bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0531_orf3
Length=186
Score E
Sequences producing significant alignments: (Bits) Value
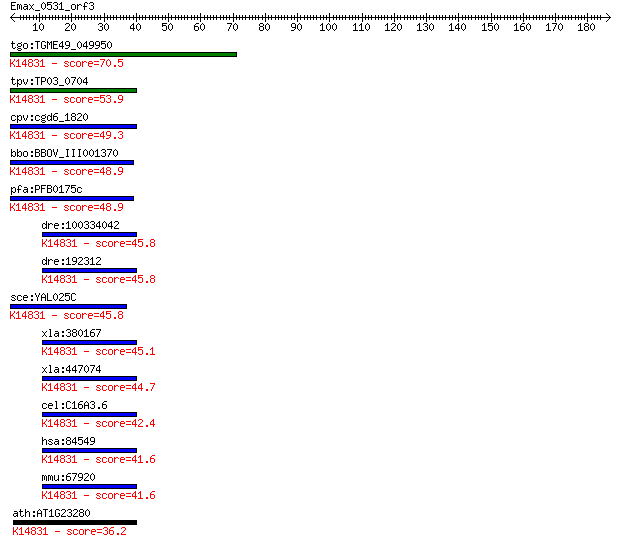
tgo:TGME49_049950 MAK16 protein, putative ; K14831 protein MAK16 70.5 3e-12
tpv:TP03_0704 hypothetical protein; K14831 protein MAK16 53.9 3e-07
cpv:cgd6_1820 hypothetical protein ; K14831 protein MAK16 49.3 8e-06
bbo:BBOV_III001370 17.m07141; Mak16 family protein; K14831 pro... 48.9 1e-05
pfa:PFB0175c nucleolar preribosomal assembly protein, putative... 48.9 1e-05
dre:100334042 Protein MAK16 homolog; K14831 protein MAK16 45.8 7e-05
dre:192312 mak16, chunp6925, fb34e01, mak16l, wu:fb34e01; MAK1... 45.8 8e-05
sce:YAL025C MAK16; Essential nuclear protein, constituent of 6... 45.8 8e-05
xla:380167 mak16-b, MGC53057, mak16, mak16l, rbm13; MAK16 homo... 45.1 2e-04
xla:447074 mak16-a, MGC85217, mak16l, rbm13; MAK16 homolog; K1... 44.7 2e-04
cel:C16A3.6 hypothetical protein; K14831 protein MAK16 42.4 0.001
hsa:84549 MAK16, MAK16L, RBM13; MAK16 homolog (S. cerevisiae);... 41.6 0.002
mmu:67920 Mak16, 2600016B03Rik, AI314911, Rbm13; MAK16 homolog... 41.6 0.002
ath:AT1G23280 MAK16 protein-related; K14831 protein MAK16 36.2 0.065
> tgo:TGME49_049950 MAK16 protein, putative ; K14831 protein MAK16
Length=333
Score = 70.5 bits (171), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 42/70 (60%), Positives = 49/70 (70%), Gaps = 0/70 (0%)
Query 1 ERREAAREKKALVAAHIEDVIEDELLKRLHQGVYGDLYNEHRLKEDTTEKEREAPEVTEA 60
ERRE AREKKA VAAH+EDVIE ELLKRL QGVYG LYN K E ++E EV +
Sbjct 142 ERREKAREKKAEVAAHLEDVIESELLKRLQQGVYGTLYNFESEKTAEAEADQEEEEVKQD 201
Query 61 KAAEKGAIRF 70
KA ++G + F
Sbjct 202 KAEKRGVVAF 211
> tpv:TP03_0704 hypothetical protein; K14831 protein MAK16
Length=232
Score = 53.9 bits (128), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 24/39 (61%), Positives = 31/39 (79%), Gaps = 0/39 (0%)
Query 1 ERREAAREKKALVAAHIEDVIEDELLKRLHQGVYGDLYN 39
ERRE+ REKKA +AA ++ I+ ELL RL QG+YG+LYN
Sbjct 141 ERRESVREKKAEIAAKLDKSIQQELLNRLSQGIYGELYN 179
> cpv:cgd6_1820 hypothetical protein ; K14831 protein MAK16
Length=258
Score = 49.3 bits (116), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 25/39 (64%), Positives = 31/39 (79%), Gaps = 0/39 (0%)
Query 1 ERREAAREKKALVAAHIEDVIEDELLKRLHQGVYGDLYN 39
ERREA+RE KAL AA IE+ IE+ELL RL +G+Y +YN
Sbjct 142 ERREASREHKALNAAKIENEIENELLLRLKEGLYDGIYN 180
> bbo:BBOV_III001370 17.m07141; Mak16 family protein; K14831 protein
MAK16
Length=226
Score = 48.9 bits (115), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 23/38 (60%), Positives = 29/38 (76%), Gaps = 0/38 (0%)
Query 1 ERREAAREKKALVAAHIEDVIEDELLKRLHQGVYGDLY 38
ERRE REKKA +AA +++ IE ELL RL +G YG+LY
Sbjct 141 ERREKIREKKAEIAARLDNAIEQELLNRLSRGTYGELY 178
> pfa:PFB0175c nucleolar preribosomal assembly protein, putative;
K14831 protein MAK16
Length=442
Score = 48.9 bits (115), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 22/38 (57%), Positives = 29/38 (76%), Gaps = 0/38 (0%)
Query 1 ERREAAREKKALVAAHIEDVIEDELLKRLHQGVYGDLY 38
ERR+ REKKAL AA++ + +E ELL RL+ G+YG LY
Sbjct 142 ERRDKTREKKALKAANLLNNVEKELLNRLNTGIYGSLY 179
> dre:100334042 Protein MAK16 homolog; K14831 protein MAK16
Length=305
Score = 45.8 bits (107), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 19/29 (65%), Positives = 23/29 (79%), Gaps = 0/29 (0%)
