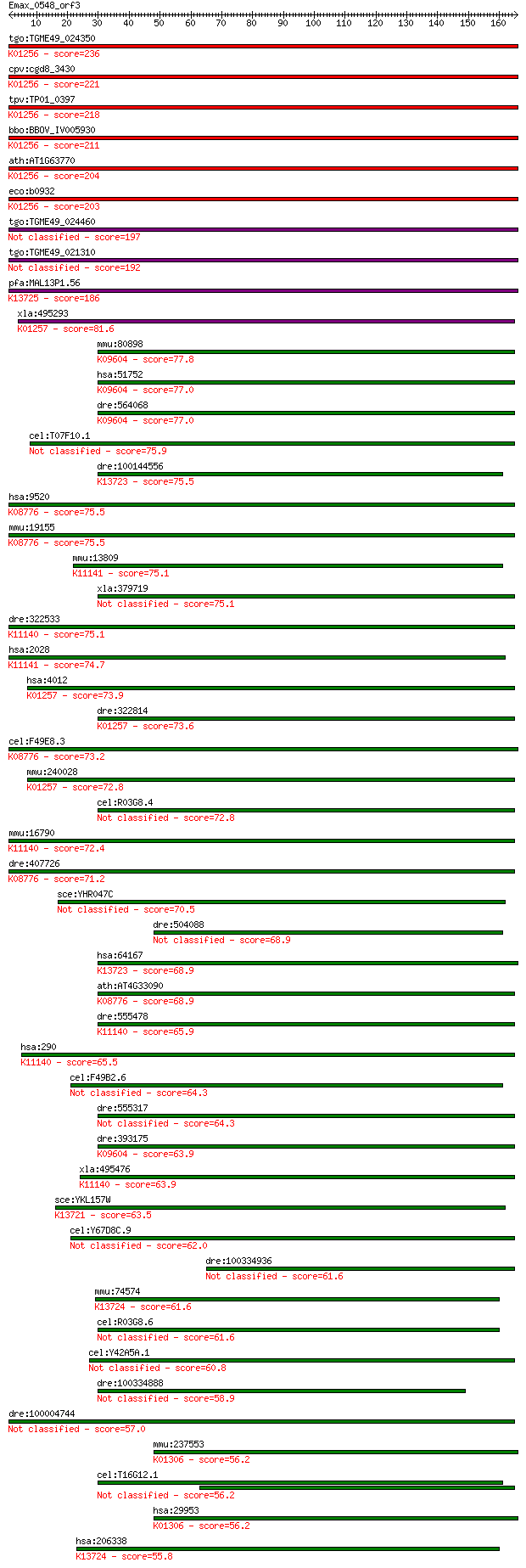
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0548_orf3
Length=165
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_024350 aminopeptidase N, putative (EC:3.4.11.2); K0... 236 3e-62
cpv:cgd8_3430 zincin/aminopeptidase N like metalloprotease ; K... 221 7e-58
tpv:TP01_0397 alpha-aminoacylpeptide hydrolase (EC:3.4.11.2); ... 218 5e-57
bbo:BBOV_IV005930 23.m06519; aminopeptidase (EC:3.4.11.2); K01... 211 7e-55
ath:AT1G63770 peptidase M1 family protein; K01256 aminopeptida... 204 1e-52
eco:b0932 pepN, ECK0923, JW0915; aminopeptidase N (EC:3.4.11.2... 203 2e-52
tgo:TGME49_024460 aminopeptidase N, putative (EC:3.4.11.2) 197 9e-51
tgo:TGME49_021310 aminopeptidase N, putative (EC:3.4.11.2) 192 5e-49
pfa:MAL13P1.56 M1-family aminopeptidase (EC:3.4.11.-); K13725 ... 186 2e-47
xla:495293 lnpep; leucyl/cystinyl aminopeptidase (EC:3.4.11.3)... 81.6 1e-15
mmu:80898 Erap1, Arts1, ERAAP, PILSA, PILSAP; endoplasmic reti... 77.8 2e-14
hsa:51752 ERAP1, A-LAP, ALAP, APPILS, ARTS-1, ARTS1, ERAAP, ER... 77.0 2e-14
dre:564068 endoplasmic reticulum aminopeptidase 1-like; K09604... 77.0 3e-14
cel:T07F10.1 hypothetical protein 75.9 6e-14
dre:100144556 zgc:172163; K13723 endoplasmic reticulum aminope... 75.5 8e-14
hsa:9520 NPEPPS, AAP-S, MP100, PSA; aminopeptidase puromycin s... 75.5 8e-14
mmu:19155 Npepps, AAP-S, MGC102199, MP100, Psa, R74825, goku; ... 75.5 8e-14
mmu:13809 Enpep, 6030431M22Rik, APA, Bp-1/6C3, Ly-51, Ly51; gl... 75.1 9e-14
xla:379719 hypothetical protein MGC69084 75.1 9e-14
dre:322533 anpep, fb64c05, wu:fb64c05, zgc:136771; alanyl (mem... 75.1 1e-13
hsa:2028 ENPEP, APA, CD249, gp160; glutamyl aminopeptidase (am... 74.7 1e-13
hsa:4012 LNPEP, CAP, IRAP, P-LAP, PLAP; leucyl/cystinyl aminop... 73.9 2e-13
dre:322814 fb73h12, wu:fb73h12; zgc:66103 (EC:3.4.11.3); K0125... 73.6 3e-13
cel:F49E8.3 pam-1; Puromycin-sensitive AMinopeptidase family m... 73.2 4e-13
mmu:240028 Lnpep, 2010309L07Rik, 4732490P18Rik, CAP, IRAP, PLA... 72.8 5e-13
cel:R03G8.4 hypothetical protein 72.8 5e-13
mmu:16790 Anpep, Apn, Cd13, Lap-1, Lap1, P150; alanyl (membran... 72.4 6e-13
dre:407726 npepps, Psa, fb68d07, sb:cb848, wu:fb68d07; aminope... 71.2 1e-12
sce:YHR047C AAP1, AAP1'; Aap1p (EC:3.4.11.-) 70.5 2e-12
dre:504088 enpep, im:7152184, si:ch211-146m5.2; glutamyl amino... 68.9 6e-12
hsa:64167 ERAP2, FLJ23633, FLJ23701, FLJ23807, L-RAP, LRAP; en... 68.9 7e-12
ath:AT4G33090 APM1; APM1 (AMINOPEPTIDASE M1); aminopeptidase; ... 68.9 8e-12
dre:555478 aminopeptidase N-like; K11140 aminopeptidase N [EC:... 65.9 5e-11
hsa:290 ANPEP, APN, CD13, GP150, LAP1, P150, PEPN; alanyl (mem... 65.5 8e-11
cel:F49B2.6 hypothetical protein 64.3 2e-10
dre:555317 aminopeptidase N-like 64.3 2e-10
dre:393175 ARTS-1, MGC56194; zgc:56194; K09604 adipocyte-deriv... 63.9 2e-10
xla:495476 anpep; alanyl (membrane) aminopeptidase; K11140 ami... 63.9 2e-10
sce:YKL157W APE2, LAP1, YKL158W; Aminopeptidase yscII; may hav... 63.5 3e-10
cel:Y67D8C.9 hypothetical protein 62.0 8e-10
dre:100334936 aminopeptidase N-like 61.6 1e-09
mmu:74574 4833403I15Rik, Aqpep, Lvrn, MGC130603; RIKEN cDNA 48... 61.6 1e-09
cel:R03G8.6 hypothetical protein 61.6 1e-09
cel:Y42A5A.1 hypothetical protein 60.8 2e-09
dre:100334888 leucyl/cystinyl aminopeptidase-like 58.9 8e-09
dre:100004744 aminopeptidase N-like 57.0 3e-08
mmu:237553 Trhde, 9330155P21Rik, MGC40831; TRH-degrading enzym... 56.2 5e-08
cel:T16G12.1 hypothetical protein 56.2 5e-08
hsa:29953 TRHDE, FLJ22381, PAP-II, PGPEP2, TRH-DE; thyrotropin... 56.2 5e-08
hsa:206338 AQPEP, APQ, FLJ90650, LVRN, MGC125378, MGC125379; l... 55.8 6e-08
> tgo:TGME49_024350 aminopeptidase N, putative (EC:3.4.11.2);
K01256 aminopeptidase N [EC:3.4.11.2]
Length=1419
Score = 236 bits (601), Expect = 3e-62, Method: Compositional matrix adjust.
