bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0446_orf3
Length=216
Score E
Sequences producing significant alignments: (Bits) Value
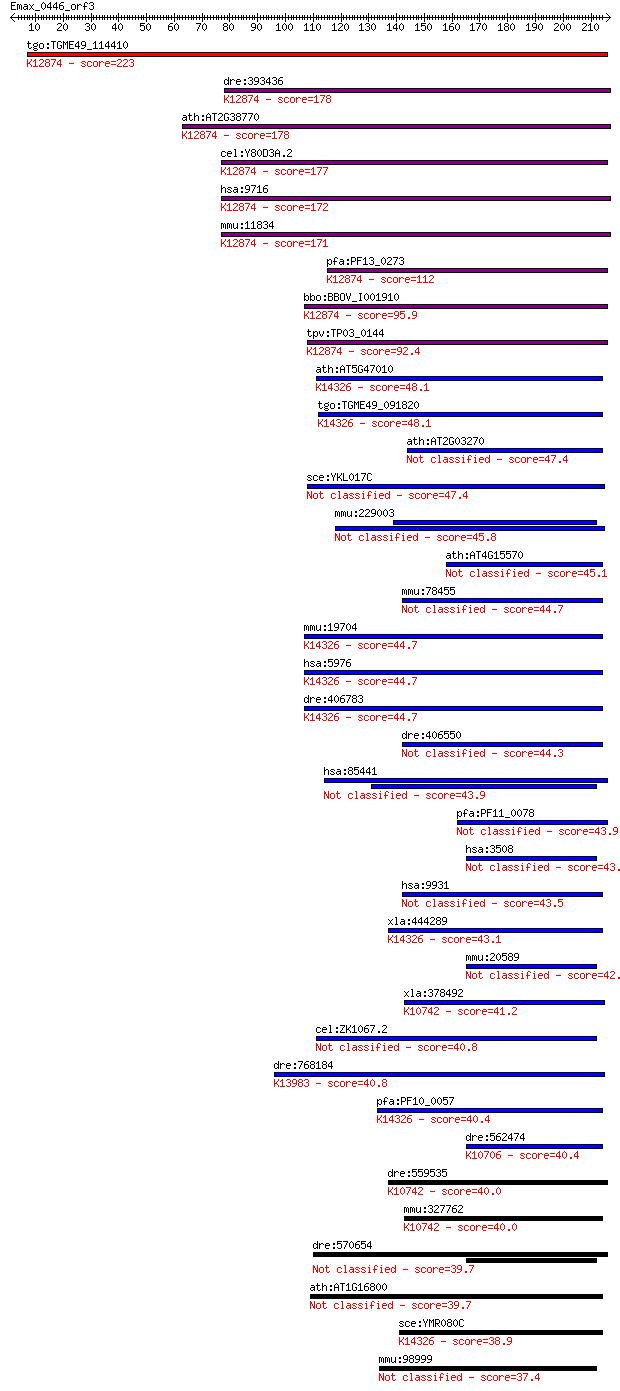
tgo:TGME49_114410 hypothetical protein ; K12874 intron-binding... 223 3e-58
dre:393436 MGC63611; zgc:63611; K12874 intron-binding protein ... 178 1e-44
ath:AT2G38770 EMB2765 (EMBRYO DEFECTIVE 2765); K12874 intron-b... 178 1e-44
cel:Y80D3A.2 emb-4; abnormal EMBroygenesis family member (emb-... 177 4e-44
hsa:9716 AQR, DKFZp686B23123, IBP160, KIAA0560, fSAP164; aquar... 172 9e-43
mmu:11834 Aqr, AW495846, mKIAA0560; aquarius; K12874 intron-bi... 171 1e-42
pfa:PF13_0273 conserved Plasmodium protein, unknown function; ... 112 1e-24
bbo:BBOV_I001910 19.m02343; hypothetical protein; K12874 intro... 95.9 9e-20
tpv:TP03_0144 hypothetical protein; K12874 intron-binding prot... 92.4 9e-19
ath:AT5G47010 LBA1; LBA1 (LOW-LEVEL BETA-AMYLASE 1); ATP bindi... 48.1 2e-05
tgo:TGME49_091820 regulator of nonsense transcripts UPF1, puta... 48.1 2e-05
ath:AT2G03270 DNA-binding protein, putative 47.4 4e-05
sce:YKL017C HCS1, DIP1; Hcs1p (EC:3.6.1.-) 47.4 4e-05
mmu:229003 MGC63178, Pric285, mKIAA1769, mPDIP1; cDNA sequence... 45.8 1e-04
ath:AT4G15570 MAA3; MAA3 (MAGATAMA 3) 45.1 2e-04
mmu:78455 Helz, 3110078M01Rik, 9430093I07Rik, 9630002H22Rik, A... 44.7 2e-04
mmu:19704 Upf1, B430202H16Rik, NORF1, PNORF-1, Rent1, Upflp; U... 44.7 3e-04
hsa:5976 UPF1, FLJ43809, FLJ46894, HUPF1, KIAA0221, NORF1, REN... 44.7 3e-04
dre:406783 upf1, rent1, wu:fi40f07, wu:fj48a01, zgc:55472; upf... 44.7 3e-04
dre:406550 helz, wu:fd51f08; zgc:77407 (EC:3.6.1.-) 44.3 4e-04
hsa:85441 PRIC285, FLJ00244, KIAA1769, MGC132634, MGC138228, P... 43.9 4e-04
pfa:PF11_0078 conserved Plasmodium protein 43.9 4e-04
hsa:3508 IGHMBP2, CATF1, FLJ34220, FLJ41171, HCSA, HMN6, SMARD... 43.9 5e-04
hsa:9931 HELZ, DHRC, DKFZp586G1924, DRHC, HUMORF5, KIAA0054, M... 43.5 5e-04
xla:444289 MGC80941 protein; K14326 regulator of nonsense tran... 43.1 6e-04
mmu:20589 Ighmbp2, AEP, Catf1, RIPE3b1, Smbp-2, Smbp2, Smubp2,... 42.0 0.002
xla:378492 dna2, XDna2, dna2-A; DNA replication helicase 2 hom... 41.2 0.002
cel:ZK1067.2 hypothetical protein 40.8 0.003
dre:768184 zgc:154086; K13983 putative helicase MOV10L1 [EC:3.... 40.8 0.004
pfa:PF10_0057 regulator of nonsense transcripts, putative; K14... 40.4 0.005
dre:562474 setx, wu:fj92h09; senataxin; K10706 senataxin [EC:3... 40.4 0.005
dre:559535 si:ch211-1n9.7; K10742 DNA replication ATP-dependen... 40.0 0.006
mmu:327762 Dna2, Dna2l, E130315B21Rik, KIAA0083; DNA replicati... 40.0 0.007
dre:570654 si:dkey-97a13.6 39.7 0.008
ath:AT1G16800 tRNA-splicing endonuclease positive effector-rel... 39.7 0.009
sce:YMR080C NAM7, IFS2, MOF4, SUP113, UPF1; ATP-dependent RNA ... 38.9 0.014
mmu:98999 Znfx1, AI481105; zinc finger, NFX1-type containing 1 37.4
> tgo:TGME49_114410 hypothetical protein ; K12874 intron-binding
protein aquarius
Length=2273
Score = 223 bits (569), Expect = 3e-58, Method: Compositional matrix adjust.
