bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0562_orf1
Length=94
Score E
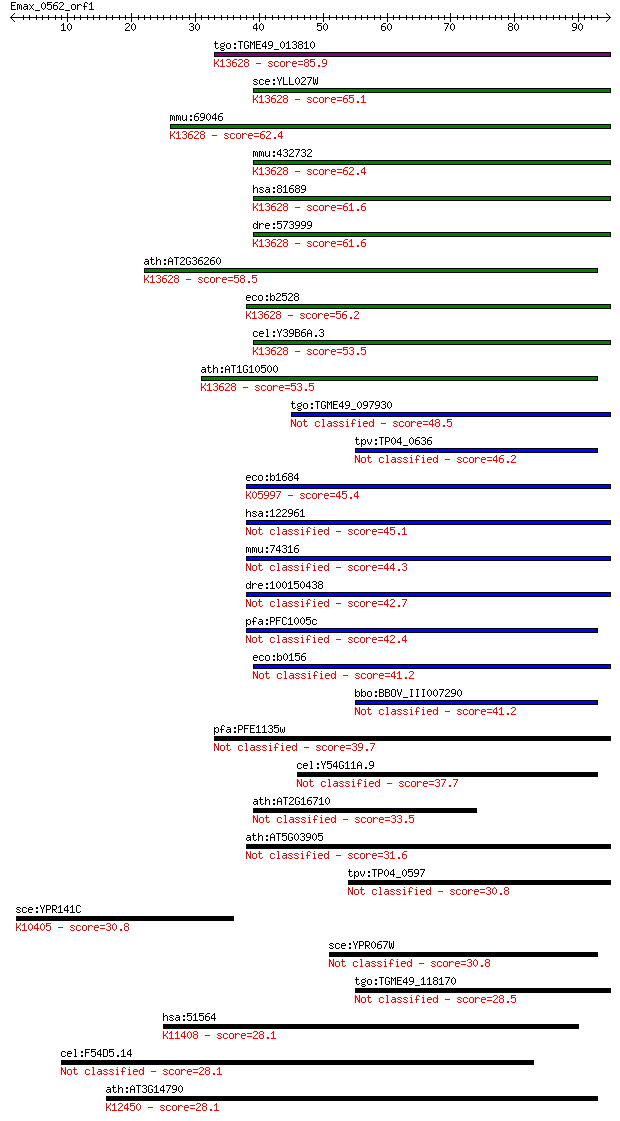
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_013810 iron-sulfur cluster assembly accessory prote... 85.9 3e-17
sce:YLL027W ISA1; Isa1p; K13628 iron-sulfur cluster assembly p... 65.1 6e-11
mmu:69046 Isca1, 1810010A06Rik, Hbld2; iron-sulfur cluster ass... 62.4 3e-10
mmu:432732 cDNA sequence AK157302; K13628 iron-sulfur cluster ... 62.4 4e-10
hsa:81689 ISCA1, HBLD2, ISA1, MGC4276, RP11-507D14.2, hIscA; i... 61.6 5e-10
dre:573999 isca1, zgc:114185; iron-sulfur cluster assembly 1 (... 61.6 6e-10
ath:AT2G36260 iron-sulfur cluster assembly complex protein, pu... 58.5 5e-09
eco:b2528 iscA, ECK2525, JW2512, yfhF; FeS cluster assembly pr... 56.2 2e-08
cel:Y39B6A.3 hypothetical protein; K13628 iron-sulfur cluster ... 53.5 1e-07
ath:AT1G10500 ATCPISCA (chloroplast-localized IscA-like protei... 53.5 2e-07
tgo:TGME49_097930 HesB-like domain containing protein 48.5 5e-06
tpv:TP04_0636 hypothetical protein 46.2 2e-05
eco:b1684 sufA, ECK1680, JW1674, ydiC; Fe-S cluster assembly p... 45.4 4e-05
hsa:122961 ISCA2, HBLD1, ISA2, c14_5557; iron-sulfur cluster a... 45.1 6e-05
mmu:74316 Isca2, 0710001C05Rik, 5730594E03Rik, Hbld1; iron-sul... 44.3 1e-04
dre:100150438 similar to Iron-sulfur cluster assembly 2 homolo... 42.7 3e-04
pfa:PFC1005c iron-sulfur cluster assembly accessory protein, p... 42.4 4e-04
eco:b0156 erpA, ECK0155, JW0152, yadR; iron-sulfur cluster ins... 41.2 8e-04
bbo:BBOV_III007290 17.m07640; HesB-like domain containing protein 41.2 8e-04
pfa:PFE1135w iron-sulfur assembly protein, putative 39.7 0.002
cel:Y54G11A.9 hypothetical protein 37.7 0.009
ath:AT2G16710 hesB-like domain-containing protein 33.5 0.17
ath:AT5G03905 hesB-like domain-containing protein 31.6 0.58
tpv:TP04_0597 hypothetical protein 30.8 0.98
sce:YPR141C KAR3, OSR11; Kar3p; K10405 kinesin family member C1 30.8 1.1
sce:YPR067W ISA2; Isa2p 30.8 1.2
tgo:TGME49_118170 iron-sulfur cluster assembly accessory domai... 28.5 4.9
hsa:51564 HDAC7, DKFZp586J0917, FLJ99588, HD7A, HDAC7A; histon... 28.1 6.4
cel:F54D5.14 hypothetical protein 28.1 7.3
ath:AT3G14790 RHM3; RHM3 (RHAMNOSE BIOSYNTHESIS 3); UDP-L-rham... 28.1 8.2
> tgo:TGME49_013810 iron-sulfur cluster assembly accessory protein,
putative ; K13628 iron-sulfur cluster assembly protein
Length=504