Identities = 106/165 (64%), Positives = 125/165 (75%), Gaps = 0/165 (0%)
Query 1 SNGNKTEEGPVQDSPDRHFVVFTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVY 60
SNGN E G V+ RHF VF DP KP YLFA++AGD +I +FTT SG+ V ++++
Sbjct 693 SNGNMVESGKVEGEKGRHFAVFEDPFQKPCYLFALVAGDLKSISQSFTTMSGRNVKVSIF 752
Query 61 SEPEQMHKLNWAMHSVKVSMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCS 120
SEPE KL WA+ SV SMKW+E+ FGREYDLD FNV C DFN GAMENKGLNIFN +
Sbjct 753 SEPEDSSKLTWALESVLKSMKWDEERFGREYDLDVFNVVCAKDFNMGAMENKGLNIFNAA 812
Query 121 LLLASMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTLK 165
LLLA T+TD +++R+L VVGHEYFHNWTGNRVT RDWFQLTLK
Sbjct 813 LLLADPSTTTDAEYQRILNVVGHEYFHNWTGNRVTCRDWFQLTLK 857
> cpv:cgd8_3430 zincin/aminopeptidase N like metalloprotease ;
K01256 aminopeptidase N [EC:3.4.11.2]
Length=936
Score = 221 bits (564), Expect = 7e-58, Method: Compositional matrix adjust.
Identities = 102/165 (61%), Positives = 122/165 (73%), Gaps = 0/165 (0%)
Query 1 SNGNKTEEGPVQDSPDRHFVVFTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVY 60
SNGN E+G VQ S +RHF +F DP PKP YLFA++AG + D F TKSGK V L VY
Sbjct 194 SNGNLLEKGDVQGSENRHFAIFVDPFPKPCYLFAVVAGVLGRLEDKFITKSGKTVRLFVY 253
Query 61 SEPEQMHKLNWAMHSVKVSMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCS 120
SEP+ + +L AM S+K++MKW+ED FG EYDL+ FN+ V FN GAMENK LNIFNCS
Sbjct 254 SEPKYVDRLRLAMESLKLAMKWDEDRFGLEYDLEIFNIVAVESFNFGAMENKSLNIFNCS 313
Query 121 LLLASMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTLK 165
LLAS + D F +L++VGHEYFHN+TGNRVT RDWFQLTLK
Sbjct 314 CLLASENITPDYFFTNILSIVGHEYFHNYTGNRVTCRDWFQLTLK 358
> tpv:TP01_0397 alpha-aminoacylpeptide hydrolase (EC:3.4.11.2);
K01256 aminopeptidase N [EC:3.4.11.2]
Length=1020
Score = 218 bits (556), Expect = 5e-57, Method: Compositional matrix adjust.
Identities = 103/165 (62%), Positives = 125/165 (75%), Gaps = 3/165 (1%)
Query 1 SNGNKTEEGPVQDSPDRHFVVFTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVY 60
SNGN+ + G D RHF F DP PKP YLFA++AG+ A+I TF T SG+ V + +
Sbjct 317 SNGNRVDSG---DLGTRHFAEFVDPFPKPCYLFALVAGNLASISTTFKTMSGRNVLVQLS 373
Query 61 SEPEQMHKLNWAMHSVKVSMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCS 120
SEPE + KL WA+ SV +MKW+E+ +GREYDLD F+V V DFN GAMENKGLNIFN +
Sbjct 374 SEPEDVGKLQWALESVVKAMKWDEEKYGREYDLDEFHVFAVRDFNFGAMENKGLNIFNTA 433
Query 121 LLLASMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTLK 165
LLLA + T+TD +F R+L+VVGHEYFHNWTGNRVT RDWFQLTLK
Sbjct 434 LLLADVNTTTDAEFVRILSVVGHEYFHNWTGNRVTCRDWFQLTLK 478
> bbo:BBOV_IV005930 23.m06519; aminopeptidase (EC:3.4.11.2); K01256
aminopeptidase N [EC:3.4.11.2]
Length=846
Score = 211 bits (538), Expect = 7e-55, Method: Compositional matrix adjust.
Identities = 100/165 (60%), Positives = 121/165 (73%), Gaps = 3/165 (1%)
Query 1 SNGNKTEEGPVQDSPDRHFVVFTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVY 60
SNGNK + G + F F DP PKPSYLFA++AG+ +I TF T SG++V + V
Sbjct 195 SNGNKVDSGI---DGSKIFAEFVDPFPKPSYLFALVAGNLKSIKKTFRTMSGRDVLVEVS 251
Query 61 SEPEQMHKLNWAMHSVKVSMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCS 120
SEPE KL WA+ SV +MKW+E+ +GREYDLD F+V C FN GAMENKGLNIFN S
Sbjct 252 SEPEDATKLEWALESVLKAMKWDEESYGREYDLDEFHVVCTRAFNFGAMENKGLNIFNSS 311
Query 121 LLLASMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTLK 165
LLLA + T+TD +F +++VVGHEYFHNWTGNRVT RDWFQLTLK
Sbjct 312 LLLADVNTTTDSEFNTIMSVVGHEYFHNWTGNRVTCRDWFQLTLK 356
> ath:AT1G63770 peptidase M1 family protein; K01256 aminopeptidase
N [EC:3.4.11.2]
Length=987
Score = 204 bits (518), Expect = 1e-52, Method: Compositional matrix adjust.
Identities = 93/165 (56%), Positives = 122/165 (73%), Gaps = 2/165 (1%)
Query 1 SNGNKTEEGPVQDSPDRHFVVFTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVY 60
SNGN +G ++ RH+ ++ DP KP YLFA++AG + DTFTT+SG++V+L ++
Sbjct 253 SNGNLISQGDIEGG--RHYALWEDPFKKPCYLFALVAGQLVSRDDTFTTRSGRQVSLKIW 310
Query 61 SEPEQMHKLNWAMHSVKVSMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCS 120
+ E + K AM+S+K +MKW+ED FG EYDLD FN+ V DFN GAMENK LNIFN
Sbjct 311 TPAEDLPKTAHAMYSLKAAMKWDEDVFGLEYDLDLFNIVAVPDFNMGAMENKSLNIFNSK 370
Query 121 LLLASMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTLK 165
L+LAS +T+TD D+ +L V+GHEYFHNWTGNRVT RDWFQL+LK
Sbjct 371 LVLASPETATDADYAAILGVIGHEYFHNWTGNRVTCRDWFQLSLK 415
> eco:b0932 pepN, ECK0923, JW0915; aminopeptidase N (EC:3.4.11.2);
K01256 aminopeptidase N [EC:3.4.11.2]
Length=870
Score = 203 bits (516), Expect = 2e-52, Method: Compositional matrix adjust.
Identities = 94/165 (56%), Positives = 121/165 (73%), Gaps = 2/165 (1%)
Query 1 SNGNKTEEGPVQDSPDRHFVVFTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVY 60
SNGN+ +G +++ RH+V + DP PKP YLFA++AGDF + DTFTT+SG+EVAL +Y
Sbjct 157 SNGNRVAQGELENG--RHWVQWQDPFPKPCYLFALVAGDFDVLRDTFTTRSGREVALELY 214
Query 61 SEPEQMHKLNWAMHSVKVSMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCS 120
+ + + WAM S+K SMKW+E+ FG EYDLD + + V FN GAMENKGLNIFN
Sbjct 215 VDRGNLDRAPWAMTSLKNSMKWDEERFGLEYDLDIYMIVAVDFFNMGAMENKGLNIFNSK 274
Query 121 LLLASMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTLK 165
+LA T+TD D+ + V+GHEYFHNWTGNRVT RDWFQL+LK
Sbjct 275 YVLARTDTATDKDYLDIERVIGHEYFHNWTGNRVTCRDWFQLSLK 319
> tgo:TGME49_024460 aminopeptidase N, putative (EC:3.4.11.2)
Length=970
Score = 197 bits (502), Expect = 9e-51, Method: Compositional matrix adjust.