Identities = 114/211 (54%), Positives = 147/211 (69%), Gaps = 2/211 (0%)
Query 7 TQAEVGNEQQSADQQNEEQEEEEEEEHQQQQLEG-EAEAQEFGEKSSVPEALFAVDVATA 65
+ E + Q D +E+E++E+ ++ E+ FG +V ++ A
Sbjct 1417 VKVEKARQVQGEDAHETNGDEQEQDENACISVDPVSVESSSFGRFCTVFSGGLSLASQRA 1476
Query 66 HYEYGKSFEELLFPFTSFFSDAPQPLFPGEAAADAAVAESCFRYMDQMQQQLQDYRAFEL 125
E+ ++ ELLFPF FF+DAPQPLF + A D +AE CFRY+D++ QQ+ D RAFEL
Sbjct 1477 MREFRHTYFELLFPFPLFFADAPQPLFVKDKAKDEEMAECCFRYIDRIFQQIDDCRAFEL 1536
Query 126 LRTAGDRADFLLTTHARMVAMTCTHAAITRERLVELNFTFDSLIIEEAAQILEVETFIPM 185
LRTAGDRA++L+T HARM+AMTCTHAA+TRE LV L F +D+LIIEEAAQILEVETFIPM
Sbjct 1537 LRTAGDRANYLITKHARMIAMTCTHAAMTRENLVGLKFQYDTLIIEEAAQILEVETFIPM 1596
Query 186 LLQKA-ADKATRLKRVVLLGDHHQLPPIVKH 215
LLQ+ +RLKRVVL+GDHHQLPPIVKH
Sbjct 1597 LLQQLERGGVSRLKRVVLIGDHHQLPPIVKH 1627
> dre:393436 MGC63611; zgc:63611; K12874 intron-binding protein
aquarius
Length=1525
Score = 178 bits (452), Expect = 1e-44, Method: Compositional matrix adjust.
Identities = 76/140 (54%), Positives = 111/140 (79%), Gaps = 1/140 (0%)
Query 78 FPFTSFFSDAPQPLFPGEA-AADAAVAESCFRYMDQMQQQLQDYRAFELLRTAGDRADFL 136
FPF +FS+APQP+F G++ D +AE C+R++ ++ QL+++RAFELLR+ DR+ +L
Sbjct 967 FPFHKYFSNAPQPVFRGQSFQEDMDIAEGCYRHIRKIFTQLEEFRAFELLRSGLDRSKYL 1026
Query 137 LTTHARMVAMTCTHAAITRERLVELNFTFDSLIIEEAAQILEVETFIPMLLQKAADKATR 196
L A+++AMTCTHAA+ R LVEL F +D++++EEAAQILE+ETFIP+LLQ D +R
Sbjct 1027 LVKEAKIIAMTCTHAALKRHDLVELGFKYDNILMEEAAQILEIETFIPLLLQNPEDGYSR 1086
Query 197 LKRVVLLGDHHQLPPIVKHM 216
LKR +++GDHHQLPP++K+M
Sbjct 1087 LKRWIMIGDHHQLPPVIKNM 1106
> ath:AT2G38770 EMB2765 (EMBRYO DEFECTIVE 2765); K12874 intron-binding
protein aquarius
Length=1509
Score = 178 bits (452), Expect = 1e-44, Method: Compositional matrix adjust.
Identities = 82/155 (52%), Positives = 114/155 (73%), Gaps = 1/155 (0%)
Query 63 ATAHYEYGKSFEELLFPFTSFFSDAPQPLFPGEA-AADAAVAESCFRYMDQMQQQLQDYR 121
A A E +SF FPF FFSD P+P+F GE+ D A+ CF ++ + Q+L++ R
Sbjct 1014 ACAGNEDNQSFVRDRFPFKDFFSDTPKPVFNGESFEKDMRAAKGCFSHLKTVFQELEECR 1073
Query 122 AFELLRTAGDRADFLLTTHARMVAMTCTHAAITRERLVELNFTFDSLIIEEAAQILEVET 181
AFELL++ DRA++L+T A++VAMTCTHAA+ R ++L F +D+L++EE+AQILE+ET
Sbjct 1074 AFELLKSTADRANYLMTKQAKIVAMTCTHAALKRRDFLQLGFKYDNLLMEESAQILEIET 1133
Query 182 FIPMLLQKAADKATRLKRVVLLGDHHQLPPIVKHM 216
FIPMLLQ+ D RLKR +L+GDHHQLPP+VK+M
Sbjct 1134 FIPMLLQRQEDGHARLKRCILIGDHHQLPPVVKNM 1168
> cel:Y80D3A.2 emb-4; abnormal EMBroygenesis family member (emb-4);
K12874 intron-binding protein aquarius
Length=1467
Score = 177 bits (448), Expect = 4e-44, Method: Compositional matrix adjust.