Score = 85.9 bits (211), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 33/62 (53%), Positives = 46/62 (74%), Gaps = 0/62 (0%)
Query 33 RKNWGEDEVVVVEGVEIRVAADAVMLLIGTQVDFVDEDVQTGFIFNNPNQKHSCGCGKSF 92
++ W DE++ G + VA+DAVM L+GT+VDFVD +++ F+F NPN+K SCGCGKSF
Sbjct 443 KQPWAADELIETAGGRLVVASDAVMFLVGTEVDFVDTELERKFVFKNPNEKQSCGCGKSF 502
Query 93 MV 94
M
Sbjct 503 MA 504
> sce:YLL027W ISA1; Isa1p; K13628 iron-sulfur cluster assembly
protein
Length=250
Score = 65.1 bits (157), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 26/56 (46%), Positives = 43/56 (76%), Gaps = 0/56 (0%)
Query 39 DEVVVVEGVEIRVAADAVMLLIGTQVDFVDEDVQTGFIFNNPNQKHSCGCGKSFMV 94
DEVV +GV+I + + A+ +IG+++D++D+ + + F+F NPN K +CGCG+SFMV
Sbjct 195 DEVVEQDGVKIVIDSKALFSIIGSEMDWIDDKLASKFVFKNPNSKGTCGCGESFMV 250
> mmu:69046 Isca1, 1810010A06Rik, Hbld2; iron-sulfur cluster assembly
1 homolog (S. cerevisiae) (EC:2.7.7.52); K13628 iron-sulfur
cluster assembly protein
Length=129
Score = 62.4 bits (150), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 29/70 (41%), Positives = 46/70 (65%), Gaps = 1/70 (1%)
Query 26 LGLSGHRRKNWGE-DEVVVVEGVEIRVAADAVMLLIGTQVDFVDEDVQTGFIFNNPNQKH 84
L S K G+ DE V+ +GV + + A + L+GT++D+V++ + + F+FNNPN K
Sbjct 60 LSYSLEYTKTKGDSDEEVIQDGVRVFIEKKAQLTLLGTEMDYVEDKLSSEFVFNNPNIKG 119
Query 85 SCGCGKSFMV 94
+CGCG+SF V
Sbjct 120 TCGCGESFHV 129
> mmu:432732 cDNA sequence AK157302; K13628 iron-sulfur cluster
assembly protein
Length=129
Score = 62.4 bits (150), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 25/56 (44%), Positives = 41/56 (73%), Gaps = 0/56 (0%)
Query 39 DEVVVVEGVEIRVAADAVMLLIGTQVDFVDEDVQTGFIFNNPNQKHSCGCGKSFMV 94
DE V+ +GV + + A + L+GT++D+V++ + + F+FNNPN K +CGCG+SF V
Sbjct 74 DEEVIQDGVRVFIEKKAQLTLLGTEMDYVEDKLSSEFVFNNPNIKGTCGCGESFHV 129
> hsa:81689 ISCA1, HBLD2, ISA1, MGC4276, RP11-507D14.2, hIscA;
iron-sulfur cluster assembly 1 homolog (S. cerevisiae) (EC:2.7.7.52);
K13628 iron-sulfur cluster assembly protein
Length=129
Score = 61.6 bits (148), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 24/56 (42%), Positives = 41/56 (73%), Gaps = 0/56 (0%)
Query 39 DEVVVVEGVEIRVAADAVMLLIGTQVDFVDEDVQTGFIFNNPNQKHSCGCGKSFMV 94
DE V+ +GV + + A + L+GT++D+V++ + + F+FNNPN K +CGCG+SF +
Sbjct 74 DEEVIQDGVRVFIEKKAQLTLLGTEMDYVEDKLSSEFVFNNPNIKGTCGCGESFNI 129
> dre:573999 isca1, zgc:114185; iron-sulfur cluster assembly 1
(EC:2.7.7.52); K13628 iron-sulfur cluster assembly protein
Length=129