Identities = 91/165 (55%), Positives = 116/165 (70%), Gaps = 2/165 (1%)
Query 1 SNGNKTEEGPVQDSPDRHFVVFTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVY 60
SNG+K G + +RHF F DP PKPSYLFA++AGDFA++ F T SG+ V + +Y
Sbjct 236 SNGDKVLSGYAGE--NRHFAKFVDPFPKPSYLFALVAGDFASVSGEFVTMSGRSVTVTIY 293
Query 61 SEPEQMHKLNWAMHSVKVSMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCS 120
++ Q ++L WA+ S+ +M+W+E+ FGREY F V CV FN GAMEN LNIF CS
Sbjct 294 AQHHQRNQLQWALRSLLRAMRWDEETFGREYQYSEFRVLCVEVFNPGAMENTSLNIFTCS 353
Query 121 LLLASMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTLK 165
LLLA + +TD D ++ VV HEYFHNWTGNRVTV+DWFQLTLK
Sbjct 354 LLLADPKLTTDADHRLIVDVVSHEYFHNWTGNRVTVQDWFQLTLK 398
> tgo:TGME49_021310 aminopeptidase N, putative (EC:3.4.11.2)
Length=966
Score = 192 bits (487), Expect = 5e-49, Method: Compositional matrix adjust.
Identities = 87/165 (52%), Positives = 118/165 (71%), Gaps = 0/165 (0%)
Query 1 SNGNKTEEGPVQDSPDRHFVVFTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVY 60
SNG E G P++HF VF+DPH KPSYLFA++AG ++ + + V ++V+
Sbjct 204 SNGELVESGDDPTDPEKHFSVFSDPHKKPSYLFALVAGKLHSVGHDYEKRDKSLVKVSVW 263
Query 61 SEPEQMHKLNWAMHSVKVSMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCS 120
S PE + KL+WA+ S+ +MK +E FGR+YD + F++ CV+ FNAGAMENKGLNIFNC
Sbjct 264 STPENVAKLSWALQSIIRAMKGDEILFGRDYDSNVFHIVCVNGFNAGAMENKGLNIFNCD 323
Query 121 LLLASMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTLK 165
LLA T+TD+++ +L VV HEYFHNW+GNRVT+RDW +LTLK
Sbjct 324 SLLADPTTTTDEEYRGILRVVAHEYFHNWSGNRVTLRDWTELTLK 368
> pfa:MAL13P1.56 M1-family aminopeptidase (EC:3.4.11.-); K13725
M1-family aminopeptidase [EC:3.4.11.-]
Length=1085
Score = 186 bits (473), Expect = 2e-47, Method: Composition-based stats.
Identities = 94/166 (56%), Positives = 117/166 (70%), Gaps = 3/166 (1%)
Query 1 SNGNKTEEGPVQDSPDRHFVVFTDPHPKPSYLFAILAGDFAAIVDTFTTK-SGKEVALAV 59
SNG+K E + RH F DPH KP YLFA++AGD + T+ TK + K+V L V
Sbjct 355 SNGDKVNEFEIPGG--RHGARFNDPHLKPCYLFAVVAGDLKHLSATYITKYTKKKVELYV 412
Query 60 YSEPEQMHKLNWAMHSVKVSMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNC 119
+SE + + KL WA+ +K SM ++ED+FG EYDL N+ VSDFN GAMENKGLNIFN
Sbjct 413 FSEEKYVSKLQWALECLKKSMAFDEDYFGLEYDLSRLNLVAVSDFNVGAMENKGLNIFNA 472
Query 120 SLLLASMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTLK 165
+ LLAS + S D + R+L VVGHEYFHN+TGNRVT+RDWFQLTLK
Sbjct 473 NSLLASKKNSIDFSYARILTVVGHEYFHNYTGNRVTLRDWFQLTLK 518
> xla:495293 lnpep; leucyl/cystinyl aminopeptidase (EC:3.4.11.3);
K01257 cystinyl aminopeptidase [EC:3.4.11.3]
Length=1024
Score = 81.6 bits (200), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 48/161 (29%), Positives = 83/161 (51%), Gaps = 12/161 (7%)
Query 4 NKTEEGPVQDSPDRHFVVFTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEP 63
+KT +G +QD ++ +YL A + GD TT+ + ++VY+ P
Sbjct 335 SKTSDGLLQDE-------YSTSVRMSTYLVAFIVGDIKN-----TTQKTNDTLVSVYAVP 382
Query 64 EQMHKLNWAMHSVKVSMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLL 123
++ ++ +A+ S + + +++G EY L+ ++ + DF AGAMEN GL F + LL
Sbjct 383 DKTDQVKYALDSTVKLLDFYSNYYGIEYPLEKLDLVAIPDFQAGAMENWGLITFRETTLL 442
Query 124 ASMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTL 164
+S+ +D + + V+ HE H W GN VT+ W L L
Sbjct 443 YKENSSSIEDKQSITTVIAHELAHQWFGNLVTMEWWNDLWL 483
> mmu:80898 Erap1, Arts1, ERAAP, PILSA, PILSAP; endoplasmic reticulum
aminopeptidase 1; K09604 adipocyte-derived leucine aminopeptidase
[EC:3.4.11.-]
Length=930
Score = 77.8 bits (190), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 45/135 (33%), Positives = 76/135 (56%), Gaps = 4/135 (2%)
Query 30 SYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQMHKLNWAMHSVKVSMKWEEDHFGR 89
+YL A + DF ++ + TKSG V ++VY+ P+++++ ++A+ + +++ ED+F
Sbjct 233 TYLVAFIISDFKSV--SKMTKSG--VKVSVYAVPDKINQADYALDAAVTLLEFYEDYFNI 288
Query 90 EYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASMQTSTDDDFERVLAVVGHEYFHNW 149
Y L ++A + DF +GAMEN GL + S LL + S+ + +V HE H W
Sbjct 289 PYPLPKQDLAAIPDFQSGAMENWGLTTYRESSLLYDKEKSSASSKLGITMIVSHELAHQW 348
Query 150 TGNRVTVRDWFQLTL 164
GN VT+ W L L
Sbjct 349 FGNLVTMEWWNDLWL 363
> hsa:51752 ERAP1, A-LAP, ALAP, APPILS, ARTS-1, ARTS1, ERAAP,
ERAAP1, KIAA0525, PILS-AP, PILSAP; endoplasmic reticulum aminopeptidase
1; K09604 adipocyte-derived leucine aminopeptidase
[EC:3.4.11.-]
Length=941
Score = 77.0 bits (188), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 45/135 (33%), Positives = 75/135 (55%), Gaps = 4/135 (2%)
Query 30 SYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQMHKLNWAMHSVKVSMKWEEDHFGR 89
+YL A + DF ++ + TKSG V ++VY+ P+++++ ++A+ + +++ ED+F
Sbjct 244 TYLVAFIISDFESV--SKITKSG--VKVSVYAVPDKINQADYALDAAVTLLEFYEDYFSI 299
Query 90 EYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASMQTSTDDDFERVLAVVGHEYFHNW 149
Y L ++A + DF +GAMEN GL + S LL + S+ + V HE H W
Sbjct 300 PYPLPKQDLAAIPDFQSGAMENWGLTTYRESALLFDAEKSSASSKLGITMTVAHELAHQW 359
Query 150 TGNRVTVRDWFQLTL 164
GN VT+ W L L
Sbjct 360 FGNLVTMEWWNDLWL 374
> dre:564068 endoplasmic reticulum aminopeptidase 1-like; K09604
adipocyte-derived leucine aminopeptidase [EC:3.4.11.-]
Length=908
Score = 77.0 bits (188), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 44/135 (32%), Positives = 74/135 (54%), Gaps = 4/135 (2%)
Query 30 SYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQMHKLNWAMHSVKVSMKWEEDHFGR 89
+YL A + DF +I + KS V ++VY+ PE++ + +A+ + + + +++F
Sbjct 208 TYLVAFIICDFHSI----SKKSQHGVEISVYTVPEKISQAEYALDTAVTMLDFYDEYFDI 263
Query 90 EYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASMQTSTDDDFERVLAVVGHEYFHNW 149
Y L ++A + DF +GAMEN GL+ + S LL + S+ D + V+ HE H W
Sbjct 264 PYPLPKHDLAAIPDFQSGAMENWGLSTYRESGLLFDPEKSSSSDKLGITKVIAHELAHQW 323
Query 150 TGNRVTVRDWFQLTL 164
GN VT++ W L L
Sbjct 324 FGNLVTMQWWNDLWL 338
> cel:T07F10.1 hypothetical protein
Length=988