Identities = 77/139 (55%), Positives = 107/139 (76%), Gaps = 1/139 (0%)
Query 77 LFPFTSFFSDAPQPLFPGEAAADAAVAESCFRYMDQMQQQLQDYRAFELLRTAGDRADFL 136
+FPFT FF D P LF G +AD VA SC+R+++Q+ ++L ++RAFELLR DR ++L
Sbjct 971 IFPFTGFFKDIPD-LFSGNNSADLKVAHSCWRHIEQIFEKLDEFRAFELLRNGRDRTEYL 1029
Query 137 LTTHARMVAMTCTHAAITRERLVELNFTFDSLIIEEAAQILEVETFIPMLLQKAADKATR 196
L A+++AMTCTHAA+ R LV+L F +D++++EEAAQILEVETFIP+LLQ D R
Sbjct 1030 LVKEAKIIAMTCTHAALRRNELVKLGFRYDNIVMEEAAQILEVETFIPLLLQNPQDGHNR 1089
Query 197 LKRVVLLGDHHQLPPIVKH 215
LKR +++GDHHQLPP+V++
Sbjct 1090 LKRWIMIGDHHQLPPVVQN 1108
> hsa:9716 AQR, DKFZp686B23123, IBP160, KIAA0560, fSAP164; aquarius
homolog (mouse); K12874 intron-binding protein aquarius
Length=1485
Score = 172 bits (435), Expect = 9e-43, Method: Compositional matrix adjust.
Identities = 74/141 (52%), Positives = 109/141 (77%), Gaps = 1/141 (0%)
Query 77 LFPFTSFFSDAPQPLFPGEAAA-DAAVAESCFRYMDQMQQQLQDYRAFELLRTAGDRADF 135
FPF +F++APQP+F G + D +AE CFR++ ++ QL+++RA ELLR+ DR+ +
Sbjct 967 FFPFHEYFANAPQPIFKGRSYEEDMEIAEGCFRHIKKIFTQLEEFRASELLRSGLDRSKY 1026
Query 136 LLTTHARMVAMTCTHAAITRERLVELNFTFDSLIIEEAAQILEVETFIPMLLQKAADKAT 195
LL A+++AMTCTHAA+ R LV+L F +D++++EEAAQILE+ETFIP+LLQ D +
Sbjct 1027 LLVKEAKIIAMTCTHAALKRHDLVKLGFKYDNILMEEAAQILEIETFIPLLLQNPQDGFS 1086
Query 196 RLKRVVLLGDHHQLPPIVKHM 216
RLKR +++GDHHQLPP++K+M
Sbjct 1087 RLKRWIMIGDHHQLPPVIKNM 1107
> mmu:11834 Aqr, AW495846, mKIAA0560; aquarius; K12874 intron-binding
protein aquarius
Length=1481
Score = 171 bits (434), Expect = 1e-42, Method: Compositional matrix adjust.
Identities = 74/141 (52%), Positives = 109/141 (77%), Gaps = 1/141 (0%)
Query 77 LFPFTSFFSDAPQPLFPGEAAA-DAAVAESCFRYMDQMQQQLQDYRAFELLRTAGDRADF 135
FPF +F++APQP+F G + D +AE CFR++ ++ QL+++RA ELLR+ DR+ +
Sbjct 971 FFPFHEYFANAPQPIFKGRSYEEDMEIAEGCFRHIKKIFTQLEEFRASELLRSGLDRSKY 1030
Query 136 LLTTHARMVAMTCTHAAITRERLVELNFTFDSLIIEEAAQILEVETFIPMLLQKAADKAT 195
LL A+++AMTCTHAA+ R LV+L F +D++++EEAAQILE+ETFIP+LLQ D +
Sbjct 1031 LLVKEAKIIAMTCTHAALKRHDLVKLGFKYDNILMEEAAQILEIETFIPLLLQNPQDGFS 1090
Query 196 RLKRVVLLGDHHQLPPIVKHM 216
RLKR +++GDHHQLPP++K+M
Sbjct 1091 RLKRWIMIGDHHQLPPVIKNM 1111
> pfa:PF13_0273 conserved Plasmodium protein, unknown function;
K12874 intron-binding protein aquarius
Length=2533
Score = 112 bits (279), Expect = 1e-24, Method: Composition-based stats.
Identities = 47/101 (46%), Positives = 75/101 (74%), Gaps = 0/101 (0%)
Query 115 QQLQDYRAFELLRTAGDRADFLLTTHARMVAMTCTHAAITRERLVELNFTFDSLIIEEAA 174
+ L+D RAFE+LR +R +++ AR++AMTCTHA+I R ++ +L F FD++II+E
Sbjct 2159 EHLKDCRAFEVLRNQRERCTYIIAKLARVIAMTCTHASINRSKIAKLQFYFDNIIIDECT 2218
Query 175 QILEVETFIPMLLQKAADKATRLKRVVLLGDHHQLPPIVKH 215
QI E +TF+P+LLQ+ ++LKR++ +GD +QLPPI+K+
Sbjct 2219 QITENDTFLPLLLQENRYYKSKLKRIIFVGDSNQLPPIIKN 2259
> bbo:BBOV_I001910 19.m02343; hypothetical protein; K12874 intron-binding
protein aquarius
Length=1554
Score = 95.9 bits (237), Expect = 9e-20, Method: Composition-based stats.