Score = 61.6 bits (148), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 25/56 (44%), Positives = 40/56 (71%), Gaps = 0/56 (0%)
Query 39 DEVVVVEGVEIRVAADAVMLLIGTQVDFVDEDVQTGFIFNNPNQKHSCGCGKSFMV 94
DE V+ +GV + + A + L+GT++DFV+ + + F+FNNPN K +CGCG+SF +
Sbjct 74 DEEVLQDGVRVFIEKKAQLTLLGTEMDFVETKLSSEFVFNNPNIKGTCGCGESFNI 129
> ath:AT2G36260 iron-sulfur cluster assembly complex protein,
putative; K13628 iron-sulfur cluster assembly protein
Length=109
Score = 58.5 bits (140), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 34/74 (45%), Positives = 48/74 (64%), Gaps = 4/74 (5%)
Query 22 ASGPLGLS---GHRRKNWGEDEVVVVEGVEIRVAADAVMLLIGTQVDFVDEDVQTGFIFN 78
A G GLS + ++ DEVV +GV+I V AVM +IGT++DFVD+ +++ F+F
Sbjct 34 AKGCNGLSYVLNYAQEKGKFDEVVEEKGVKILVDPKAVMHVIGTEMDFVDDKLRSEFVFV 93
Query 79 NPNQKHSCGCGKSF 92
NPN CGCG+SF
Sbjct 94 NPNAT-KCGCGESF 106
> eco:b2528 iscA, ECK2525, JW2512, yfhF; FeS cluster assembly
protein; K13628 iron-sulfur cluster assembly protein
Length=107
Score = 56.2 bits (134), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 28/57 (49%), Positives = 36/57 (63%), Gaps = 0/57 (0%)
Query 38 EDEVVVVEGVEIRVAADAVMLLIGTQVDFVDEDVQTGFIFNNPNQKHSCGCGKSFMV 94
ED V +GV++ V ++ L GTQ+DFV E + GF F NPN K CGCG+SF V
Sbjct 51 EDIVFEDKGVKVVVDGKSLQFLDGTQLDFVKEGLNEGFKFTNPNVKDECGCGESFHV 107
> cel:Y39B6A.3 hypothetical protein; K13628 iron-sulfur cluster
assembly protein
Length=129
Score = 53.5 bits (127), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 21/56 (37%), Positives = 39/56 (69%), Gaps = 0/56 (0%)
Query 39 DEVVVVEGVEIRVAADAVMLLIGTQVDFVDEDVQTGFIFNNPNQKHSCGCGKSFMV 94
DE V +G+++ + A + L+G+++D+V + + + F+F NPN K +CGCG+SF +
Sbjct 74 DEEVEQDGIKVWIEPKAQLSLLGSEMDYVTDKLSSEFVFRNPNIKGTCGCGESFSI 129
> ath:AT1G10500 ATCPISCA (chloroplast-localized IscA-like protein);
structural molecule; K13628 iron-sulfur cluster assembly
protein
Length=180
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 22/62 (35%), Positives = 34/62 (54%), Gaps = 0/62 (0%)
Query 31 HRRKNWGEDEVVVVEGVEIRVAADAVMLLIGTQVDFVDEDVQTGFIFNNPNQKHSCGCGK 90
+R +D + +G I +++ L G Q+D+ D + GF F+NPN +CGCGK
Sbjct 115 NRANARPDDSTIEYQGFTIVCDPKSMLFLFGMQLDYSDALIGGGFSFSNPNATQTCGCGK 174
Query 91 SF 92
SF
Sbjct 175 SF 176
> tgo:TGME49_097930 HesB-like domain containing protein
Length=265
Score = 48.5 bits (114), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 21/50 (42%), Positives = 31/50 (62%), Gaps = 0/50 (0%)