Score = 75.9 bits (185), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 51/158 (32%), Positives = 78/158 (49%), Gaps = 5/158 (3%)
Query 8 EGPVQDSPDRHFVVFTDPHPK-PSYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQM 66
E V+D + P P+ SYL AI +F + TTKSG V V+S PE+
Sbjct 264 EDKVEDGQPGFIISTFKPTPRMSSYLLAIFISEFE--YNEATTKSG--VRFRVWSRPEEK 319
Query 67 HKLNWAMHSVKVSMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASM 126
+ +A+ + +++ E ++ + L ++ + DF+AGAMEN GL + S LL
Sbjct 320 NSTMYAVEAGVKCLEYYEKYYNISFPLPKQDMVALPDFSAGAMENWGLITYRESALLYDP 379
Query 127 QTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTL 164
+ + RV V+ HE H W GN VT++ W L L
Sbjct 380 RIYSGSQKRRVAVVIAHELAHQWFGNLVTLKWWNDLWL 417
> dre:100144556 zgc:172163; K13723 endoplasmic reticulum aminopeptidase
2 [EC:3.4.11.-]
Length=931
Score = 75.5 bits (184), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 43/131 (32%), Positives = 73/131 (55%), Gaps = 5/131 (3%)
Query 30 SYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQMHKLNWAMHSVKVSMKWEEDHFGR 89
SYL A + DF ++ + T +G + +++Y+ PE+ H+ ++A+ + +++ E +F
Sbjct 235 SYLLAFIVCDFKSV--SGLTATG--INISIYAVPEKWHQTHYALEAALRLLEFYEQYFNI 290
Query 90 EYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASMQTSTDDDFERVLAVVGHEYFHNW 149
Y L ++ + DF +GAMEN GL + + LL S+ D V V+GHE H W
Sbjct 291 LYPLPKLDLIAIPDFESGAMENWGLTTYRETSLLYDPDISSASDKLWVTMVIGHELAHQW 350
Query 150 TGNRVTVRDWF 160
GN VT+ DW+
Sbjct 351 FGNLVTM-DWW 360
> hsa:9520 NPEPPS, AAP-S, MP100, PSA; aminopeptidase puromycin
sensitive (EC:3.4.11.14); K08776 puromycin-sensitive aminopeptidase
[EC:3.4.11.-]
Length=919
Score = 75.5 bits (184), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 50/164 (30%), Positives = 77/164 (46%), Gaps = 4/164 (2%)
Query 1 SNGNKTEEGPVQDSPDRHFVVFTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVY 60
SN N + P D + V F +YL A + G++ D T+S V + VY
Sbjct 214 SNMNVIDRKPYPDDENLVEVKFARTPVMSTYLVAFVVGEY----DFVETRSKDGVCVRVY 269
Query 61 SEPEQMHKLNWAMHSVKVSMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCS 120
+ + + +A+ ++ + +D+F Y L ++ ++DF AGAMEN GL + +
Sbjct 270 TPVGKAEQGKFALEVAAKTLPFYKDYFNVPYPLPKIDLIAIADFAAGAMENWGLVTYRET 329
Query 121 LLLASMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTL 164
LL + S + V VVGHE H W GN VT+ W L L
Sbjct 330 ALLIDPKNSCSSSRQWVALVVGHELAHQWFGNLVTMEWWTHLWL 373
> mmu:19155 Npepps, AAP-S, MGC102199, MP100, Psa, R74825, goku;
aminopeptidase puromycin sensitive (EC:3.4.11.14); K08776
puromycin-sensitive aminopeptidase [EC:3.4.11.-]
Length=920
Score = 75.5 bits (184), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 50/164 (30%), Positives = 77/164 (46%), Gaps = 4/164 (2%)
Query 1 SNGNKTEEGPVQDSPDRHFVVFTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVY 60
SN N + P D + V F +YL A + G++ D T+S V + VY
Sbjct 215 SNMNVIDRKPYPDDENLVEVKFARTPVMSTYLVAFVVGEY----DFVETRSKDGVCVRVY 270
Query 61 SEPEQMHKLNWAMHSVKVSMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCS 120
+ + + +A+ ++ + +D+F Y L ++ ++DF AGAMEN GL + +
Sbjct 271 TPVGKAEQGKFALEVAAKTLPFYKDYFNVPYPLPKIDLIAIADFAAGAMENWGLVTYRET 330
Query 121 LLLASMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTL 164
LL + S + V VVGHE H W GN VT+ W L L
Sbjct 331 ALLIDPKNSCSSSRQWVALVVGHELAHQWFGNLVTMEWWTHLWL 374
> mmu:13809 Enpep, 6030431M22Rik, APA, Bp-1/6C3, Ly-51, Ly51;
glutamyl aminopeptidase (EC:3.4.11.7); K11141 glutamyl aminopeptidase
[EC:3.4.11.7]
Length=945
Score = 75.1 bits (183), Expect = 9e-14, Method: Compositional matrix adjust.
Identities = 48/139 (34%), Positives = 66/139 (47%), Gaps = 5/139 (3%)
Query 22 FTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQMHKLNWAMHSVKVSMK 81
F P +YL F AI ++SGK L VY +P Q +A + +
Sbjct 268 FVKSVPMSTYLVCFAVHRFTAI--ERKSRSGK--PLKVYVQPNQKETAEYAANITQAVFD 323
Query 82 WEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASMQTSTDDDFERVLAVV 141
+ ED+F EY L + + DF GAMEN GL + + LL S + +RV +VV
Sbjct 324 YFEDYFAMEYALPKLDKIAIPDFGTGAMENWGLVTYRETNLLYDPLLSASSNQQRVASVV 383
Query 142 GHEYFHNWTGNRVTVRDWF 160
HE H W GN VT+ DW+
Sbjct 384 AHELVHQWFGNTVTM-DWW 401
> xla:379719 hypothetical protein MGC69084
Length=997
Score = 75.1 bits (183), Expect = 9e-14, Method: Compositional matrix adjust.
Identities = 41/135 (30%), Positives = 71/135 (52%), Gaps = 5/135 (3%)
Query 30 SYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQMHKLNWAMHSVKVSMKWEEDHFGR 89
+YL A + GD TT+ + ++VY+ PE+ ++ +A+ S + + +++G
Sbjct 352 TYLVAFIVGDIKN-----TTRETNDTLVSVYTVPEKTDQVKYALDSAVKLLDFYSNYYGI 406
Query 90 EYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASMQTSTDDDFERVLAVVGHEYFHNW 149
+Y L+ ++ + DF A AMEN GL F + LL + +S+ D + + + HE H W
Sbjct 407 KYPLEKLDLVAIPDFQAAAMENWGLITFRETSLLYNEDSSSIKDKQTITIAIAHELTHQW 466
Query 150 TGNRVTVRDWFQLTL 164
GN VT+ W L L
Sbjct 467 FGNLVTMEWWNDLWL 481
> dre:322533 anpep, fb64c05, wu:fb64c05, zgc:136771; alanyl (membrane)
aminopeptidase (EC:3.4.11.2); K11140 aminopeptidase
N [EC:3.4.11.2]
Length=965
Score = 75.1 bits (183), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 49/167 (29%), Positives = 82/167 (49%), Gaps = 8/167 (4%)
Query 1 SNGNKTEEGPVQ-DSPDRHFVVFTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAV 59
SNG EE PV D F +YL A + +F T+ + ++ + +
Sbjct 247 SNGVVIEEIPVTVDGISLTKTTFAPTEKMSTYLLAFIVSEF-----TYIEQKLDDLQIRI 301
Query 60 YSEPEQM--HKLNWAMHSVKVSMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIF 117
++ E + ++ +A+ +++ E+++ Y L + + DFNAGAMEN GL +
Sbjct 302 FARKEAIDANQGEYALSVTGKILRFFEEYYNSSYPLPKSDQIALPDFNAGAMENWGLITY 361
Query 118 NCSLLLASMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTL 164
+ LL + S++ + ERV+ V+ HE H W GN VT+R W L L
Sbjct 362 RETALLYDEEMSSNGNKERVVTVIAHELAHQWFGNLVTIRWWNDLWL 408
> hsa:2028 ENPEP, APA, CD249, gp160; glutamyl aminopeptidase (aminopeptidase
A) (EC:3.4.11.7); K11141 glutamyl aminopeptidase
[EC:3.4.11.7]
Length=957