Identities = 51/109 (46%), Positives = 70/109 (64%), Gaps = 13/109 (11%)
Query 107 FRYMDQMQQQLQDYRAFELLRTAGDRADFLLTTHARMVAMTCTHAAITRERLVELNFTFD 166
RY M ++LQ FE+LR DR +L+ +AR+VAMTCTHAAI RE L N +
Sbjct 1210 LRYFHSMLKELQ---PFEVLRNNYDRGRYLVEKYARLVAMTCTHAAIARETLS--NLRYS 1264
Query 167 SLIIEEAAQILEVETFIPMLLQKAADKATRLKRVVLLGDHHQLPPIVKH 215
+L++EEAAQ++E ETF A A LKR++L GDH+QLPP++ +
Sbjct 1265 NLVMEEAAQVMEAETF--------ALLAHPLKRIILSGDHYQLPPVINN 1305
> tpv:TP03_0144 hypothetical protein; K12874 intron-binding protein
aquarius
Length=1766
Score = 92.4 bits (228), Expect = 9e-19, Method: Composition-based stats.
Identities = 45/108 (41%), Positives = 74/108 (68%), Gaps = 10/108 (9%)
Query 108 RYMDQMQQQLQDYRAFELLRTAGDRADFLLTTHARMVAMTCTHAAITRERLVELNFTFDS 167
++++++ + L + FE+LR DR +L+ ++R+VAMTCTHA+I++E L LN+ +
Sbjct 1382 KFIERLYEMLFELLPFEILRNNRDRMKYLVENYSRIVAMTCTHASISQEELSTLNYK--T 1439
Query 168 LIIEEAAQILEVETFIPMLLQKAADKATRLKRVVLLGDHHQLPPIVKH 215
L+ EEAAQILE+E+FIP+ +KR++L GDH QL PI+++
Sbjct 1440 LVFEEAAQILEIESFIPI--------CNNIKRLILCGDHLQLSPIIQN 1479
> ath:AT5G47010 LBA1; LBA1 (LOW-LEVEL BETA-AMYLASE 1); ATP binding
/ DNA binding / RNA helicase/ hydrolase; K14326 regulator
of nonsense transcripts 1 [EC:3.6.4.-]
Length=1254
Score = 48.1 bits (113), Expect = 2e-05, Method: Composition-based stats.
Identities = 33/103 (32%), Positives = 54/103 (52%), Gaps = 15/103 (14%)
Query 111 DQMQQQLQDYRAFELLRTAGDRADFLLTTHARMVAMTCTHAAITRERLVELNFTFDSLII 170
+Q + D + ++ L+ A +R +T A ++ TC AA R NF F ++I
Sbjct 603 EQGELSSSDEKKYKNLKRATERE---ITQSADVICCTCVGAADLRLS----NFRFRQVLI 655
Query 171 EEAAQILEVETFIPMLLQKAADKATRLKRVVLLGDHHQLPPIV 213
+E+ Q E E IP++L +K+VVL+GDH QL P++
Sbjct 656 DESTQATEPECLIPLVLG--------VKQVVLVGDHCQLGPVI 690
> tgo:TGME49_091820 regulator of nonsense transcripts UPF1, putative
(EC:3.1.11.5); K14326 regulator of nonsense transcripts
1 [EC:3.6.4.-]
Length=1449
Score = 48.1 bits (113), Expect = 2e-05, Method: Composition-based stats.
Identities = 35/104 (33%), Positives = 53/104 (50%), Gaps = 14/104 (13%)
Query 112 QMQQQLQDYRAFE--LLRTAGDRADFLLTTHARMVAMTCTHAAITRERLVELNFTFDSLI 169
Q+++Q + A + LR RA+ + A ++ TC A R + F F ++
Sbjct 734 QLKEQTGELAAADERRLRLLISRAEMEILQTADVICTTCVGAGDNRLQ----GFRFRQVV 789
Query 170 IEEAAQILEVETFIPMLLQKAADKATRLKRVVLLGDHHQLPPIV 213
I+EAAQ E E IP++L K+VVL+GDH QL P+V
Sbjct 790 IDEAAQATEPECLIPIVLGA--------KQVVLIGDHCQLGPVV 825
> ath:AT2G03270 DNA-binding protein, putative
Length=639
Score = 47.4 bits (111), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 28/70 (40%), Positives = 38/70 (54%), Gaps = 12/70 (17%)
Query 144 VAMTCTHAAITRERLVELNFTFDSLIIEEAAQILEVETFIPMLLQKAADKATRLKRVVLL 203
V +T A+TR+ N TFD +II+E AQ LEV +I +L + R +L
Sbjct 345 VILTTLTGALTRKLD---NRTFDLVIIDEGAQALEVACWIALL---------KGSRCILA 392
Query 204 GDHHQLPPIV 213
GDH QLPP +
Sbjct 393 GDHLQLPPTI 402
> sce:YKL017C HCS1, DIP1; Hcs1p (EC:3.6.1.-)
Length=683
Score = 47.4 bits (111), Expect = 4e-05, Method: Composition-based stats.
