bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0532_orf2
Length=142
Score E
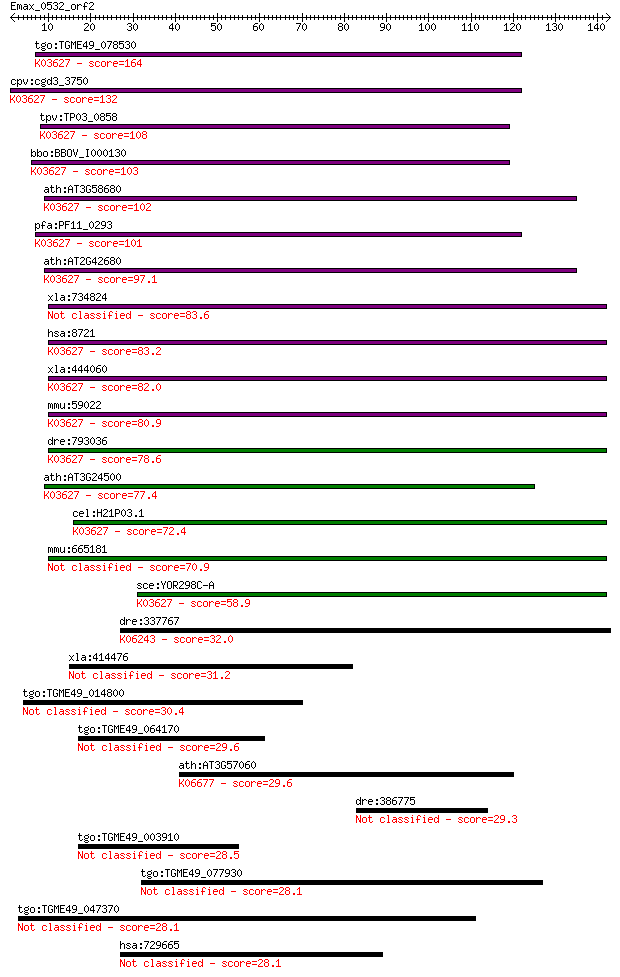
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_078530 multiprotein bridging factor type 1, putativ... 164 1e-40
cpv:cgd3_3750 multiprotein bridging factor type 1 like transcr... 132 4e-31
tpv:TP03_0858 multiprotein bridging factor type 1; K03627 puta... 108 8e-24
bbo:BBOV_I000130 16.m00729; multiprotein bridging factor type ... 103 2e-22
ath:AT3G58680 MBF1B; MBF1B (MULTIPROTEIN BRIDGING FACTOR 1B); ... 102 4e-22
pfa:PF11_0293 multiprotein bridging factor type 1, putative; K... 101 7e-22
ath:AT2G42680 MBF1A; MBF1A (MULTIPROTEIN BRIDGING FACTOR 1A); ... 97.1 1e-20
xla:734824 edf1, MGC131240, edf-1, mbf1; endothelial different... 83.6 2e-16
hsa:8721 EDF1, EDF-1, MBF1, MGC9058; endothelial differentiati... 83.2 3e-16
xla:444060 edf1, MGC82687; endothelial differentiation-related... 82.0 6e-16
mmu:59022 Edf1, 0610008L11Rik, AA409425; endothelial different... 80.9 1e-15
dre:793036 edf1, MGC73192, MGC86829, zgc:73192, zgc:86829; end... 78.6 6e-15
ath:AT3G24500 MBF1C; MBF1C (MULTIPROTEIN BRIDGING FACTOR 1C); ... 77.4 1e-14
cel:H21P03.1 mbf-1; MBF (multiprotein bridging factor) transcr... 72.4 4e-13
mmu:665181 Gm11964, OTTMUSG00000005089; predicted gene 11964 70.9
sce:YOR298C-A MBF1, SUF13; Mbf1p; K03627 putative transcriptio... 58.9 4e-09
dre:337767 hm:zehs0001; K06243 laminin, beta 2 32.0
xla:414476 eif4ebp2, MGC78987; eukaryotic translation initiati... 31.2 1.0
tgo:TGME49_014800 hypothetical protein 30.4 2.0
tgo:TGME49_064170 hypothetical protein 29.6 3.0
ath:AT3G57060 binding; K06677 condensin complex subunit 1 29.6
dre:386775 gltscr1, sb:cb888, si:dkey-224e22.6, wu:fc28g11; gl... 29.3 4.2
tgo:TGME49_003910 TBC domain-containing protein 28.5 7.4
tgo:TGME49_077930 hypothetical protein 28.1 8.3
tgo:TGME49_047370 hypothetical protein 28.1 8.3
hsa:729665 C14orf38, c14_5395; chromosome 14 open reading fram... 28.1 9.1
> tgo:TGME49_078530 multiprotein bridging factor type 1, putative
; K03627 putative transcription factor
Length=144
Score = 164 bits (414), Expect = 1e-40, Method: Compositional matrix adjust.