Score = 74.7 bits (182), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 52/161 (32%), Positives = 74/161 (45%), Gaps = 7/161 (4%)
Query 1 SNGNKTEEGPVQDSPDRHFVVFTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVY 60
SN +E V D R F P +YL F ++ + SGK L +Y
Sbjct 257 SNMPVAKEESVDDKWTR--TTFEKSVPMSTYLVCFAVHQFDSV--KRISNSGK--PLTIY 310
Query 61 SEPEQMHKLNWAMHSVKVSMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCS 120
+PEQ H +A + K + E++F Y L + + DF GAMEN GL + +
Sbjct 311 VQPEQKHTAEYAANITKSVFDYFEEYFAMNYSLPKLDKIAIPDFGTGAMENWGLITYRET 370
Query 121 LLLASMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQ 161
LL + S + +RV VV HE H W GN VT+ DW++
Sbjct 371 NLLYDPKESASSNQQRVATVVAHELVHQWFGNIVTM-DWWE 410
> hsa:4012 LNPEP, CAP, IRAP, P-LAP, PLAP; leucyl/cystinyl aminopeptidase
(EC:3.4.11.3); K01257 cystinyl aminopeptidase [EC:3.4.11.3]
Length=1025
Score = 73.9 bits (180), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 44/158 (27%), Positives = 82/158 (51%), Gaps = 12/158 (7%)
Query 7 EEGPVQDSPDRHFVVFTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQM 66
++G VQD F++ +YL A + G+ + ++ +++Y+ PE++
Sbjct 340 DDGLVQDE-------FSESVKMSTYLVAFIVGEMKNL-----SQDVNGTLVSIYAVPEKI 387
Query 67 HKLNWAMHSVKVSMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASM 126
++++A+ + +++ +++F +Y L ++ + DF AGAMEN GL F LL
Sbjct 388 GQVHYALETTVKLLEFFQNYFEIQYPLKKLDLVAIPDFEAGAMENWGLLTFREETLLYDS 447
Query 127 QTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTL 164
TS+ D + V ++ HE H W GN VT++ W L L
Sbjct 448 NTSSMADRKLVTKIIAHELAHQWFGNLVTMKWWNDLWL 485
> dre:322814 fb73h12, wu:fb73h12; zgc:66103 (EC:3.4.11.3); K01257
cystinyl aminopeptidase [EC:3.4.11.3]
Length=1003
Score = 73.6 bits (179), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 44/135 (32%), Positives = 74/135 (54%), Gaps = 6/135 (4%)
Query 30 SYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQMHKLNWAMHSVKVSMKWEEDHFGR 89
+YL A + +F++ +K+ + ++VY+ P++ ++++A+ + +K+ F
Sbjct 337 TYLVAFIVAEFSS-----HSKNVSKTTVSVYAVPDKKDQVHYALETACKLLKFYNTFFEI 391
Query 90 EYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASMQTSTDDDFERVLAVVGHEYFHNW 149
EY L ++ + DF AGAMEN GL F + LL Q+S D + V +V+ HE H W
Sbjct 392 EYPLSKLDLVAIPDFLAGAMENWGLITFRETTLLVGNQSSRFDK-QLVTSVIAHELAHQW 450
Query 150 TGNRVTVRDWFQLTL 164
GN VT+R W L L
Sbjct 451 FGNLVTMRWWNDLWL 465
> cel:F49E8.3 pam-1; Puromycin-sensitive AMinopeptidase family
member (pam-1); K08776 puromycin-sensitive aminopeptidase [EC:3.4.11.-]
Length=948
Score = 73.2 bits (178), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 54/165 (32%), Positives = 75/165 (45%), Gaps = 5/165 (3%)
Query 1 SNGNKTEEGPVQDSPDRHFVVFTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVY 60
SN N E P D R V F SYL A G+ I + TKSG V + VY
Sbjct 240 SNMNVISETPTADG-KRKAVTFATSPKMSSYLVAFAVGELEYI--SAQTKSG--VEMRVY 294
Query 61 SEPEQMHKLNWAMHSVKVSMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCS 120
+ P + + +++ + W + F +Y L ++ + DF+ GAMEN GL +
Sbjct 295 TVPGKKEQGQYSLDLSVKCIDWYNEWFDIKYPLPKCDLIAIPDFSMGAMENWGLVTYREI 354
Query 121 LLLASMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTLK 165
LL ++ RV VV HE H W GN VT++ W L LK
Sbjct 355 ALLVDPGVTSTRQKSRVALVVAHELAHLWFGNLVTMKWWTDLWLK 399
> mmu:240028 Lnpep, 2010309L07Rik, 4732490P18Rik, CAP, IRAP, PLAP,
gp160, vp165; leucyl/cystinyl aminopeptidase (EC:3.4.11.3);
K01257 cystinyl aminopeptidase [EC:3.4.11.3]
Length=1025
Score = 72.8 bits (177), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 46/158 (29%), Positives = 80/158 (50%), Gaps = 12/158 (7%)
Query 7 EEGPVQDSPDRHFVVFTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQM 66
EEG +QD F++ +YL A + G+ + ++ ++VY+ PE++
Sbjct 340 EEGLIQDE-------FSESVKMSTYLVAFIVGEMRNL-----SQDVNGTLVSVYAVPEKI 387
Query 67 HKLNWAMHSVKVSMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASM 126
+++ A+ + +++ + +F +Y L ++ + DF AGAMEN GL F LL
Sbjct 388 GQVHHALDTTIKLLEFYQTYFEIQYPLKKLDLVAIPDFEAGAMENWGLLTFREETLLYDN 447
Query 127 QTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTL 164
TS+ D + V ++ HE H W GN VT++ W L L
Sbjct 448 ATSSVADRKLVTKIIAHELAHQWFGNLVTMQWWNDLWL 485
> cel:R03G8.4 hypothetical protein
Length=786
Score = 72.8 bits (177), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 45/135 (33%), Positives = 70/135 (51%), Gaps = 7/135 (5%)
Query 30 SYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQMHKLNWAMHSVKVSMKWEEDHFGR 89
SY+ AI GD TK+G V + VYS+P + ++ A++ ++ ++ E FG
Sbjct 225 SYILAIFVGDVQ--FKEAVTKNG--VRIRVYSDPGHIDSVDHALNVSRIVLEGFEKQFGY 280
Query 90 EYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASMQTSTDDDFERVLAVVGHEYFHNW 149
Y++D ++ V +F GAMEN GL + L+ ++ D + VV HE H W
Sbjct 281 PYEMDKLDLIAVYNFRYGAMENWGLIVHQAYTLIENLMPGNTD---IISEVVAHEIAHQW 337
Query 150 TGNRVTVRDWFQLTL 164
GN VT++ W QL L
Sbjct 338 FGNLVTMKFWDQLWL 352
> mmu:16790 Anpep, Apn, Cd13, Lap-1, Lap1, P150; alanyl (membrane)
aminopeptidase (EC:3.4.11.2); K11140 aminopeptidase N [EC:3.4.11.2]
Length=966
Score = 72.4 bits (176), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 49/166 (29%), Positives = 77/166 (46%), Gaps = 6/166 (3%)
Query 1 SNGNKTEEGPVQDSPDRHFVVFTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVY 60
SN E P + P F +YL A + +F I ++ S V + ++
Sbjct 247 SNMLPKESKPYPEDPSCTMTEFHSTPKMSTYLLAYIVSEFKNI----SSVSANGVQIGIW 302
Query 61 SEPEQMH--KLNWAMHSVKVSMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFN 118
+ P + + ++A++ + + H+ Y L + + DFNAGAMEN GL +
Sbjct 303 ARPSAIDEGQGDYALNVTGPILNFFAQHYNTSYPLPKSDQIALPDFNAGAMENWGLVTYR 362
Query 119 CSLLLASMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTL 164
S L+ Q+S+ + ERV+ V+ HE H W GN VTV W L L
Sbjct 363 ESSLVFDSQSSSISNKERVVTVIAHELAHQWFGNLVTVAWWNDLWL 408
> dre:407726 npepps, Psa, fb68d07, sb:cb848, wu:fb68d07; aminopeptidase
puromycin sensitive (EC:3.4.11.-); K08776 puromycin-sensitive
aminopeptidase [EC:3.4.11.-]
Length=872