Identities = 35/117 (29%), Positives = 64/117 (54%), Gaps = 17/117 (14%)
Query 108 RYMDQMQQQLQDYRAFELLRTAGDRADFL----LTTHARMVAMTCTHAAITRERLVEL-- 161
+ + +Q+ +++ +LLR + +F L +R+V +T H + +RE L L
Sbjct 324 KKLKNYKQRKENWNEIKLLRKDLKKREFKTIKDLIIQSRIV-VTTLHGSSSRE-LCSLYR 381
Query 162 ---NFT-FDSLIIEEAAQILEVETFIPMLLQKAADKATRLKRVVLLGDHHQLPPIVK 214
NF FD+LII+E +Q +E + +IP++ + + ++VL GD+ QLPP +K
Sbjct 382 DDPNFQLFDTLIIDEVSQAMEPQCWIPLIAHQ-----NQFHKLVLAGDNKQLPPTIK 433
> mmu:229003 MGC63178, Pric285, mKIAA1769, mPDIP1; cDNA sequence
BC006779
Length=2947
Score = 45.8 bits (107), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 31/73 (42%), Positives = 38/73 (52%), Gaps = 10/73 (13%)
Query 139 THARMVAMTCTHAAITRERLVELNFTFDSLIIEEAAQILEVETFIPMLLQKAADKATRLK 198
H R+V T + A RE V F F + I+EAAQ+LE E IP+ A L
Sbjct 885 VHHRLVVTTTSQA---RELQVPAGF-FSHIFIDEAAQMLECEALIPL------SYALSLT 934
Query 199 RVVLLGDHHQLPP 211
RVVL GDH Q+ P
Sbjct 935 RVVLAGDHMQVTP 947
Score = 42.4 bits (98), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 30/97 (30%), Positives = 50/97 (51%), Gaps = 12/97 (12%)
Query 118 QDYRAFELLRTAGDRADFLLTTHARMVAMTCTHAAITRERLVELNFTFDSLIIEEAAQIL 177
+D R + R G L H+ ++ TC+ AA +++ + ++I+EA
Sbjct 2611 EDLRVYR--RVLGKARKHELERHS-VILCTCSCAASKSLKILNVR----QILIDEAGMAT 2663
Query 178 EVETFIPMLLQKAADKATRLKRVVLLGDHHQLPPIVK 214
E ET IP++ + +++VVLLGDH QL P+VK
Sbjct 2664 EPETLIPLVC-----FSKTVEKVVLLGDHKQLRPVVK 2695
> ath:AT4G15570 MAA3; MAA3 (MAGATAMA 3)
Length=818
Score = 45.1 bits (105), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 26/56 (46%), Positives = 33/56 (58%), Gaps = 8/56 (14%)
Query 158 LVELNFTFDSLIIEEAAQILEVETFIPMLLQKAADKATRLKRVVLLGDHHQLPPIV 213
L + N FD +II+EAAQ +E T IP+ ATR K+V L+GD QLP V
Sbjct 477 LAKSNRGFDVVIIDEAAQAVEPATLIPL--------ATRCKQVFLVGDPKQLPATV 524
> mmu:78455 Helz, 3110078M01Rik, 9430093I07Rik, 9630002H22Rik,
AI851979, DRHC, KIAA0054; helicase with zinc finger domain
Length=1965
Score = 44.7 bits (104), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 28/75 (37%), Positives = 43/75 (57%), Gaps = 13/75 (17%)
Query 142 RMVAMTCTHAAITRERLVELNFT---FDSLIIEEAAQILEVETFIPMLLQKAADKATRLK 198
R+V +T + T + L +L+ F ++++EAAQ +E ET +P+ L AT+
Sbjct 767 RVVVVTLS----TSQYLCQLDLEPGFFTHVLLDEAAQAMECETIMPLAL------ATKNT 816
Query 199 RVVLLGDHHQLPPIV 213
R+VL GDH QL P V
Sbjct 817 RIVLAGDHMQLSPFV 831
> mmu:19704 Upf1, B430202H16Rik, NORF1, PNORF-1, Rent1, Upflp;
UPF1 regulator of nonsense transcripts homolog (yeast); K14326
regulator of nonsense transcripts 1 [EC:3.6.4.-]
Length=1124
Score = 44.7 bits (104), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 40/124 (32%), Positives = 60/124 (48%), Gaps = 35/124 (28%)
Query 107 FRYMDQMQ-----QQLQD------------YRAFELLRTAGDRADFLLTTHARMVAMTCT 149
R MD M QQL+D YRA + RTA +R L +A ++ TC
Sbjct 571 IRNMDSMPELQKLQQLKDETGELSSADEKRYRALK--RTA-ERE---LLMNADVICCTCV 624
Query 150 HAAITRERLVELNFTFDSLIIEEAAQILEVETFIPMLLQKAADKATRLKRVVLLGDHHQL 209
A RL ++ F S++I+E+ Q E E +P++L K+++L+GDH QL
Sbjct 625 GAG--DPRLAKMQFR--SILIDESTQATEPECMVPVVLGA--------KQLILVGDHCQL 672
Query 210 PPIV 213
P+V
Sbjct 673 GPVV 676
> hsa:5976 UPF1, FLJ43809, FLJ46894, HUPF1, KIAA0221, NORF1, RENT1,
pNORF1; UPF1 regulator of nonsense transcripts homolog
(yeast); K14326 regulator of nonsense transcripts 1 [EC:3.6.4.-]
Length=1118
Score = 44.7 bits (104), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 40/124 (32%), Positives = 60/124 (48%), Gaps = 35/124 (28%)
Query 107 FRYMDQMQ-----QQLQD------------YRAFELLRTAGDRADFLLTTHARMVAMTCT 149
R MD M QQL+D YRA + RTA +R L +A ++ TC
Sbjct 565 IRNMDSMPELQKLQQLKDETGELSSADEKRYRALK--RTA-ERE---LLMNADVICCTCV 618
Query 150 HAAITRERLVELNFTFDSLIIEEAAQILEVETFIPMLLQKAADKATRLKRVVLLGDHHQL 209
A RL ++ F S++I+E+ Q E E +P++L K+++L+GDH QL
Sbjct 619 GAG--DPRLAKMQFR--SILIDESTQATEPECMVPVVLGA--------KQLILVGDHCQL 666
Query 210 PPIV 213
P+V
Sbjct 667 GPVV 670
> dre:406783 upf1, rent1, wu:fi40f07, wu:fj48a01, zgc:55472; upf1
regulator of nonsense transcripts homolog (yeast); K14326
regulator of nonsense transcripts 1 [EC:3.6.4.-]
Length=1100
Score = 44.7 bits (104), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 40/124 (32%), Positives = 60/124 (48%), Gaps = 35/124 (28%)
Query 107 FRYMDQMQ-----QQLQD------------YRAFELLRTAGDRADFLLTTHARMVAMTCT 149
R MD M QQL+D YRA + RTA +R L +A ++ TC
Sbjct 545 IRNMDSMPELQKLQQLKDETGELSSSDEKRYRALK--RTA-ERE---LLMNADVICCTCV 598