Identities = 78/115 (67%), Positives = 95/115 (82%), Gaps = 0/115 (0%)
Query 7 NFQDWTPVTFQKTSPGGRRVVKEQEVNKTRRAGEVIETEKKFLGGRNKTTKANLIANAKK 66
+FQDWTPV++ KT + V KEQE+N+ RR GE +ETEKKFLGG+NK TK L NA+K
Sbjct 2 SFQDWTPVSWNKTGQRQKGVTKEQEINQARRRGEELETEKKFLGGQNKATKGGLCPNARK 61
Query 67 VEEDTGDYHVDRVSTDFSRALAQARRNKGMTQAQLAQAINEKPSVVSEYESGKHL 121
+EEDTGDYHV+RVS DFSRAL QAR+ K MTQA+LAQAINEKPSVV++YESG+ +
Sbjct 62 IEEDTGDYHVERVSADFSRALQQARQAKKMTQAELAQAINEKPSVVNDYESGRAI 116
> cpv:cgd3_3750 multiprotein bridging factor type 1 like transcriptional
co-activator ; K03627 putative transcription factor
Length=158
Score = 132 bits (331), Expect = 4e-31, Method: Compositional matrix adjust.
Identities = 70/121 (57%), Positives = 87/121 (71%), Gaps = 3/121 (2%)
Query 1 FGKMSGNFQDWTPVTFQKTSPGGRRVVKEQEVNKTRRAGEVIETEKKFLGGRNKTTKANL 60
KMS QDW V ++K + + KEQ++N+ RR GE I TEKKFLGGRN +TK N+
Sbjct 9 ISKMS---QDWNQVVWKKGGSRPKGISKEQDLNQARRKGEEIITEKKFLGGRNASTKQNI 65
Query 61 IANAKKVEEDTGDYHVDRVSTDFSRALAQARRNKGMTQAQLAQAINEKPSVVSEYESGKH 120
NA K++EDTGDY + RVS +FSRAL QAR K +TQAQLAQ INEK SVV++YESGK
Sbjct 66 PQNAAKLDEDTGDYRIFRVSGEFSRALQQARVAKKLTQAQLAQMINEKASVVNDYESGKA 125
Query 121 L 121
+
Sbjct 126 I 126
> tpv:TP03_0858 multiprotein bridging factor type 1; K03627 putative
transcription factor
Length=119
Score = 108 bits (269), Expect = 8e-24, Method: Compositional matrix adjust.