Score = 71.2 bits (173), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 48/164 (29%), Positives = 76/164 (46%), Gaps = 4/164 (2%)
Query 1 SNGNKTEEGPVQDSPDRHFVVFTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVY 60
SN N + P + V F +YL A + G++ D ++S V + VY
Sbjct 169 SNMNVVDRKPYAEDQSLVEVKFATTPIMSTYLVAFVIGEY----DFVESQSSDGVTVRVY 224
Query 61 SEPEQMHKLNWAMHSVKVSMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCS 120
+ + + +A+ ++ + +D+F Y L ++ ++DF AGAMEN GL + +
Sbjct 225 TPVGKAEQGKFALEVATKTLPFYKDYFSVPYPLPKIDLIAIADFAAGAMENWGLVTYRET 284
Query 121 LLLASMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTL 164
LL + S + V VVGHE H W GN VT+ W L L
Sbjct 285 ALLIDPKNSCASSRQWVALVVGHELAHQWFGNLVTMEWWTHLWL 328
> sce:YHR047C AAP1, AAP1'; Aap1p (EC:3.4.11.-)
Length=856
Score = 70.5 bits (171), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 43/145 (29%), Positives = 70/145 (48%), Gaps = 6/145 (4%)
Query 17 RHFVVFTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQMHKLNWAMHSV 76
+ + F +YL A + D + + + + VYS P +A +
Sbjct 179 KKYTTFNTTPKMSTYLVAFIVADL-----RYVESNNFRIPVRVYSTPGDEKFGQFAANLA 233
Query 77 KVSMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASMQTSTDDDFER 136
++++ ED F EY L ++ V +F+AGAMEN GL + LL ++ S+ D +R
Sbjct 234 ARTLRFFEDTFNIEYPLPKMDMVAVHEFSAGAMENWGLVTYRVIDLLLDIENSSLDRIQR 293
Query 137 VLAVVGHEYFHNWTGNRVTVRDWFQ 161
V V+ HE H W GN VT+ DW++
Sbjct 294 VAEVIQHELAHQWFGNLVTM-DWWE 317
> dre:504088 enpep, im:7152184, si:ch211-146m5.2; glutamyl aminopeptidase
(EC:3.4.11.7)
Length=951
Score = 68.9 bits (167), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 38/113 (33%), Positives = 62/113 (54%), Gaps = 3/113 (2%)
Query 48 TTKSGKEVALAVYSEPEQMHKLNWAMHSVKVSMKWEEDHFGREYDLDTFNVACVSDFNAG 107
T+K G + L +Y++P Q+ +A +V + E++F EY + + + DF G
Sbjct 291 TSKRG--IPLRIYAQPLQISTAAYAADVTQVIFDYFEEYFDMEYSIQKLDKIAIPDFGTG 348
Query 108 AMENKGLNIFNCSLLLASMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWF 160
AMEN GL + + LL + S+ + +RV +V+ HE H W GN VT+ DW+
Sbjct 349 AMENWGLITYRETNLLFDEKESSSVNKQRVASVIAHELVHQWFGNIVTM-DWW 400
> hsa:64167 ERAP2, FLJ23633, FLJ23701, FLJ23807, L-RAP, LRAP;
endoplasmic reticulum aminopeptidase 2 (EC:3.4.11.-); K13723
endoplasmic reticulum aminopeptidase 2 [EC:3.4.11.-]
Length=960
Score = 68.9 bits (167), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 43/136 (31%), Positives = 73/136 (53%), Gaps = 4/136 (2%)
Query 30 SYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQMHKLNWAMHSVKVSMKWEEDHFGR 89
+YL A + DF ++ + T SG V +++Y+ P++ ++ ++A+ + + + E +F
Sbjct 261 TYLVAYIVCDFHSL--SGFTSSG--VKVSIYASPDKRNQTHYALQASLKLLDFYEKYFDI 316
Query 90 EYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASMQTSTDDDFERVLAVVGHEYFHNW 149
Y L ++ + DF GAMEN GL + + LL +TS+ D V V+ HE H W
Sbjct 317 YYPLSKLDLIAIPDFAPGAMENWGLITYRETSLLFDPKTSSASDKLWVTRVIAHELAHQW 376
Query 150 TGNRVTVRDWFQLTLK 165
GN VT+ W + LK
Sbjct 377 FGNLVTMEWWNDIWLK 392
> ath:AT4G33090 APM1; APM1 (AMINOPEPTIDASE M1); aminopeptidase;
K08776 puromycin-sensitive aminopeptidase [EC:3.4.11.-]
Length=879
Score = 68.9 bits (167), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 45/135 (33%), Positives = 68/135 (50%), Gaps = 4/135 (2%)
Query 30 SYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQMHKLNWAMHSVKVSMKWEEDHFGR 89
+YL AI+ G F + D T G + + VY + + + +A+H ++ +++F
Sbjct 198 TYLVAIVVGLFDYVEDH--TSDG--IKVRVYCQVGKADQGKFALHVGAKTLDLFKEYFAV 253
Query 90 EYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASMQTSTDDDFERVLAVVGHEYFHNW 149
Y L ++ + DF AGAMEN GL + + LL Q S + +RV VV HE H W
Sbjct 254 PYPLPKMDMIAIPDFAAGAMENYGLVTYRETALLYDEQHSAASNKQRVATVVAHELAHQW 313
Query 150 TGNRVTVRDWFQLTL 164
GN VT+ W L L
Sbjct 314 FGNLVTMEWWTHLWL 328
> dre:555478 aminopeptidase N-like; K11140 aminopeptidase N [EC:3.4.11.2]
Length=960
Score = 65.9 bits (159), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 42/135 (31%), Positives = 70/135 (51%), Gaps = 2/135 (1%)
Query 30 SYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQMHKLNWAMHSVKVSMKWEEDHFGR 89
+YL A + DF+ I + K+G V + + + ++A+ + +++ E ++
Sbjct 269 TYLVAFVVSDFSYINNE--DKAGVLVRIWARKKAIDDGQGDYALSITQPILEFFESYYNT 326
Query 90 EYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASMQTSTDDDFERVLAVVGHEYFHNW 149
Y L + + DFN+GAMEN GL + + LL QTS + + +R+ VV HE H W
Sbjct 327 SYPLSKSDQVALPDFNSGAMENWGLVTYRETALLYDPQTSANGNKQRIATVVSHELAHMW 386
Query 150 TGNRVTVRDWFQLTL 164
GN VT++ W L L
Sbjct 387 FGNLVTLKWWNDLWL 401
> hsa:290 ANPEP, APN, CD13, GP150, LAP1, P150, PEPN; alanyl (membrane)
aminopeptidase (EC:3.4.11.2); K11140 aminopeptidase
N [EC:3.4.11.2]
Length=967
Score = 65.5 bits (158), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 44/162 (27%), Positives = 75/162 (46%), Gaps = 6/162 (3%)
Query 5 KTEEGPVQDSPDRHFVVFTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPE 64
K P+ + P+ + F +YL A + +F D ++ V + +++ P
Sbjct 252 KGPSTPLPEDPNWNVTEFHTTPKMSTYLLAFIVSEF----DYVEKQASNGVLIRIWARPS 307
Query 65 QMHKL--NWAMHSVKVSMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCSLL 122
+ ++A++ + + H+ Y L + + DFNAGAMEN GL + + L
Sbjct 308 AIAAGHGDYALNVTGPILNFFAGHYDTPYPLPKSDQIGLPDFNAGAMENWGLVTYRENSL 367
Query 123 LASMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTL 164
L +S+ + ERV+ V+ HE H W GN VT+ W L L
Sbjct 368 LFDPLSSSSSNKERVVTVIAHELAHQWFGNLVTIEWWNDLWL 409
> cel:F49B2.6 hypothetical protein
Length=1082
Score = 64.3 bits (155), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 41/142 (28%), Positives = 72/142 (50%), Gaps = 4/142 (2%)
Query 21 VFTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQM--HKLNWAMHSVKV 78
VF +YL A+ D + T + K + + +Y+ P+ M + + + +
Sbjct 377 VFEKSVKMSTYLLAVAVLDGYGYIKRLTRNTQKAIEVRLYA-PQDMLTGQSEFGLDTTIR 435
Query 79 SMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASMQTSTDDDFERVL 138
++++ ED+F Y LD ++ + DF+ GAMEN GL F S LL + + ++ E +
