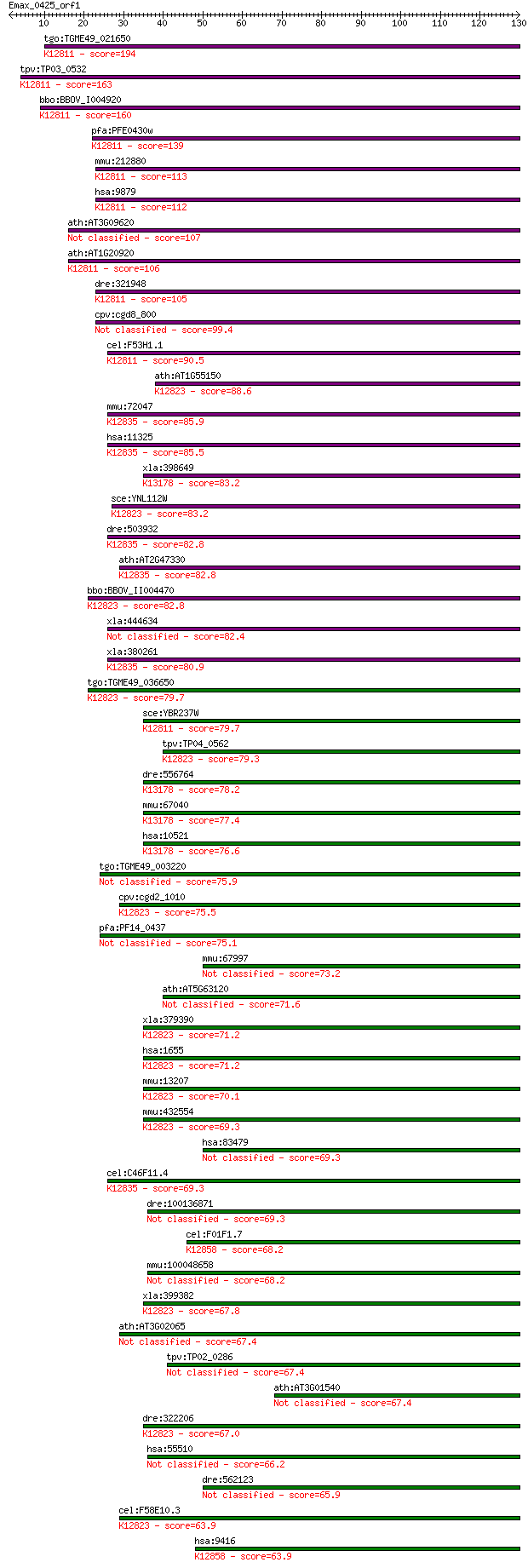
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0425_orf1
Length=129
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_021650 ATP-dependent RNA helicase, putative (EC:3.1... 194 8e-50
tpv:TP03_0532 ATP-dependent RNA helicase; K12811 ATP-dependent... 163 2e-40
bbo:BBOV_I004920 19.m02284; DEAD/DEAH box helicase and helicas... 160 8e-40
pfa:PFE0430w ATP-dependent RNA Helicase, putative; K12811 ATP-... 139 2e-33
mmu:212880 Ddx46, 2200005K02Rik, 8430438J23Rik, AI325430, AI95... 113 2e-25
hsa:9879 DDX46, FLJ25329, KIAA0801, MGC9936, PRPF5, Prp5; DEAD... 112 2e-25
ath:AT3G09620 DEAD/DEAH box helicase, putative 107 8e-24
ath:AT1G20920 DEAD box RNA helicase, putative; K12811 ATP-depe... 106 1e-23
dre:321948 ddx46, fb39a03, wu:fb39a03; DEAD (Asp-Glu-Ala-Asp) ... 105 3e-23
cpv:cgd8_800 Prp5p C terminal KH. eIF4A-1-family RNA SFII heli... 99.4 2e-21
cel:F53H1.1 hypothetical protein; K12811 ATP-dependent RNA hel... 90.5 1e-18
ath:AT1G55150 DEAD box RNA helicase, putative (RH20); K12823 A... 88.6 5e-18
mmu:72047 Ddx42, 1810047H21Rik, AW319508, AW556242, B430002H05... 85.9 3e-17
hsa:11325 DDX42, FLJ43179, RHELP, RNAHP, SF3b125; DEAD (Asp-Gl... 85.5 4e-17
xla:398649 ddx17, MGC80019; DEAD (Asp-Glu-Ala-Asp) box polypep... 83.2 2e-16
sce:YNL112W DBP2; Dbp2p (EC:3.6.1.-); K12823 ATP-dependent RNA... 83.2 2e-16
dre:503932 ddx42, im:7148194, zgc:111815; DEAD (Asp-Glu-Ala-As... 82.8 2e-16
ath:AT2G47330 DEAD/DEAH box helicase, putative; K12835 ATP-dep... 82.8 3e-16
bbo:BBOV_II004470 18.m06373; p68-like protein; K12823 ATP-depe... 82.8 3e-16
xla:444634 MGC84147 protein 82.4 3e-16
xla:380261 ddx42, MGC54025; DEAD (Asp-Glu-Ala-Asp) box polypep... 80.9 8e-16
tgo:TGME49_036650 DEAD/DEAH box helicase, putative (EC:5.99.1.... 79.7 2e-15
sce:YBR237W PRP5, RNA5; Prp5p (EC:3.6.1.-); K12811 ATP-depende... 79.7 2e-15
tpv:TP04_0562 RNA helicase; K12823 ATP-dependent RNA helicase ... 79.3 3e-15
dre:556764 similar to Probable RNA-dependent helicase p72 (DEA... 78.2 5e-15
mmu:67040 Ddx17, 2610007K22Rik, A430025E01Rik, AI047725, C8092... 77.4 1e-14
hsa:10521 DDX17, DKFZp761H2016, P72, RH70; DEAD (Asp-Glu-Ala-A... 76.6 2e-14
tgo:TGME49_003220 ATP-dependent RNA helicase, putative 75.9 3e-14