Identities = 56/111 (50%), Positives = 75/111 (67%), Gaps = 1/111 (0%)
Query 8 FQDWTPVTFQKTSPGGRRVVKEQEVNKTRRAGEVIETEKKFLGGRNKTTKANLIANAKKV 67
+QDW PV + K + KE +NK RRAG ++T+KKFLGG+NKTTK+ L NA K+
Sbjct 3 YQDWKPVVWTKHE-NFKGPNKESALNKARRAGVELDTQKKFLGGQNKTTKSFLPPNAAKI 61
Query 68 EEDTGDYHVDRVSTDFSRALAQARRNKGMTQAQLAQAINEKPSVVSEYESG 118
E + +H++RVS F AL +AR K MTQ QLA+AINE S++ EYE+G
Sbjct 62 ENENESFHIERVSFAFRTALQKARMAKNMTQLQLARAINESESLIKEYENG 112
> bbo:BBOV_I000130 16.m00729; multiprotein bridging factor type
1; K03627 putative transcription factor
Length=143
Score = 103 bits (257), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 55/113 (48%), Positives = 73/113 (64%), Gaps = 1/113 (0%)
Query 6 GNFQDWTPVTFQKTSPGGRRVVKEQEVNKTRRAGEVIETEKKFLGGRNKTTKANLIANAK 65
+ QDWTPV + K R ++ +N RR GE I TEKKFLGG NK+TKA L +NA
Sbjct 2 ASHQDWTPVVWSKKD-NCRGPQRKAALNDARRTGEDISTEKKFLGGLNKSTKAYLPSNAA 60
Query 66 KVEEDTGDYHVDRVSTDFSRALAQARRNKGMTQAQLAQAINEKPSVVSEYESG 118
K++ +T D+ ++RV F +AL +AR KG+TQ LA+ INE S V EYE+G
Sbjct 61 KIDNETEDFRIERVEFHFRQALQKARMAKGLTQQSLARLINEPESTVKEYENG 113
> ath:AT3G58680 MBF1B; MBF1B (MULTIPROTEIN BRIDGING FACTOR 1B);
DNA binding / transcription coactivator; K03627 putative transcription
factor
Length=142
Score = 102 bits (254), Expect = 4e-22, Method: Compositional matrix adjust.
Identities = 56/131 (42%), Positives = 75/131 (57%), Gaps = 5/131 (3%)
Query 9 QDWTPVTFQKTSPGGRRVVKEQEVNKTRRAGEVIETEKKFLGGRNKTTKANLIANAKKVE 68
QDW PV +K +P E+ VN RR+G IET +KF G NK + N KK++
Sbjct 9 QDWEPVVIRKRAPNAAAKRDEKTVNAARRSGADIETVRKFNAGSNKAASSGTSLNTKKLD 68
Query 69 EDTGDYHVDRVSTDFSRALAQARRNKGMTQAQLAQAINEKPSVVSEYESGKHLDGE---- 124
+DT + DRV T+ +A+ QAR K +TQ+QLA INEKP V+ EYESGK + +
Sbjct 69 DDTENLSHDRVPTELKKAIMQARGEKKLTQSQLAHLINEKPQVIQEYESGKAIPNQQILS 128
Query 125 -LHRGTGRFLK 134
L R G L+
Sbjct 129 KLERALGAKLR 139
> pfa:PF11_0293 multiprotein bridging factor type 1, putative;
K03627 putative transcription factor
Length=136
Score = 101 bits (252), Expect = 7e-22, Method: Compositional matrix adjust.