Sbjct 436 ALEFFEDYFNISYPLDKIDLLALDDFSEGAMENWGLVTFRDSALLFNERKASVVAKEHIA 495
Query 139 AVVGHEYFHNWTGNRVTVRDWF 160
++ HE H W GN VT+ DW+
Sbjct 496 LIICHEIAHQWFGNLVTM-DWW 516
> dre:555317 aminopeptidase N-like
Length=956
Score = 64.3 bits (155), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 39/137 (28%), Positives = 69/137 (50%), Gaps = 5/137 (3%)
Query 30 SYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQMH--KLNWAMHSVKVSMKWEEDHF 87
+YL A + +F +I ++ + ++ E + + ++A++ +K+ E ++
Sbjct 272 TYLLAFVISEFPSIQSPL---GANKILVRIWGRREAIENGEGDYALNVTFPVLKYLESYY 328
Query 88 GREYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASMQTSTDDDFERVLAVVGHEYFH 147
Y L + + DF+AGAMEN GL + + L + S+ +D E V+ V+ HE H
Sbjct 329 NTTYPLSKSDQIALPDFSAGAMENWGLVTYRETFLFYEPKVSSHEDKEGVITVISHELAH 388
Query 148 NWTGNRVTVRDWFQLTL 164
W GN VT+R W L L
Sbjct 389 MWFGNLVTMRWWNDLWL 405
> dre:393175 ARTS-1, MGC56194; zgc:56194; K09604 adipocyte-derived
leucine aminopeptidase [EC:3.4.11.-]
Length=963
Score = 63.9 bits (154), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 46/155 (29%), Positives = 73/155 (47%), Gaps = 24/155 (15%)
Query 30 SYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQMHKLNWAMHSVKVSMKWEEDHFGR 89
+YL A + DF +I T S V ++VY+ PE++ + +A+ + + + +D+F
Sbjct 236 TYLVAYIVSDFLSISKT----SQHGVQISVYAVPEKIDQAEFALDAAVKLLDFYDDYFDI 291
Query 90 EYDL------------DTFNV--------ACVSDFNAGAMENKGLNIFNCSLLLASMQTS 129
Y L + F+V A + DF +GAMEN GL + S LL S
Sbjct 292 PYPLPKQEKPTIKHAKEEFSVYIICFVDLAAIPDFQSGAMENWGLTTYRESALLFDPHKS 351
Query 130 TDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTL 164
+ D + ++ HE H W GN VT++ W L L
Sbjct 352 SASDKLGITMIIAHELAHQWFGNLVTMQWWNDLWL 386
> xla:495476 anpep; alanyl (membrane) aminopeptidase; K11140 aminopeptidase
N [EC:3.4.11.2]
Length=963
Score = 63.9 bits (154), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 50/148 (33%), Positives = 72/148 (48%), Gaps = 13/148 (8%)
Query 24 DPHPKPS-YLFAILAGDFAAIV----DTFTTKS--GKEVALAVYSEPEQMHKLNWAMHSV 76
D PK S YL A + +F +I DT T G++ A+ + E +A+
Sbjct 263 DKTPKMSTYLVAFIVSEFESIGNDGNDTVTGVKIWGRKKAIVDEKQGE------YALSVT 316
Query 77 KVSMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASMQTSTDDDFER 136
K + + E ++ Y L + + DF+AGAMEN GL + + LL S+ + ER
Sbjct 317 KPILDFFEKYYRTPYPLPKSDQVALPDFSAGAMENWGLVTYRETALLFDENVSSIGNKER 376
Query 137 VLAVVGHEYFHNWTGNRVTVRDWFQLTL 164
V+ VV HE H W GN VT+R W L L
Sbjct 377 VVTVVAHELAHQWFGNLVTIRWWNDLWL 404
> sce:YKL157W APE2, LAP1, YKL158W; Aminopeptidase yscII; may have
a role in obtaining leucine from dipeptide substrates; sequence
coordinates have changed since RT-PCR analysis showed
that the adjacent ORF YKL158W comprises the 5' exon of APE2/YKL157W;
K13721 aminopeptidase 2 [EC:3.4.11.-]
Length=935
Score = 63.5 bits (153), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 46/147 (31%), Positives = 66/147 (44%), Gaps = 7/147 (4%)
Query 16 DRHFVVFTDPHPKPS-YLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQMHKLNWAMH 74
D V + PK S YL A + + + + + VY+ P +A
Sbjct 273 DGKKVTLFNTTPKMSTYLVAFIVAELK-----YVESKNFRIPVRVYATPGNEKHGQFAAD 327
Query 75 SVKVSMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASMQTSTDDDF 134
++ + E FG +Y L + V +F+AGAMEN GL + LL ST D
Sbjct 328 LTAKTLAFFEKTFGIQYPLPKMDNVAVHEFSAGAMENWGLVTYRVVDLLLDKDNSTLDRI 387
Query 135 ERVLAVVGHEYFHNWTGNRVTVRDWFQ 161
+RV VV HE H W GN VT+ DW++
Sbjct 388 QRVAEVVQHELAHQWFGNLVTM-DWWE 413
> cel:Y67D8C.9 hypothetical protein
Length=1087
Score = 62.0 bits (149), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 42/153 (27%), Positives = 69/153 (45%), Gaps = 14/153 (9%)
Query 21 VFTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQMHKLNWAMHSVKVSM 80
VF P +YL A G+F + +++ + + + V++ PE + + + + V
Sbjct 312 VFQTTPPMSTYLLAFAIGEFVKL----ESRTERGIPVTVWTYPEDVMSMKFTLEYAPVIF 367
Query 81 KWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASMQTSTDDDFE----- 135
ED Y L ++ +F+ G MEN GL +F + + A TD E
Sbjct 368 DRLEDALEIPYPLPKVDLIAARNFHVGGMENWGLVVFEFASI-AYTPPITDHVNETVDRM 426
Query 136 ----RVLAVVGHEYFHNWTGNRVTVRDWFQLTL 164
R+ ++ HE H W GN VT+RDW +L L
Sbjct 427 YNEFRIGKLIAHEAAHQWFGNLVTMRDWSELFL 459
> dre:100334936 aminopeptidase N-like
Length=166
Score = 61.6 bits (148), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 35/100 (35%), Positives = 54/100 (54%), Gaps = 0/100 (0%)
Query 65 QMHKLNWAMHSVKVSMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLA 124
Q K ++A++ +K+ E ++ Y L + + DF+AGAMEN GL + + L
Sbjct 45 QEIKGDYALNVTFPVLKYLESYYNTTYPLSKSDQIALPDFSAGAMENWGLVTYRETFLFY 104
Query 125 SMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTL 164
+ S+ +D E V+ V+ HE H W GN VT+R W L L
Sbjct 105 EPKVSSHEDKEGVITVISHELAHMWFGNLVTMRWWNDLWL 144
> mmu:74574 4833403I15Rik, Aqpep, Lvrn, MGC130603; RIKEN cDNA
4833403I15 gene; K13724 aminopeptidase Q [EC:3.4.11.-]
Length=991
Score = 61.6 bits (148), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 40/131 (30%), Positives = 63/131 (48%), Gaps = 3/131 (2%)
Query 29 PSYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQMHKLNWAMHSVKVSMKWEEDHFG 88
P+YL A++ D I T+ GKE+ + + L++A + + ED F
Sbjct 301 PTYLVALVVCDLDHIS---RTERGKEIRVWARKDDIASGYLDFAANITGPIFSFLEDLFN 357
Query 89 REYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASMQTSTDDDFERVLAVVGHEYFHN 148
Y L ++ + F +GAMEN GL IF+ S LL + + +L+++ HE H
Sbjct 358 ISYRLPKTDIVALPIFASGAMENWGLLIFDESSLLLEPEDELTEKRAMILSIIAHEVGHQ 417
Query 149 WTGNRVTVRDW 159
W GN VT+ W
Sbjct 418 WFGNLVTMSWW 428
> cel:R03G8.6 hypothetical protein
Length=747
Score = 61.6 bits (148), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 39/130 (30%), Positives = 63/130 (48%), Gaps = 12/130 (9%)
Query 30 SYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQMHKLNWAMHSVKVSMKWEEDHFGR 89
SY+ A+ GD + T V + VY++P + +++ A++ ++ ++ E FG
Sbjct 227 SYILALFIGD----IQFKETILNNGVRIRVYTDPVNIDRVDHALNISRIVLEGFERQFGI 282