Query 11 ALVAAHIEDVIEDELLKRLHQGVYGDLYN 39
AL+AA +E+ IE ELL RL QG YGD+YN
Sbjct 152 ALIAAQLENAIEKELLDRLKQGTYGDIYN 180
> dre:192312 mak16, chunp6925, fb34e01, mak16l, wu:fb34e01; MAK16
homolog (S. cerevisiae); K14831 protein MAK16
Length=303
Score = 45.8 bits (107), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 19/29 (65%), Positives = 23/29 (79%), Gaps = 0/29 (0%)
Query 11 ALVAAHIEDVIEDELLKRLHQGVYGDLYN 39
AL+AA +E+ IE ELL RL QG YGD+YN
Sbjct 152 ALIAAQLENAIEKELLDRLKQGTYGDIYN 180
> sce:YAL025C MAK16; Essential nuclear protein, constituent of
66S pre-ribosomal particles; required for maturation of 25S
and 5.8S rRNAs; required for maintenance of M1 satellite double-stranded
RNA of the L-A virus; K14831 protein MAK16
Length=306
Score = 45.8 bits (107), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 23/36 (63%), Positives = 26/36 (72%), Gaps = 0/36 (0%)
Query 1 ERREAAREKKALVAAHIEDVIEDELLKRLHQGVYGD 36
+RRE RE+KALVAA IE IE EL+ RL G YGD
Sbjct 142 KRREQNRERKALVAAKIEKAIEKELMDRLKSGAYGD 177
> xla:380167 mak16-b, MGC53057, mak16, mak16l, rbm13; MAK16 homolog;
K14831 protein MAK16
Length=304
Score = 45.1 bits (105), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 19/29 (65%), Positives = 24/29 (82%), Gaps = 0/29 (0%)
Query 11 ALVAAHIEDVIEDELLKRLHQGVYGDLYN 39
ALVAA +++ IE ELL+RL QG YGD+YN
Sbjct 152 ALVAAQLDNAIEKELLERLKQGAYGDIYN 180
> xla:447074 mak16-a, MGC85217, mak16l, rbm13; MAK16 homolog;
K14831 protein MAK16
Length=301
Score = 44.7 bits (104), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 19/29 (65%), Positives = 24/29 (82%), Gaps = 0/29 (0%)
Query 11 ALVAAHIEDVIEDELLKRLHQGVYGDLYN 39
ALVAA +++ IE ELL+RL QG YGD+YN
Sbjct 152 ALVAAQLDNAIEKELLERLKQGAYGDIYN 180
> cel:C16A3.6 hypothetical protein; K14831 protein MAK16
Length=323
Score = 42.4 bits (98), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 19/29 (65%), Positives = 22/29 (75%), Gaps = 0/29 (0%)
Query 11 ALVAAHIEDVIEDELLKRLHQGVYGDLYN 39
ALVAA ++ IE ELL RL QG YGD+YN
Sbjct 152 ALVAAKLDHAIEKELLARLKQGTYGDVYN 180
> hsa:84549 MAK16, MAK16L, RBM13; MAK16 homolog (S. cerevisiae);
K14831 protein MAK16
Length=300
Score = 41.6 bits (96), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 17/29 (58%), Positives = 23/29 (79%), Gaps = 0/29 (0%)
Query 11 ALVAAHIEDVIEDELLKRLHQGVYGDLYN 39
AL+AA +++ IE ELL+RL Q YGD+YN
Sbjct 152 ALIAAQLDNAIEKELLERLKQDTYGDIYN 180
> mmu:67920 Mak16, 2600016B03Rik, AI314911, Rbm13; MAK16 homolog
(S. cerevisiae); K14831 protein MAK16
Length=296
Score = 41.6 bits (96), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 17/29 (58%), Positives = 23/29 (79%), Gaps = 0/29 (0%)
Query 11 ALVAAHIEDVIEDELLKRLHQGVYGDLYN 39
AL+AA +++ IE ELL+RL Q YGD+YN
Sbjct 152 ALIAAQLDNAIEKELLERLKQDTYGDIYN 180
> ath:AT1G23280 MAK16 protein-related; K14831 protein MAK16
Length=303
Score = 36.2 bits (82), Expect = 0.065, Method: Compositional matrix adjust.
Identities = 18/39 (46%), Positives = 30/39 (76%), Gaps = 1/39 (2%)
Query 2 RREAAREKKALVAAHIEDVIEDELLKRLHQGVY-GDLYN 39
+RE+ RE+KA+ AA ++ IE EL++RL +G+Y ++YN
Sbjct 142 KRESRREEKAIKAAQLDKAIETELMERLKKGIYPTEIYN 180
Lambda K H
0.302 0.122 0.316
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5170784960
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40