Identities = 52/116 (44%), Positives = 80/116 (68%), Gaps = 5/116 (4%)
Query 7 NFQDWTPVTFQKTSPGGRRVVKEQEVNKTRRAGEVIETEKKFLGGRNKTTKANLIA-NAK 65
+ QD PV + KT + K +++++ R+ G +E EKK+ GG+NK++K NLI N
Sbjct 2 DHQDLKPVIWHKTE----KKTKPKDIHEARKLGIDVEVEKKYFGGKNKSSKGNLIIENKA 57
Query 66 KVEEDTGDYHVDRVSTDFSRALAQARRNKGMTQAQLAQAINEKPSVVSEYESGKHL 121
K+E++T ++ +DRV+ FSRAL QAR +K +TQAQLA+ +NE SV+ EYE+GK +
Sbjct 58 KIEQETENFKIDRVTPAFSRALQQARISKKLTQAQLARLVNESESVIKEYENGKAI 113
> ath:AT2G42680 MBF1A; MBF1A (MULTIPROTEIN BRIDGING FACTOR 1A);
DNA binding / transcription coactivator; K03627 putative transcription
factor
Length=142
Score = 97.1 bits (240), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 54/131 (41%), Positives = 74/131 (56%), Gaps = 5/131 (3%)
Query 9 QDWTPVTFQKTSPGGRRVVKEQEVNKTRRAGEVIETEKKFLGGRNKTTKANLIANAKKVE 68
QDW PV +K E+ VN RR+G IET +KF G NK + N K ++
Sbjct 9 QDWEPVVIRKKPANAAAKRDEKTVNAARRSGADIETVRKFNAGTNKAASSGTSLNTKMLD 68
Query 69 EDTGDYHVDRVSTDFSRALAQARRNKGMTQAQLAQAINEKPSVVSEYESGKHLDGE---- 124
+DT + +RV T+ +A+ QAR +K +TQ+QLAQ INEKP V+ EYESGK + +
Sbjct 69 DDTENLTHERVPTELKKAIMQARTDKKLTQSQLAQIINEKPQVIQEYESGKAIPNQQILS 128
Query 125 -LHRGTGRFLK 134
L R G L+
Sbjct 129 KLERALGAKLR 139
> xla:734824 edf1, MGC131240, edf-1, mbf1; endothelial differentiation-related
factor 1
Length=148
Score = 83.6 bits (205), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 49/138 (35%), Positives = 75/138 (54%), Gaps = 8/138 (5%)
Query 10 DWTPVT-FQKTSPGGRRVVKEQEVNKTRRAGEVIETEKKFLGGRNKTTKANLIANAKKVE 68
DW VT +K P + +Q + +R GE +ET KK+ G+NK + + N K++
Sbjct 5 DWDTVTVLRKKGPTAAQAKSKQAITAAQRRGEELETSKKWSAGQNK--QHTITKNTAKLD 62
Query 69 EDTGDYHVDRVSTDFSRALAQARRNKGMTQAQLAQAINEKPSVVSEYESGKHLD-----G 123
+T + H DRV + + + Q R+ KGMTQ LA INEKP V+++YESGK + G
Sbjct 63 RETEELHHDRVPLEVGKVIQQGRQGKGMTQKDLATKINEKPQVIADYESGKAIPNNQVMG 122
Query 124 ELHRGTGRFLKADSLQTP 141
++ R G L+ + P
Sbjct 123 KIERVIGMKLRGKDIGKP 140
> hsa:8721 EDF1, EDF-1, MBF1, MGC9058; endothelial differentiation-related
factor 1; K03627 putative transcription factor
Length=148
Score = 83.2 bits (204), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 47/138 (34%), Positives = 78/138 (56%), Gaps = 8/138 (5%)
Query 10 DWTPVT-FQKTSPGGRRVVKEQEVNKTRRAGEVIETEKKFLGGRNKTTKANLIANAKKVE 68
DW VT +K P + +Q + +R GE +ET KK+ G+NK + ++ N K++
Sbjct 5 DWDTVTVLRKKGPTAAQAKSKQAILAAQRRGEDVETSKKWAAGQNK--QHSITKNTAKLD 62
Query 69 EDTGDYHVDRVSTDFSRALAQARRNKGMTQAQLAQAINEKPSVVSEYESGKHLD-----G 123
+T + H DRV+ + + + Q R++KG+TQ LA INEKP V+++YESG+ + G
Sbjct 63 RETEELHHDRVTLEVGKVIQQGRQSKGLTQKDLATKINEKPQVIADYESGRAIPNNQVLG 122
Query 124 ELHRGTGRFLKADSLQTP 141
++ R G L+ + P
Sbjct 123 KIERAIGLKLRGKDIGKP 140
> xla:444060 edf1, MGC82687; endothelial differentiation-related
factor 1 homolog; K03627 putative transcription factor
Length=147
Score = 82.0 bits (201), Expect = 6e-16, Method: Compositional matrix adjust.
