bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0507_orf1
Length=104
Score E
Sequences producing significant alignments: (Bits) Value
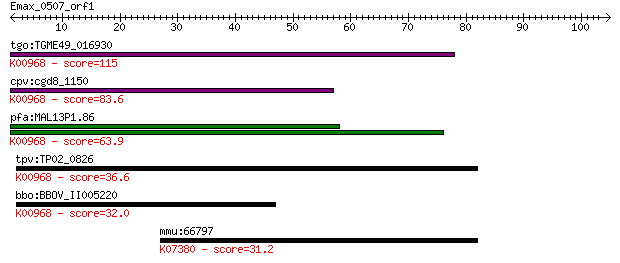
tgo:TGME49_016930 cholinephosphate cytidylyltransferase, putat... 115 3e-26
cpv:cgd8_1150 choline-phosphate cytidylyltransferase ; K00968 ... 83.6 1e-16
pfa:MAL13P1.86 ctp; cholinephosphate cytidylyltransferase (EC:... 63.9 1e-10
tpv:TP02_0826 cholinephosphate cytidylyltransferase; K00968 ch... 36.6 0.020
bbo:BBOV_II005220 18.m06431; choline-phosphate cytidylyltransf... 32.0 0.47
mmu:66797 Cntnap2, 5430425M22Rik, Caspr2, mKIAA0868; contactin... 31.2 0.75
> tgo:TGME49_016930 cholinephosphate cytidylyltransferase, putative
(EC:2.7.7.15); K00968 choline-phosphate cytidylyltransferase
[EC:2.7.7.15]
Length=329
Score = 115 bits (288), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 55/79 (69%), Positives = 63/79 (79%), Gaps = 2/79 (2%)
Query 1 DELTKVTLTDRPLGVHFDESVEKLRNSIHQTYDTWRGRSHRLLRGFARKFEPMKSLIGIH 60
DELTKVTLTDRPLGV+FDESVEKLRNSIH+ YD+WR SH+ L+GFAR FEPMKS IG+H
Sbjct 250 DELTKVTLTDRPLGVNFDESVEKLRNSIHEKYDSWRAHSHKFLKGFARTFEPMKSFIGLH 309
Query 61 VN--HHQSSDDDAAISDGS 77
H S ++D A SD S
Sbjct 310 HKSPRHTSDEEDGAASDAS 328
> cpv:cgd8_1150 choline-phosphate cytidylyltransferase ; K00968
choline-phosphate cytidylyltransferase [EC:2.7.7.15]
Length=341
Score = 83.6 bits (205), Expect = 1e-16, Method: Composition-based stats.
Identities = 37/56 (66%), Positives = 47/56 (83%), Gaps = 0/56 (0%)
Query 1 DELTKVTLTDRPLGVHFDESVEKLRNSIHQTYDTWRGRSHRLLRGFARKFEPMKSL 56
+ELTKVTLTDRPLG+ FDESV+ +RN IH+++D WRG S + L GFAR F+PM+SL
Sbjct 271 NELTKVTLTDRPLGITFDESVDNIRNQIHKSFDAWRGVSKKYLEGFARTFDPMRSL 326
> pfa:MAL13P1.86 ctp; cholinephosphate cytidylyltransferase (EC:2.7.7.15);
K00968 choline-phosphate cytidylyltransferase [EC:2.7.7.15]
Length=896
Score = 63.9 bits (154), Expect = 1e-10, Method: Composition-based stats.
Identities = 28/57 (49%), Positives = 41/57 (71%), Gaps = 0/57 (0%)
Query 1 DELTKVTLTDRPLGVHFDESVEKLRNSIHQTYDTWRGRSHRLLRGFARKFEPMKSLI 57
DELTKVTLTD+PLG FD+ ++ +R+ +H + WR S +LL+ FA+ F+PM +I
Sbjct 229 DELTKVTLTDKPLGTDFDQGIDIIRDKVHDLFKLWRYHSKKLLKDFAKSFDPMFIII 285
Score = 57.8 bits (138), Expect = 8e-09, Method: Composition-based stats.
Identities = 31/76 (40%), Positives = 46/76 (60%), Gaps = 1/76 (1%)
Query 1 DELTKVTLTDRPLGVHFDESVEKLRNSIHQTYDTWRGRSHRLLRGFARKFEPMKSLIGI- 59
DELTKVTLTD+PLG FD+ VE L+ + + W+ S++L+ F RK E L I
Sbjct 814 DELTKVTLTDKPLGTDFDQGVENLQVKFKELFKIWKNASNKLITDFTRKLEATSYLTSIQ 873
Query 60 HVNHHQSSDDDAAISD 75
++ ++ +DD A S+
Sbjct 874 NIIDYEIENDDYASSN 889
> tpv:TP02_0826 cholinephosphate cytidylyltransferase; K00968
choline-phosphate cytidylyltransferase [EC:2.7.7.15]
Length=523
Score = 36.6 bits (83), Expect = 0.020, Method: Composition-based stats.
Identities = 21/95 (22%), Positives = 44/95 (46%), Gaps = 15/95 (15%)
Query 2 ELTKVTLTDRPLGVHFDESVEKLRNSIHQTYDTWRGRSHRLLRGFAR------------- 48
++T++TLTD PLG F++ ++ +R+S+ + W + +L F
Sbjct 198 QITQLTLTDEPLGAGFNDRIDLVRSSLFKAIAIWLNKHKIVLEDFKEIPYVDPAETVYSK 257
Query 49 --KFEPMKSLIGIHVNHHQSSDDDAAISDGSGLSD 81
EP++S+ +++ DD+ + G+ D
Sbjct 258 SVTMEPVESIPKLNLPSGNLKDDEEVVVYTYGVFD 292
> bbo:BBOV_II005220 18.m06431; choline-phosphate cytidylyltransferase
(EC:2.7.7.15); K00968 choline-phosphate cytidylyltransferase
[EC:2.7.7.15]
Length=546
Score = 32.0 bits (71), Expect = 0.47, Method: Compositional matrix adjust.
Identities = 17/45 (37%), Positives = 24/45 (53%), Gaps = 4/45 (8%)
Query 2 ELTKVTLTDRPLGVHFDESVEKLRNSIHQTYDTWRGRSHRLLRGF 46
E+ K TLTD P+G FD ++K I + D WR S ++ F
Sbjct 496 EVHKATLTDHPVGHEFDRLIDK----IVEVVDGWRKDSKVMIENF 536
> mmu:66797 Cntnap2, 5430425M22Rik, Caspr2, mKIAA0868; contactin
associated protein-like 2; K07380 contactin associated protein-like
2
Length=1332
Score = 31.2 bits (69), Expect = 0.75, Method: Composition-based stats.
Identities = 18/60 (30%), Positives = 30/60 (50%), Gaps = 5/60 (8%)
Query 27 SIHQTYDTWRGRSHRLLRGFAR-----KFEPMKSLIGIHVNHHQSSDDDAAISDGSGLSD 81
S+ + TW L FA + + ++S +G+H+N+ Q+ IS GSGL+D
Sbjct 396 SVSFQFRTWNPSGLLLFSHFADNLGNVEIDLVESKVGVHINNTQTKTSQIDISSGSGLND 455
Lambda K H
0.311 0.128 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2022291452
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40