Query 150 HAAITRERLVELNFTFDSLIIEEAAQILEVETFIPMLLQKAADKATRLKRVVLLGDHHQL 209
A RL ++ F S++I+E+ Q E E +P++L K+++L+GDH QL
Sbjct 599 GAG--DPRLAKMQFR--SILIDESTQATEPECMVPVVLGA--------KQLILVGDHCQL 646
Query 210 PPIV 213
P+V
Sbjct 647 GPVV 650
> dre:406550 helz, wu:fd51f08; zgc:77407 (EC:3.6.1.-)
Length=1860
Score = 44.3 bits (103), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 28/75 (37%), Positives = 42/75 (56%), Gaps = 13/75 (17%)
Query 142 RMVAMTCTHAAITRERLVELNF---TFDSLIIEEAAQILEVETFIPMLLQKAADKATRLK 198
R+V +T + T + L +L+ F ++++EAAQ +E ET +P+ L A +
Sbjct 759 RVVVVTLS----TSQYLCQLDLEPGIFTHILLDEAAQAMECETIMPLAL------AVKST 808
Query 199 RVVLLGDHHQLPPIV 213
RVVL GDH QL P V
Sbjct 809 RVVLAGDHMQLSPFV 823
> hsa:85441 PRIC285, FLJ00244, KIAA1769, MGC132634, MGC138228,
PDIP-1; peroxisomal proliferator-activated receptor A interacting
complex 285
Length=2649
Score = 43.9 bits (102), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 33/102 (32%), Positives = 53/102 (51%), Gaps = 23/102 (22%)
Query 114 QQQLQDYRAFELLRTAGDRADFLLTTHARMVAMTCTHAAITRERLVELNFTFDSLIIEEA 173
++ L + R FEL DR + +L T C+ AA +++++ ++++EA
Sbjct 2320 KKVLWEARKFEL-----DRHEVILCT--------CSCAASASLKILDVR----QILVDEA 2362
Query 174 AQILEVETFIPMLLQKAADKATRLKRVVLLGDHHQLPPIVKH 215
E ET IP++ A+K VVLLGDH QL P+VK+
Sbjct 2363 GMATEPETLIPLVQFPQAEK------VVLLGDHKQLRPVVKN 2398
Score = 40.8 bits (94), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 34/86 (39%), Positives = 43/86 (50%), Gaps = 15/86 (17%)
Query 131 DRADFLLTTHA-----RMVAMTCTHAAITRERLVELNFTFDSLIIEEAAQILEVETFIPM 185
DR F T A R+V T + A RE V + F F ++I+EAAQ+LE E P+
Sbjct 626 DRQAFRPPTRAELARHRVVVTTTSQA---RELRVPVGF-FSHILIDEAAQMLECEALTPL 681
Query 186 LLQKAADKATRLKRVVLLGDHHQLPP 211
A TRL VL GDH Q+ P
Sbjct 682 ---AYASHGTRL---VLAGDHMQVTP 701
> pfa:PF11_0078 conserved Plasmodium protein
Length=1024
Score = 43.9 bits (102), Expect = 4e-04, Method: Composition-based stats.
Identities = 22/54 (40%), Positives = 30/54 (55%), Gaps = 8/54 (14%)
Query 162 NFTFDSLIIEEAAQILEVETFIPMLLQKAADKATRLKRVVLLGDHHQLPPIVKH 215
NF FD++ I+E Q E +IP+ L K K V L GDH QL P++K+
Sbjct 545 NFLFDAVCIDECCQCTEPLCYIPISLSK--------KNVFLFGDHKQLSPLIKY 590
> hsa:3508 IGHMBP2, CATF1, FLJ34220, FLJ41171, HCSA, HMN6, SMARD1,
SMUBP2; immunoglobulin mu binding protein 2 (EC:3.6.4.13
3.6.4.12)
Length=993
Score = 43.9 bits (102), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 19/47 (40%), Positives = 28/47 (59%), Gaps = 9/47 (19%)
Query 165 FDSLIIEEAAQILEVETFIPMLLQKAADKATRLKRVVLLGDHHQLPP 211
FD ++I+E AQ LE +IP+L + ++ +L GDH QLPP
Sbjct 369 FDVVVIDECAQALEASCWIPLL---------KARKCILAGDHKQLPP 406
> hsa:9931 HELZ, DHRC, DKFZp586G1924, DRHC, HUMORF5, KIAA0054,
MGC163454; helicase with zinc finger
Length=1942
Score = 43.5 bits (101), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 27/72 (37%), Positives = 41/72 (56%), Gaps = 7/72 (9%)
Query 142 RMVAMTCTHAAITRERLVELNFTFDSLIIEEAAQILEVETFIPMLLQKAADKATRLKRVV 201
R+V +T + + +E F F ++++EAAQ +E ET +P+ L AT+ R+V
Sbjct 766 RVVVVTLNTSQYLCQLDLEPGF-FTHILLDEAAQAMECETIMPLAL------ATQNTRIV 818
Query 202 LLGDHHQLPPIV 213
L GDH QL P V
Sbjct 819 LAGDHMQLSPFV 830
> xla:444289 MGC80941 protein; K14326 regulator of nonsense transcripts
1 [EC:3.6.4.-]
Length=1098
Score = 43.1 bits (100), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 25/77 (32%), Positives = 42/77 (54%), Gaps = 12/77 (15%)
Query 137 LTTHARMVAMTCTHAAITRERLVELNFTFDSLIIEEAAQILEVETFIPMLLQKAADKATR 196
L +A ++ TC A RL ++ F S++I+E+ Q E E +P++L
Sbjct 585 LLMNADVICCTCVGAG--DPRLAKMQFR--SILIDESTQATEPECMVPVVLGA------- 633
Query 197 LKRVVLLGDHHQLPPIV 213
K+++L+GDH QL P+V
Sbjct 634 -KQLILVGDHCQLGPVV 649
> mmu:20589 Ighmbp2, AEP, Catf1, RIPE3b1, Smbp-2, Smbp2, Smubp2,
nmd, sma; immunoglobulin mu binding protein 2 (EC:3.6.4.13
3.6.4.12)
Length=993
Score = 42.0 bits (97), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 18/47 (38%), Positives = 27/47 (57%), Gaps = 9/47 (19%)
Query 165 FDSLIIEEAAQILEVETFIPMLLQKAADKATRLKRVVLLGDHHQLPP 211
FD ++++E AQ LE +IP+L + + +L GDH QLPP
Sbjct 368 FDVVVVDECAQALEASCWIPLL---------KAPKCILAGDHRQLPP 405
> xla:378492 dna2, XDna2, dna2-A; DNA replication helicase 2 homolog
(EC:3.6.4.12); K10742 DNA replication ATP-dependent helicase
Dna2 [EC:3.6.4.12]
Length=1053
Score = 41.2 bits (95), Expect = 0.002, Method: Composition-based stats.