Identities = 48/138 (34%), Positives = 74/138 (53%), Gaps = 8/138 (5%)
Query 10 DWTPVT-FQKTSPGGRRVVKEQEVNKTRRAGEVIETEKKFLGGRNKTTKANLIANAKKVE 68
DW VT +K P + +Q + +R GE +ET KK+ G+NK + + N K++
Sbjct 5 DWDTVTVLRKKGPTAAQAKSKQAITAAQRRGEEVETSKKWSAGQNK--QHTITRNTAKLD 62
Query 69 EDTGDYHVDRVSTDFSRALAQARRNKGMTQAQLAQAINEKPSVVSEYESGKHLD-----G 123
+T + H DRV + + + Q R+ KGMTQ LA INEKP V+++YE GK + G
Sbjct 63 RETEELHHDRVPLEVGKVIQQGRQGKGMTQKDLATKINEKPQVIADYECGKAIPNNQVMG 122
Query 124 ELHRGTGRFLKADSLQTP 141
++ R G L+ + P
Sbjct 123 KIERVIGLKLRGKDIGKP 140
> mmu:59022 Edf1, 0610008L11Rik, AA409425; endothelial differentiation-related
factor 1; K03627 putative transcription factor
Length=148
Score = 80.9 bits (198), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 46/138 (33%), Positives = 78/138 (56%), Gaps = 8/138 (5%)
Query 10 DWTPVT-FQKTSPGGRRVVKEQEVNKTRRAGEVIETEKKFLGGRNKTTKANLIANAKKVE 68
DW VT +K P + +Q + +R GE +ET KK+ G+NK + ++ N K++
Sbjct 5 DWDTVTVLRKKGPTAAQAKSKQAILAAQRRGEDVETSKKWAAGQNK--QHSITKNTAKLD 62
Query 69 EDTGDYHVDRVSTDFSRALAQARRNKGMTQAQLAQAINEKPSVVSEYESGKHLD-----G 123
+T + H DRV+ + + + + R++KG+TQ LA INEKP V+++YESG+ + G
Sbjct 63 RETEELHHDRVTLEVGKVIQRGRQSKGLTQKDLATKINEKPQVIADYESGRAIPNNQVLG 122
Query 124 ELHRGTGRFLKADSLQTP 141
++ R G L+ + P
Sbjct 123 KIERAIGLKLRGKDIGKP 140
> dre:793036 edf1, MGC73192, MGC86829, zgc:73192, zgc:86829; endothelial
differentiation-related factor 1; K03627 putative
transcription factor
Length=146
Score = 78.6 bits (192), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 45/137 (32%), Positives = 71/137 (51%), Gaps = 7/137 (5%)
Query 10 DWTPVTFQKTSPGGRRVVKEQEVNKTRRAGEVIETEKKFLGGRNKTTKANLIANAKKVEE 69
DW VT + + +Q V +R GE +ET KK+ G+NK + + N K++
Sbjct 5 DWDTVTVLRKKGSAAQSKSKQAVTAAQRKGEAVETSKKWAAGQNK--QHVVTKNTAKLDR 62
Query 70 DTGDYHVDRVSTDFSRALAQARRNKGMTQAQLAQAINEKPSVVSEYESGKHLD-----GE 124
+T + RV + + + Q R+NKG+TQ LA INEKP +++EYE GK + G+
Sbjct 63 ETEELSHQRVPLEVGKVIQQGRQNKGLTQKDLATKINEKPQIIAEYECGKAIPNNQVMGK 122
Query 125 LHRGTGRFLKADSLQTP 141
+ R G L+ + P
Sbjct 123 IERAIGLKLRGKDIGLP 139
> ath:AT3G24500 MBF1C; MBF1C (MULTIPROTEIN BRIDGING FACTOR 1C);
DNA binding / transcription coactivator/ transcription factor;
K03627 putative transcription factor
Length=148
Score = 77.4 bits (189), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 45/118 (38%), Positives = 65/118 (55%), Gaps = 2/118 (1%)
Query 9 QDWTPVTFQKTSPGGRRVVKEQEVNKTRRAGEVIETEKKFLGGRNKTTKANLIA--NAKK 66