Query 45 EGVEIRVAADAVMLLIGTQVDFVDEDVQTGFIFNNPNQKHSCGCGKSFMV 94
+G I V +++ +IGT++D+ D+ + GF NPN SCGCG SF V
Sbjct 194 DGFSIVVDPQSLLYVIGTKLDYADDLIGGGFRVINPNASRSCGCGMSFGV 243
> tpv:TP04_0636 hypothetical protein
Length=157
Score = 46.2 bits (108), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 18/38 (47%), Positives = 24/38 (63%), Gaps = 0/38 (0%)
Query 55 AVMLLIGTQVDFVDEDVQTGFIFNNPNQKHSCGCGKSF 92
A L+G+ VD+ +D+ GF F+NPN CGCG SF
Sbjct 118 AAFYLLGSYVDYSKDDLSEGFTFDNPNVTSKCGCGMSF 155
> eco:b1684 sufA, ECK1680, JW1674, ydiC; Fe-S cluster assembly
protein; K05997 Fe-S cluster assembly protein SufA
Length=122
Score = 45.4 bits (106), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 22/57 (38%), Positives = 35/57 (61%), Gaps = 0/57 (0%)
Query 38 EDEVVVVEGVEIRVAADAVMLLIGTQVDFVDEDVQTGFIFNNPNQKHSCGCGKSFMV 94
+D + +G ++ V A+ + GT+VDFV E + F F+NP ++ CGCG+SF V
Sbjct 66 DDLLFEHDGAKLFVPLQAMPFIDGTEVDFVREGLNQIFKFHNPKAQNECGCGESFGV 122
> hsa:122961 ISCA2, HBLD1, ISA2, c14_5557; iron-sulfur cluster
assembly 2 homolog (S. cerevisiae)
Length=154
Score = 45.1 bits (105), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 20/58 (34%), Positives = 32/58 (55%), Gaps = 1/58 (1%)
Query 38 EDEVVVVEGVEIRVAADAVMLLIGTQVDFVDEDVQTGF-IFNNPNQKHSCGCGKSFMV 94
+D V G + V +D++ + G QVDF E +++ F + NNP + C CG SF +
Sbjct 95 DDRVFEQGGARVVVDSDSLAFVKGAQVDFSQELIRSSFQVLNNPQAQQGCSCGSSFSI 152
> mmu:74316 Isca2, 0710001C05Rik, 5730594E03Rik, Hbld1; iron-sulfur
cluster assembly 2 homolog (S. cerevisiae)
Length=154
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 21/58 (36%), Positives = 32/58 (55%), Gaps = 1/58 (1%)
Query 38 EDEVVVVEGVEIRVAADAVMLLIGTQVDFVDEDVQTGF-IFNNPNQKHSCGCGKSFMV 94
+D V G + V +D++ + G QVDF E +++ F + NNP + C CG SF V
Sbjct 95 DDRVFEQGGARVVVDSDSLAFVKGAQVDFSQELIRSSFQVLNNPQAQQGCSCGSSFSV 152
> dre:100150438 similar to Iron-sulfur cluster assembly 2 homolog,
mitochondrial precursor (HESB-like domain-containing protein
1)
Length=155
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 22/58 (37%), Positives = 31/58 (53%), Gaps = 1/58 (1%)
Query 38 EDEVVVVEGVEIRVAADAVMLLIGTQVDFVDEDVQTGF-IFNNPNQKHSCGCGKSFMV 94
+D V GV I V D++ + G+ VDF E ++ F + NP +H C CG SF V
Sbjct 96 DDRVFEKNGVGIVVDQDSLEFVKGSTVDFSQELIRASFQVLKNPQAEHGCSCGSSFSV 153
> pfa:PFC1005c iron-sulfur cluster assembly accessory protein,
putative
Length=404
Score = 42.4 bits (98), Expect = 4e-04, Method: Composition-based stats.
