJ11212, PPNAD3; phosphodiesterase 8B
(EC:3.1.4.17); K01120 3',5'-cyclic-nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=788
Score = 29.6 bits (65), Expect = 2.2, Method: Composition-based stats.
Identities = 23/74 (31%), Positives = 36/74 (48%), Gaps = 11/74 (14%)
Query 2 LIQLGTAEYVYVIDALKLFDHLHILNAITAN---PTIVKVFHNGDNDIAWLQRDFNVFVV 58
L +L EYV+ + + H H+ IT N P I ++ DN+ +W DFN+F +
Sbjct 416 LRRLSGNEYVFTKNVHQ--SHSHLAMPITINDVPPCISQLL---DNEESW---DFNIFEL 467
Query 59 NAFDTGKAAKYLGV 72
A + YLG+
Sbjct 468 EAITHKRPLVYLGL 481
> cpv:cgd2_1340 possible phosphatidylinositol 3- and 4-kinase
family protein
Length=678
Score = 29.6 bits (65), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 16/44 (36%), Positives = 24/44 (54%), Gaps = 2/44 (4%)
Query 68 KYLGVPGGTSLRNI--LQREFKVIKNERMSTCDWSRRPLTWDMR 109
KYL +P GTS+R+I R ++ M T + R PL + +R
Sbjct 63 KYLDLPEGTSIRDIQLFYRGVEIPNGRFMHTFEKQRHPLHYSLR 106
> tgo:TGME49_050890 3'-5' exonuclease domain-containing protein
Length=672
Score = 28.9 bits (63), Expect = 4.3, Method: Composition-based stats.
Identities = 24/91 (26%), Positives = 38/91 (41%), Gaps = 1/91 (1%)
Query 26 LNAITANPTIVKVFHNGDNDIAWLQRDFNVFVVNAFDTGKAAKYLGVPGGT-SLRNILQR 84
L A+ +VKV ++ LQR+F V N AA LG + SL+ +
Sbjct 225 LTALLLRADVVKVTQGATGEVEALQREFGVSPRNFLCLHAAAIALGCATNSRSLQALCGL 284
Query 85 EFKVIKNERMSTCDWSRRPLTWDMRKYAVKD 115
+ ++ + WSR L+ + YA D
Sbjct 285 FLERFLDKSLQLSTWSRDALSPEQCMYAATD 315
> ath:AT4G23560 AtGH9B15 (Arabidopsis thaliana glycosyl hydrolase
9B15); catalytic/ hydrolase, hydrolyzing O-glycosyl compounds
Length=479
Score = 28.5 bits (62), Expect = 5.0, Method: Composition-based stats.
Identities = 14/47 (29%), Positives = 21/47 (44%), Gaps = 0/47 (0%)
Query 29 ITANPTIVKVFHNGDNDIAWLQRDFNVFVVNAFDTGKAAKYLGVPGG 75
+ A +V F+NG ND+A + D FV + + PGG
Sbjct 271 VGAQALLVSEFYNGANDLAKFKSDVESFVCAMMPGSSSQQIKPTPGG 317
Lambda K H
0.327 0.141 0.451
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2054672932
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40