Identities = 30/75 (40%), Positives = 38/75 (50%), Gaps = 19/75 (25%)
Query 143 MVAMTC---THAAITRERLVELNFTFDSLIIEEAAQILEVETFIPMLLQKAADKATRLKR 199
+VA TC H TR R FD I++EA+QI + P+ AD R
Sbjct 739 VVATTCMGVNHPIFTRRR-------FDFCIVDEASQISQPICLGPLFF---AD------R 782
Query 200 VVLLGDHHQLPPIVK 214
VL+GDH QLPP+VK
Sbjct 783 FVLVGDHQQLPPLVK 797
> cel:ZK1067.2 hypothetical protein
Length=2219
Score = 40.8 bits (94), Expect = 0.003, Method: Composition-based stats.
Identities = 29/102 (28%), Positives = 52/102 (50%), Gaps = 11/102 (10%)
Query 111 DQMQQQLQDYR-AFELLRTAGDRADFLLTTHARMVAMTCTHAAITRERLVELNFTFDSLI 169
+ + +Q+++YR A E + A +R D + ++ T T + R L ++ LI
Sbjct 1177 ENLPRQIREYREACENFKNAQNRVDAEIMRMTMIIGATTTGCSRLRPTLEKVGPRI--LI 1234
Query 170 IEEAAQILEVETFIPMLLQKAADKATRLKRVVLLGDHHQLPP 211
+EEAA++LE M+ + ++ VV++GDH QL P
Sbjct 1235 VEEAAEVLEAHIISAMI--------STVEHVVMIGDHKQLRP 1268
> dre:768184 zgc:154086; K13983 putative helicase MOV10L1 [EC:3.6.4.13]
Length=1106
Score = 40.8 bits (94), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 38/121 (31%), Positives = 62/121 (51%), Gaps = 19/121 (15%)
Query 96 AAADAAVAESCFRYMDQMQQQLQDY-RAFELLRTAGDRADFLLTTHARMVAMTCTHAAIT 154
+A+ A V +C R + M ++L+ Y RA E +R A + R+V TC+ A +
Sbjct 749 SASLARVNATC-RPEESMSEELRQYARAGEDIRHA---------SFHRIVVSTCSSAGMF 798
Query 155 RERLVELNFTFDSLIIEEAAQILEVETFIPM-LLQKAADKATRLKRVVLLGDHHQLPPIV 213
+ + + F + ++EA Q E ET IP+ LL + + + +VL GD QL P+V
Sbjct 799 YQIGLRVGH-FTHVFVDEAGQATEPETLIPLSLLSETSGQ------IVLAGDPKQLGPVV 851
Query 214 K 214
K
Sbjct 852 K 852
> pfa:PF10_0057 regulator of nonsense transcripts, putative; K14326
regulator of nonsense transcripts 1 [EC:3.6.4.-]
Length=1554
Score = 40.4 bits (93), Expect = 0.005, Method: Composition-based stats.
Identities = 25/81 (30%), Positives = 41/81 (50%), Gaps = 12/81 (14%)
Query 133 ADFLLTTHARMVAMTCTHAAITRERLVELNFTFDSLIIEEAAQILEVETFIPMLLQKAAD 192
A+ + A ++ TC A R + F F ++++EA Q E E +P++
Sbjct 920 AEHEILIEADVICCTCVGAMDKRLK----KFRFRQVLVDEATQSTEPECLVPLV------ 969
Query 193 KATRLKRVVLLGDHHQLPPIV 213
T K++VL+GDH QL PI+
Sbjct 970 --TGAKQIVLVGDHCQLGPII 988
> dre:562474 setx, wu:fj92h09; senataxin; K10706 senataxin [EC:3.6.4.-]
Length=2046
Score = 40.4 bits (93), Expect = 0.005, Method: Composition-based stats.
Identities = 23/49 (46%), Positives = 29/49 (59%), Gaps = 8/49 (16%)
Query 165 FDSLIIEEAAQILEVETFIPMLLQKAADKATRLKRVVLLGDHHQLPPIV 213
F +II+EA+Q E ET IPML R V+L+GD +QLPP V
Sbjct 1633 FSCVIIDEASQAKETETLIPMLY--------RCPSVILVGDPNQLPPTV 1673
> dre:559535 si:ch211-1n9.7; K10742 DNA replication ATP-dependent
helicase Dna2 [EC:3.6.4.12]
Length=1397
Score = 40.0 bits (92), Expect = 0.006, Method: Composition-based stats.