Query 90 EYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASMQTSTDDDFERVLAVVGHEYFHNW 149
Y ++ + V +F GAMEN GL I N L+ D V +V HE H W
Sbjct 283 RYPMEKLDFVSVQNFKFGAMENWGLVIHNAYSLIG--------DPMDVTEIVIHEIAHQW 334
Query 150 TGNRVTVRDW 159
GN VT++ W
Sbjct 335 FGNLVTMKYW 344
> cel:Y42A5A.1 hypothetical protein
Length=1045
Score = 60.8 bits (146), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 43/138 (31%), Positives = 63/138 (45%), Gaps = 4/138 (2%)
Query 27 PKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQMHKLNWAMHSVKVSMKWEEDH 86
P +YLFA D+ + +TF SG+ V VY +P ++ S+ + + ED+
Sbjct 346 PMSTYLFAFSVSDYPYL-ETF---SGRGVRSRVYCDPTKLVDAQLITKSIGPVLDFYEDY 401
Query 87 FGREYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASMQTSTDDDFERVLAVVGHEYF 146
FG Y L+ +V V + AMEN GL + L + V +V HE
Sbjct 402 FGIPYPLEKLDVVIVPALSVTAMENWGLITIRQTNGLYTEGRFAISQKHDVQEIVAHELA 461
Query 147 HNWTGNRVTVRDWFQLTL 164
H W GN VT++ W L L
Sbjct 462 HQWFGNLVTMKWWNDLWL 479
> dre:100334888 leucyl/cystinyl aminopeptidase-like
Length=478
Score = 58.9 bits (141), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 35/119 (29%), Positives = 64/119 (53%), Gaps = 6/119 (5%)
Query 30 SYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQMHKLNWAMHSVKVSMKWEEDHFGR 89
+YL A + +F++ +K+ + ++VY+ P++ ++++A+ + +K+ F
Sbjct 351 TYLVAFIVAEFSS-----HSKNVSKTTVSVYAVPDKKDQVHYALETACKLLKFYNTFFEI 405
Query 90 EYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASMQTSTDDDFERVLAVVGHEYFHN 148
EY L ++ + DF AGAMEN GL F + LL Q+S D + V +V+ HE H
Sbjct 406 EYPLSKLDLVAIPDFLAGAMENWGLITFRETTLLVGNQSSRFDK-QLVTSVIAHELAHQ 463
> dre:100004744 aminopeptidase N-like
Length=936
Score = 57.0 bits (136), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 42/167 (25%), Positives = 78/167 (46%), Gaps = 8/167 (4%)
Query 1 SNGNKTEE-GPVQDSPDRHFVVFTDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAV 59
SNG + E + D F SYL A++ D+ + T + + + +
Sbjct 219 SNGMEIENVDTIVDGQPVTVTTFEPTKIMSSYLLALVVSDYTNV-----TSADGTLQIRI 273
Query 60 YSEPEQMHKL--NWAMHSVKVSMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIF 117
++ + + ++A++ +K+ E+++ Y L + + DF GAMEN GL ++
Sbjct 274 WARKKAIEDGHGDYALNITGPILKFFENYYNVPYPLSKSDQIALPDFYFGAMENWGLVMY 333
Query 118 NCSLLLASMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTL 164
S LL S++ + ER ++ HE H W GN VT++ W ++ L
Sbjct 334 RESNLLYDPTVSSNANKERTATIIAHELAHMWFGNLVTLKWWNEVWL 380
> mmu:237553 Trhde, 9330155P21Rik, MGC40831; TRH-degrading enzyme
(EC:3.4.19.6); K01306 pyroglutamyl-peptidase II [EC:3.4.19.6]
Length=1025
Score = 56.2 bits (134), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 39/120 (32%), Positives = 60/120 (50%), Gaps = 4/120 (3%)
Query 48 TTKSGKEVALAVYSEPEQMHKL--NWAMHSVKVSMKWEEDHFGREYDLDTFNVACVSDFN 105
TTKSG V + +Y+ P+ + + ++A+H K +++ ED+F Y L ++ V
Sbjct 346 TTKSG--VVVRLYARPDAIRRGSGDYALHITKRLIEFYEDYFKVPYSLPKLDLLAVPKHP 403
Query 106 AGAMENKGLNIFNCSLLLASMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTLK 165
AMEN GL+IF +L S+ V V+ HE H W G+ VT W + LK
Sbjct 404 YAAMENWGLSIFVEQRILLDPSVSSISYLLDVTMVIVHEICHQWFGDLVTPVWWEDVWLK 463
> cel:T16G12.1 hypothetical protein
Length=1890
Score = 56.2 bits (134), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 39/133 (29%), Positives = 66/133 (49%), Gaps = 7/133 (5%)
Query 30 SYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQMHKLNWAMHSVKVSMKWEEDHFGR 89
+YL A+ G F+ + T++G + +S EQ + A++ ++ + E++F
Sbjct 1205 TYLLALCVGHFSNLATV--TRTGVLTRVWTWSGMEQYGEF--ALNVTAGTIDFMENYFSY 1260
Query 90 EYDLDTFNVACVSDF--NAGAMENKGLNIFNCSLLLASMQTSTDDDFERVLAVVGHEYFH 147
++ L +V + ++ NAGAMEN GL I SL + +T D V HE H
Sbjct 1261 DFPLKKLDVMALPEYTMNAGAMENWGLIIGEYSLFMFDPDYATTRDITEVAETTAHEVVH 1320
Query 148 NWTGNRVTVRDWF 160
W G+ VT+ DW+
Sbjct 1321 QWFGDIVTL-DWW 1332
Score = 42.0 bits (97), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 29/102 (28%), Positives = 45/102 (44%), Gaps = 0/102 (0%)
Query 63 PEQMHKLNWAMHSVKVSMKWEEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCSLL 122
P L +A + + ++ G ++ L + + +F AGAMEN GL I+ +
Sbjct 310 PGTEQYLQFAAQNAGECLYQLGEYTGIKFPLSKADQLGMPEFLAGAMENWGLIIYKYQYI 369
Query 123 LASMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTL 164
+ T T + E V+ HE H W G+ VT W L L
Sbjct 370 AYNPTTMTTRNMEAAAKVMCHELAHQWFGDLVTTAWWDDLFL 411
> hsa:29953 TRHDE, FLJ22381, PAP-II, PGPEP2, TRH-DE; thyrotropin-releasing
hormone degrading enzyme (EC:3.4.19.6); K01306
pyroglutamyl-peptidase II [EC:3.4.19.6]
Length=1024
Score = 56.2 bits (134), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 39/120 (32%), Positives = 60/120 (50%), Gaps = 4/120 (3%)
Query 48 TTKSGKEVALAVYSEPEQMHKL--NWAMHSVKVSMKWEEDHFGREYDLDTFNVACVSDFN 105
TTKSG V + +Y+ P+ + + ++A+H K +++ ED+F Y L ++ V
Sbjct 345 TTKSG--VVVRLYARPDAIRRGSGDYALHITKRLIEFYEDYFKVPYSLPKLDLLAVPKHP 402
Query 106 AGAMENKGLNIFNCSLLLASMQTSTDDDFERVLAVVGHEYFHNWTGNRVTVRDWFQLTLK 165
AMEN GL+IF +L S+ V V+ HE H W G+ VT W + LK
Sbjct 403 YAAMENWGLSIFVEQRILLDPSVSSISYLLDVTMVIVHEICHQWFGDLVTPVWWEDVWLK 462
> hsa:206338 AQPEP, APQ, FLJ90650, LVRN, MGC125378, MGC125379;
laeverin; K13724 aminopeptidase Q [EC:3.4.11.-]
Length=990
Score = 55.8 bits (133), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 41/137 (29%), Positives = 63/137 (45%), Gaps = 4/137 (2%)
Query 23 TDPHPKPSYLFAILAGDFAAIVDTFTTKSGKEVALAVYSEPEQMHKLNWAMHSVKVSMKW 82
T PH P+YL A + D+ + T+ GKE+ + + ++A++ +
Sbjct 299 TTPH-MPTYLVAFVICDYDHVN---RTERGKEIRIWARKDAIANGSADFALNITGPIFSF 354
Query 83 EEDHFGREYDLDTFNVACVSDFNAGAMENKGLNIFNCSLLLASMQTSTDDDFERVLAVVG 142
ED F Y L ++ + F+ AMEN GL IF+ S LL + + + VV
Sbjct 355 LEDLFNISYSLPKTDIIALPSFDNHAMENWGLMIFDESGLLLEPKDQLTEKKTLISYVVS 414
Query 143 HEYFHNWTGNRVTVRDW 159
HE H W GN VT+ W
Sbjct 415 HEIGHQWFGNLVTMNWW 431
Lambda K H
0.319 0.133 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3962792044
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40