QDW PV K+ + + + VN R G ++T KKF G NK K+ + N KK
Sbjct 11 QDWEPVVLHKSKQKSQDLRDPKAVNAALRNGVAVQTVKKFDAGSNKKGKSTAVPVINTKK 70
Query 67 VEEDTGDYHVDRVSTDFSRALAQARRNKGMTQAQLAQAINEKPSVVSEYESGKHLDGE 124
+EE+T +DRV + + +AR K M+QA LA+ INE+ VV EYE+GK + +
Sbjct 71 LEEETEPAAMDRVKAEVRLMIQKARLEKKMSQADLAKQINERTQVVQEYENGKAVPNQ 128
> cel:H21P03.1 mbf-1; MBF (multiprotein bridging factor) transcriptional
coactivator family member (mbf-1); K03627 putative
transcription factor
Length=156
Score = 72.4 bits (176), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 45/131 (34%), Positives = 67/131 (51%), Gaps = 7/131 (5%)
Query 16 FQKTSPGGRRVVKEQEVNKTRRAGEVIETEKKFLGGRNKTTKANLIANAKKVEEDTGDYH 75
K P + + ++N +RAG I TEKK + G N+ AN N +++E+T + H
Sbjct 19 ITKRGPVNKTLKSAAQLNAAQRAGVDISTEKKTMSGGNRQHSAN--KNTLRLDEETEELH 76
Query 76 VDRVSTDFSRALAQARRNKGMTQAQLAQAINEKPSVVSEYESGK-----HLDGELHRGTG 130
+V+ + + QAR KG TQ L+ INEKP VV EYESGK + ++ R G
Sbjct 77 HQKVALSLGKVMQQARATKGWTQKDLSTQINEKPQVVGEYESGKAVPNQQIMAKMERALG 136
Query 131 RFLKADSLQTP 141
L+ + P
Sbjct 137 VKLRGKDIGMP 147
> mmu:665181 Gm11964, OTTMUSG00000005089; predicted gene 11964
Length=147
Score = 70.9 bits (172), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 45/138 (32%), Positives = 75/138 (54%), Gaps = 9/138 (6%)
Query 10 DWTPVT-FQKTSPGGRRVVKEQEVNKTRRAGEVIETEKKFLGGRNKTTKANLIANAKKVE 68
DW VT K P + +Q + +R E +ET KK+ G+NK + ++ N K++
Sbjct 5 DWDTVTVLCKKGPTAAQAKSKQAILAAQRR-EDVETSKKWAAGQNK--QHSITKNTAKLD 61
Query 69 EDTGDYHVDRVSTDFSRALAQARRNKGMTQAQLAQAINEKPSVVSEYESGKHLD-----G 123
+T + H DRV+ + + + + R++KG+TQ LA INEKP V+++YESG+ + G
Sbjct 62 WETEELHHDRVALEVGKVIQRGRQSKGLTQKDLATKINEKPQVIADYESGRAIPNNQVLG 121
Query 124 ELHRGTGRFLKADSLQTP 141
+ R G L+ + P
Sbjct 122 KTERAIGLKLRGKDVGKP 139
> sce:YOR298C-A MBF1, SUF13; Mbf1p; K03627 putative transcription
factor
Length=151
Score = 58.9 bits (141), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 36/116 (31%), Positives = 63/116 (54%), Gaps = 7/116 (6%)
Query 31 EVNKTRRAGEVIETEKKFLGGRNKTTKANLIANAKKVEEDTGDYHVDRVSTDFSRALAQA 90
++N RR G V+ +KK+ G T N KV+ +T ++ + RA+++A
Sbjct 31 QINAARRQGLVVSVDKKY--GSTNTRGDNEGQRLTKVDRETDIVKPKKLDPNVGRAISRA 88
Query 91 RRNKGMTQAQLAQAINEKPSVVSEYESGKHLDGE-----LHRGTGRFLKADSLQTP 141
R +K M+Q LA INEKP+VV++YE+ + + + L R G L+ +++ +P
Sbjct 89 RTDKKMSQKDLATKINEKPTVVNDYEAARAIPNQQVLSKLERALGVKLRGNNIGSP 144
> dre:337767 hm:zehs0001; K06243 laminin, beta 2
Length=1782
Score = 32.0 bits (71), Expect = 0.59, Method: Composition-based stats.