Identities = 20/57 (35%), Positives = 31/57 (54%), Gaps = 2/57 (3%)
Query 38 EDEVVVVEGVEIRVAAD--AVMLLIGTQVDFVDEDVQTGFIFNNPNQKHSCGCGKSF 92
EDE++ + I + D ++ +I T +D+ +D+ FIF NPN C CG SF
Sbjct 342 EDEIIYDDQNNILLVIDKNCILYVINTTLDYYKDDLTEKFIFKNPNITSICPCGTSF 398
> eco:b0156 erpA, ECK0155, JW0152, yadR; iron-sulfur cluster insertion
protein
Length=114
Score = 41.2 bits (95), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 20/56 (35%), Positives = 31/56 (55%), Gaps = 0/56 (0%)
Query 39 DEVVVVEGVEIRVAADAVMLLIGTQVDFVDEDVQTGFIFNNPNQKHSCGCGKSFMV 94
D + +GV + V ++ L+G VD+ + + FI NPN K +CGCG SF +
Sbjct 59 DMTIEKQGVGLVVDPMSLQYLVGGSVDYTEGLEGSRFIVTNPNAKSTCGCGSSFSI 114
> bbo:BBOV_III007290 17.m07640; HesB-like domain containing protein
Length=200
Score = 41.2 bits (95), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 15/38 (39%), Positives = 24/38 (63%), Gaps = 0/38 (0%)
Query 55 AVMLLIGTQVDFVDEDVQTGFIFNNPNQKHSCGCGKSF 92
A ++G +D+ ++ GF+F+NPN CGCG+SF
Sbjct 161 AAFHMLGCHIDYEISPLEEGFVFSNPNVTSKCGCGQSF 198
> pfa:PFE1135w iron-sulfur assembly protein, putative
Length=177
Score = 39.7 bits (91), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 20/66 (30%), Positives = 37/66 (56%), Gaps = 4/66 (6%)
Query 33 RKNWGEDEVVVVEGVEIR----VAADAVMLLIGTQVDFVDEDVQTGFIFNNPNQKHSCGC 88
+KN E++ + + E++ + + +V+ + +D+ ++ + GF F NPN CGC
Sbjct 112 KKNEIEEDDYIQQFDELKFILSIDSTSVIYIYNNTLDYSNDLISGGFKFINPNATKKCGC 171
Query 89 GKSFMV 94
GKSF V
Sbjct 172 GKSFNV 177
> cel:Y54G11A.9 hypothetical protein
Length=134
Score = 37.7 bits (86), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 20/48 (41%), Positives = 28/48 (58%), Gaps = 1/48 (2%)
Query 46 GVEIRVAADAVMLLIGTQVDFVDEDVQTGF-IFNNPNQKHSCGCGKSF 92
G EI V ++ L G VDFV++ +++ F I NNP + C CG SF
Sbjct 82 GAEIVVDELSLGFLKGATVDFVEDLMKSSFRIVNNPIAEKGCSCGSSF 129
> ath:AT2G16710 hesB-like domain-containing protein
Length=110
Score = 33.5 bits (75), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 16/35 (45%), Positives = 25/35 (71%), Gaps = 0/35 (0%)
Query 39 DEVVVVEGVEIRVAADAVMLLIGTQVDFVDEDVQT 73
DE+V +GV I V A+M +IGT++DFVD+ ++
Sbjct 71 DELVEEKGVRILVEPKALMHVIGTKMDFVDDKLRV 105
> ath:AT5G03905 hesB-like domain-containing protein
Length=158
Score = 31.6 bits (70), Expect = 0.58, Method: Compositional matrix adjust.
Identities = 18/58 (31%), Positives = 30/58 (51%), Gaps = 1/58 (1%)
Query 38 EDEVVVVEGVEIRVAADAVMLLIGTQVDFVDEDVQTGFIFN-NPNQKHSCGCGKSFMV 94
+D V GV++ V + L+ G +D+ +E ++ F+ NP+ C C SFMV
Sbjct 99 DDRVFEKNGVKLVVDNVSYDLVKGATIDYEEELIRAAFVVAVNPSAVGGCSCKSSFMV 156
> tpv:TP04_0597 hypothetical protein
Length=90
Score = 30.8 bits (68), Expect = 0.98, Method: Compositional matrix adjust.