Identities = 28/82 (34%), Positives = 40/82 (48%), Gaps = 19/82 (23%)
Query 137 LTTHARMVAMTCT---HAAITRERLVELNFTFDSLIIEEAAQILEVETFIPMLLQKAADK 193
L + +VA TC H +R R FD I++EA+QI + P+ +
Sbjct 1049 LYSRELIVATTCMGVKHPIFSRRR-------FDFCIVDEASQISQPVCIGPLFYAQ---- 1097
Query 194 ATRLKRVVLLGDHHQLPPIVKH 215
R VL+GDH QLPPIV++
Sbjct 1098 -----RFVLVGDHQQLPPIVQN 1114
> mmu:327762 Dna2, Dna2l, E130315B21Rik, KIAA0083; DNA replication
helicase 2 homolog (yeast) (EC:3.6.4.12); K10742 DNA replication
ATP-dependent helicase Dna2 [EC:3.6.4.12]
Length=1062
Score = 40.0 bits (92), Expect = 0.007, Method: Composition-based stats.
Identities = 26/74 (35%), Positives = 37/74 (50%), Gaps = 19/74 (25%)
Query 143 MVAMTC---THAAITRERLVELNFTFDSLIIEEAAQILEVETFIPMLLQKAADKATRLKR 199
+VA TC H +R+ TFD I++EA+QI + P+ + R
Sbjct 739 IVATTCMGINHPIFSRK-------TFDFCIVDEASQISQPVCLGPLFFSR---------R 782
Query 200 VVLLGDHHQLPPIV 213
VL+GDH QLPP+V
Sbjct 783 FVLVGDHQQLPPLV 796
> dre:570654 si:dkey-97a13.6
Length=2781
Score = 39.7 bits (91), Expect = 0.008, Method: Composition-based stats.
Identities = 32/106 (30%), Positives = 54/106 (50%), Gaps = 23/106 (21%)
Query 110 MDQMQQQLQDYRAFELLRTAGDRADFLLTTHARMVAMTCTHAAITRERLVELNFTFDSLI 169
+++ ++ L D R EL +R D +L T CT AA ++ + ++
Sbjct 2451 IEEYKKLLNDARLHEL-----ERHDIILCT--------CTAAASPN---LKKTLSARQIL 2494
Query 170 IEEAAQILEVETFIPMLLQKAADKATRLKRVVLLGDHHQLPPIVKH 215
I+E A E +T +P++ + + ++VVLLGDH QL PIVK+
Sbjct 2495 IDECAMATEPQTLVPLV-------SFKPEKVVLLGDHKQLRPIVKN 2533
Score = 35.8 bits (81), Expect = 0.11, Method: Composition-based stats.
Identities = 22/47 (46%), Positives = 30/47 (63%), Gaps = 6/47 (12%)
Query 165 FDSLIIEEAAQILEVETFIPMLLQKAADKATRLKRVVLLGDHHQLPP 211
F ++I+EA+Q+LE E + + L ADK TR VVL GDH Q+ P
Sbjct 681 FTHILIDEASQMLEGEALMALGL---ADKHTR---VVLAGDHMQMAP 721
> ath:AT1G16800 tRNA-splicing endonuclease positive effector-related
Length=1939
Score = 39.7 bits (91), Expect = 0.009, Method: Composition-based stats.
Identities = 34/117 (29%), Positives = 54/117 (46%), Gaps = 17/117 (14%)
Query 109 YMDQMQQQLQDYRAFELLRTAGDRADFLLTTHARMVAMTCTHA-----AITRERLVELNF 163
Y D Q Q+ +A +RT + + A++V T + ++ E L F
Sbjct 1361 YKDLSAVQAQERKANYEMRTLKQKLRKSILKEAQIVVTTLSGCGGDLYSVCAESLAAHKF 1420
Query 164 -------TFDSLIIEEAAQILEVETFIPMLLQKAADKATRLKRVVLLGDHHQLPPIV 213
FD+++I+EAAQ LE T IP+ L K +R + +++GD QLP V
Sbjct 1421 GSPSEDNLFDAVVIDEAAQALEPATLIPLQLLK-----SRGTKCIMVGDPKQLPATV 1472
> sce:YMR080C NAM7, IFS2, MOF4, SUP113, UPF1; ATP-dependent RNA
helicase of the SFI superfamily involved in nonsense mediated
mRNA decay; required for efficient translation termination
at nonsense codons and targeting of NMD substrates to P-bodies;
involved in telomere maintenance (EC:3.6.1.-); K14326
regulator of nonsense transcripts 1 [EC:3.6.4.-]
Length=971
Score = 38.9 bits (89), Expect = 0.014, Method: Composition-based stats.
Identities = 26/73 (35%), Positives = 40/73 (54%), Gaps = 13/73 (17%)
Query 141 ARMVAMTCTHAAITRERLVELNFTFDSLIIEEAAQILEVETFIPMLLQKAADKATRLKRV 200
A +V TC A R L+ F +++I+E+ Q E E IP++ K A K+V
Sbjct 547 ADVVCCTCVGAGDKR-----LDTKFRTVLIDESTQASEPECLIPIV--KGA------KQV 593
Query 201 VLLGDHHQLPPIV 213
+L+GDH QL P++
Sbjct 594 ILVGDHQQLGPVI 606
> mmu:98999 Znfx1, AI481105; zinc finger, NFX1-type containing
1
Length=1909
Score = 37.4 bits (85), Expect = 0.045, Method: Compositional matrix adjust.
Identities = 28/78 (35%), Positives = 41/78 (52%), Gaps = 10/78 (12%)
Query 134 DFLLTTHARMVAMTCTHAAITRERLVELNFTFDSLIIEEAAQILEVETFIPMLLQKAADK 193
D + A +V MT T AA R+ L ++ +I+EEAA++LE T L KA
Sbjct 964 DLHILKDAEVVGMTTTGAAKYRQILQQVEPRI--VIVEEAAEVLEAHTI--ATLSKAC-- 1017
Query 194 ATRLKRVVLLGDHHQLPP 211
+ ++L+GDH QL P
Sbjct 1018 ----QHLILIGDHQQLRP 1031
Lambda K H
0.317 0.129 0.356
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6945028560
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40