Identities = 31/117 (26%), Positives = 51/117 (43%), Gaps = 15/117 (12%)
Query 27 VKEQEVNKTRRAGEVIETEKKFLGGRNKTTKA-NLIANAKKVEEDTGDYHVDRVSTDFSR 85
+ EQ N R+A +++ KK RNK N + KK D R +
Sbjct 1556 ILEQTQNDVRKAEQLLLDAKK---ARNKAEGVKNTAESVKKALNDA-----SRAQAAAEK 1607
Query 86 ALAQARRNKGMTQAQLAQAINEKPSVVSEYESGKHLDGELHRGTGRFLKADSLQTPR 142
A+ +A+ + G+TQ QLAQ +E + + G+L R + ++L+T R
Sbjct 1608 AIQKAKNDIGLTQNQLAQIQSETSASERDLNDAVDRLGDLER------QIEALKTKR 1658
> xla:414476 eif4ebp2, MGC78987; eukaryotic translation initiation
factor 4E binding protein 2
Length=113
Score = 31.2 bits (69), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 17/67 (25%), Positives = 31/67 (46%), Gaps = 0/67 (0%)
Query 15 TFQKTSPGGRRVVKEQEVNKTRRAGEVIETEKKFLGGRNKTTKANLIANAKKVEEDTGDY 74
T T+PGG R++ +++ RR + +T + L T N + KVE + +
Sbjct 37 TLFSTTPGGTRIIYDRKFLLDRRTSPLAQTPPRRLPDIPGVTSPNTVVEEPKVETNNLNN 96
Query 75 HVDRVST 81
H + +T
Sbjct 97 HETKTAT 103
> tgo:TGME49_014800 hypothetical protein
Length=759
Score = 30.4 bits (67), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 23/73 (31%), Positives = 39/73 (53%), Gaps = 7/73 (9%)
Query 4 MSGNFQDWTPVTFQKTSPGGRRVVKEQEVNKTRRAG-----EVIETEKKFLGGRNK--TT 56
+SGNFQ P + + G RVVK+++ +++ ++G E EKK +G R TT
Sbjct 66 LSGNFQHGPPAKALQPAGGTSRVVKKRKTDRSSQSGSASPKESSRQEKKAVGSRCSAATT 125
Query 57 KANLIANAKKVEE 69
+L + A++ E
Sbjct 126 PPSLPSWAQRERE 138
> tgo:TGME49_064170 hypothetical protein
Length=2165
Score = 29.6 bits (65), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 18/44 (40%), Positives = 27/44 (61%), Gaps = 6/44 (13%)
Query 17 QKTSPGGRRVVKEQEVNKTRRAGEVIETEKKFLGGRNKTTKANL 60
Q+ S G RR++ +E +TRRAG IE E RN+ ++A+L
Sbjct 2105 QENSNGERRIITRREKPQTRRAGNDIEVE------RNEHSEASL 2142
> ath:AT3G57060 binding; K06677 condensin complex subunit 1
Length=1396
Score = 29.6 bits (65), Expect = 3.3, Method: Composition-based stats.
Identities = 20/79 (25%), Positives = 33/79 (41%), Gaps = 0/79 (0%)
Query 41 VIETEKKFLGGRNKTTKANLIANAKKVEEDTGDYHVDRVSTDFSRALAQARRNKGMTQAQ 100
V E + + K K L A ++ EE +H+++ + + A+ R K T
Sbjct 1170 VTENFRSIINKGKKFAKPELKACIEEFEEKINKFHMEKKEQEETARNAEVHREKTKTMES 1229
Query 101 LAQAINEKPSVVSEYESGK 119
LA K V EY+ G+
Sbjct 1230 LAVLSKVKEEPVEEYDEGE 1248
> dre:386775 gltscr1, sb:cb888, si:dkey-224e22.6, wu:fc28g11;
glioma tumor suppressor candidate region gene 1
Length=1800
Score = 29.3 bits (64), Expect = 4.2, Method: Composition-based stats.