Identities = 12/41 (29%), Positives = 22/41 (53%), Gaps = 0/41 (0%)
Query 54 DAVMLLIGTQVDFVDEDVQTGFIFNNPNQKHSCGCGKSFMV 94
+++ L+ +DF +E + + F + PN C CG SF +
Sbjct 49 ESIELIKSCTLDFQEELIGSKFTLDIPNSTRKCSCGNSFEI 89
> sce:YPR141C KAR3, OSR11; Kar3p; K10405 kinesin family member
C1
Length=729
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 14/35 (40%), Positives = 22/35 (62%), Gaps = 1/35 (2%)
Query 2 ERQPR-PHDGSATEASATPSHASGPLGLSGHRRKN 35
E PR P G +T+ +TPS + L ++GH+R+N
Sbjct 2 ESLPRTPTKGRSTQHLSTPSPKNDILAMNGHKRRN 36
> sce:YPR067W ISA2; Isa2p
Length=185
Score = 30.8 bits (68), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 13/42 (30%), Positives = 24/42 (57%), Gaps = 0/42 (0%)
Query 51 VAADAVMLLIGTQVDFVDEDVQTGFIFNNPNQKHSCGCGKSF 92
+ + ++ +L T + + +E + + F N + K SCGCG SF
Sbjct 140 IDSKSLNILNNTTLTYTNELIGSSFKIINGSLKSSCGCGSSF 181
> tgo:TGME49_118170 iron-sulfur cluster assembly accessory domain
containing protein
Length=274
Score = 28.5 bits (62), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 15/41 (36%), Positives = 22/41 (53%), Gaps = 1/41 (2%)
Query 55 AVMLLIGTQVDFVDEDVQTGF-IFNNPNQKHSCGCGKSFMV 94
+V LL + VD+ D + F I NN ++ C CG SF +
Sbjct 230 SVPLLENSVVDYSDTLAASEFRIVNNEQAENVCSCGHSFAI 270
> hsa:51564 HDAC7, DKFZp586J0917, FLJ99588, HD7A, HDAC7A; histone
deacetylase 7 (EC:3.5.1.98); K11408 histone deacetylase
7 [EC:3.5.1.98]
Length=954
Score = 28.1 bits (61), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 15/65 (23%), Positives = 24/65 (36%), Gaps = 0/65 (0%)
Query 25 PLGLSGHRRKNWGEDEVVVVEGVEIRVAADAVMLLIGTQVDFVDEDVQTGFIFNNPNQKH 84
PL GHR + + + A +L ++ TG I+++ KH
Sbjct 474 PLAQGGHRPLSRAQSSPAAPASLSAPEPASQARVLSSSETPARTLPFTTGLIYDSVMLKH 533
Query 85 SCGCG 89
C CG
Sbjct 534 QCSCG 538
> cel:F54D5.14 hypothetical protein
Length=1130
Score = 28.1 bits (61), Expect = 7.3, Method: Composition-based stats.
Identities = 23/76 (30%), Positives = 34/76 (44%), Gaps = 3/76 (3%)
Query 9 DGSATEASATPSHASGPLGLSGHRRKNWGEDEVVVVEGVEIRVAAD--AVMLLIGTQVDF 66
DGS A+ S G GH R +G D+ V EG R+ D + + + TQ D
Sbjct 626 DGSQAYANGPNSQYRFYSGRGGHARGTFGNDQGDVDEGALARLIEDTKSEAMRLETQ-DL 684
Query 67 VDEDVQTGFIFNNPNQ 82
+D + I+N +Q
Sbjct 685 RKQDHELKVIYNERDQ 700
> ath:AT3G14790 RHM3; RHM3 (RHAMNOSE BIOSYNTHESIS 3); UDP-L-rhamnose
synthase/ catalytic (EC:4.2.1.76); K12450 UDP-glucose
4,6-dehydratase [EC:4.2.1.76]
Length=664
Score = 28.1 bits (61), Expect = 8.2, Method: Composition-based stats.
Identities = 21/77 (27%), Positives = 31/77 (40%), Gaps = 3/77 (3%)
Query 16 SATPSHASGPLGLSGHRRKNWGEDEVVVVEGVEIRVAADAVMLLIGTQVDFVDEDVQTGF 75
S PSH GL+G +W E E + + VA + + + D + + TG
Sbjct 425 SIKPSHVFNAAGLTGRPNVDWCESH--KTETIRVNVAGTLTLADVCRENDLLMMNFATGC 482
Query 76 IFNNPNQKHSCGCGKSF 92
IF + H G G F
Sbjct 483 IFEY-DAAHPEGSGIGF 498
Lambda K H
0.315 0.133 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2069995292
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40