Identities = 15/31 (48%), Positives = 18/31 (58%), Gaps = 0/31 (0%)
Query 83 FSRALAQARRNKGMTQAQLAQAINEKPSVVS 113
FS A A N G+T LAQA+ KP V+S
Sbjct 1047 FSAVSAGATVNTGITTPNLAQAVQAKPGVIS 1077
> tgo:TGME49_003910 TBC domain-containing protein
Length=1629
Score = 28.5 bits (62), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 17/41 (41%), Positives = 23/41 (56%), Gaps = 5/41 (12%)
Query 17 QKTSP---GGRRVVKEQEVNKTRRAGEVIETEKKFLGGRNK 54
Q SP GGRR +E E+ +T GE ++ E +L GR K
Sbjct 1576 QSASPETTGGRR--REGELERTGEEGEALQKEPSYLAGRKK 1614
> tgo:TGME49_077930 hypothetical protein
Length=1573
Score = 28.1 bits (61), Expect = 8.3, Method: Compositional matrix adjust.
Identities = 26/97 (26%), Positives = 49/97 (50%), Gaps = 13/97 (13%)
Query 32 VNKTRRAGEVIETEKKFLGGRNKTTKANLIANAKKVEEDTGDYHVDRVSTDFSRAL-AQA 90
+ + RR ++E EK+ L A A+++EE T D H+ RV+ + + L A A
Sbjct 1366 IKEFRRKIVMVEWEKQVLD-----------AEARELEEKTKDVHMLRVTKEIQKYLAADA 1414
Query 91 RRNKGMTQAQLA-QAINEKPSVVSEYESGKHLDGELH 126
+ + +QA + +A + P+ + E+ + G+LH
Sbjct 1415 DKKERRSQAGVGKEATPKVPAESTVTENSGKVSGQLH 1451
> tgo:TGME49_047370 hypothetical protein
Length=1600
Score = 28.1 bits (61), Expect = 8.3, Method: Composition-based stats.
Identities = 25/108 (23%), Positives = 47/108 (43%), Gaps = 10/108 (9%)
Query 3 KMSGNFQDWTPVTFQKTSPGGRRVVKEQEVNKTRRAGEVIETEKKFLGGRNKTTKANLIA 62
K++ ++W V + S RR+ + E+ + + V +TEKK+ L A
Sbjct 1476 KLAPKVREWAVVEAGEFSDLFRRLCMQLELAGSEKVAFVQQTEKKYF--------EQLSA 1527
Query 63 NAKKVEEDTGDYHVDRVSTDFSRALAQARRNKGMTQAQLAQAINEKPS 110
AK+V++ + H+ + L +R L QA++E+ S
Sbjct 1528 LAKEVQQQ--EEHLSHRLAVYKNNLFVEQRKLATYTTYLEQALDERKS 1573
> hsa:729665 C14orf38, c14_5395; chromosome 14 open reading frame
38
Length=793
Score = 28.1 bits (61), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 20/69 (28%), Positives = 37/69 (53%), Gaps = 8/69 (11%)
Query 27 VKEQEVNKTRRAGEVIE-------TEKKFLGGRNKTTKANLIANAKKVEEDTGDYHVDRV 79
VKE ++NK + E++E E+++ R K + + I A++ EED + H+
Sbjct 545 VKETQINK-EKEEELVEYLPQLQVAEQEYKEKRRKLEELSNIITAQRQEEDLLNNHIFLF 603
Query 80 STDFSRALA 88
+ DFSR ++
Sbjct 604 TRDFSRYIS 612
Lambda K H
0.312 0.128 0.358
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2683748972
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40