cpv:cgd2_1010 hypothetical protein ; K12823 ATP-dependent RNA ... 75.5 4e-14
pfa:PF14_0437 helicase, putative 75.1 5e-14
mmu:67997 Ddx59, 1210002B07Rik, 4833411G06Rik; DEAD (Asp-Glu-A... 73.2 2e-13
ath:AT5G63120 ethylene-responsive DEAD box RNA helicase, putat... 71.6 5e-13
xla:379390 MGC53795; similar to DEAD/H (Asp-Glu-Ala-Asp/His) b... 71.2 6e-13
hsa:1655 DDX5, DKFZp434E109, DKFZp686J01190, G17P1, HLR1, HUMP... 71.2 8e-13
mmu:13207 Ddx5, 2600009A06Rik, G17P1, HUMP68, Hlr1, MGC118083,... 70.1 2e-12
mmu:432554 Gm12183, OTTMUSG00000005521; predicted gene 12183; ... 69.3 3e-12
hsa:83479 DDX59, DKFZp564B1023, ZNHIT5; DEAD (Asp-Glu-Ala-Asp)... 69.3 3e-12
cel:C46F11.4 hypothetical protein; K12835 ATP-dependent RNA he... 69.3 3e-12
dre:100136871 ddx43, zgc:174910; DEAD (Asp-Glu-Ala-Asp) box po... 69.3 3e-12
cel:F01F1.7 ddx-23; DEAD boX helicase homolog family member (d... 68.2 5e-12
mmu:100048658 Ddx43, OTTMUSG00000019690; DEAD (Asp-Glu-Ala-Asp... 68.2 7e-12
xla:399382 ddx5, MGC81559; DEAD (Asp-Glu-Ala-Asp) box polypept... 67.8 8e-12
ath:AT3G02065 DEAD/DEAH box helicase family protein 67.4 1e-11
tpv:TP02_0286 ATP-dependent RNA Helicase 67.4 1e-11
ath:AT3G01540 DRH1; DRH1 (DEAD BOX RNA HELICASE 1); ATP-depend... 67.4 1e-11
dre:322206 ddx5, wu:fa56a07, wu:fb11e01, wu:fb16c10, wu:fb53b0... 67.0 1e-11
hsa:55510 DDX43, CT13, DKFZp434H2114, HAGE; DEAD (Asp-Glu-Ala-... 66.2 2e-11
dre:562123 ddx59, si:dkey-39e8.2; DEAD (Asp-Glu-Ala-Asp) box p... 65.9 3e-11
cel:F58E10.3 hypothetical protein; K12823 ATP-dependent RNA he... 63.9 1e-10
hsa:9416 DDX23, MGC8416, PRPF28, U5-100K, U5-100KD, prp28; DEA... 63.9 1e-10
> tgo:TGME49_021650 ATP-dependent RNA helicase, putative (EC:3.1.3.69
3.4.21.72); K12811 ATP-dependent RNA helicase DDX46/PRP5
[EC:3.6.4.13]
Length=1544
Score = 194 bits (492), Expect = 8e-50, Method: Compositional matrix adjust.
Identities = 84/120 (70%), Positives = 105/120 (87%), Gaps = 0/120 (0%)
Query 10 NLSYFDLVKRVGLKKELPLVDHASCKFPPIKKNLYVQVKELTDLKDHEVEAIRKTNGNIK 69
NLSYFDL+ +VG KK+LP VDH + +PPIKKNLY+QVKE+T +KDHEV+A+RKT+GNIK
Sbjct 903 NLSYFDLLMKVGAKKQLPTVDHEASAYPPIKKNLYIQVKEITCMKDHEVDALRKTHGNIK 962
Query 70 VRGKQCPRPVSSFHQCGLPEKILRHLEARGFEKPFPVQSQCIPILMCGRDLIAVAETGSG 129
VRGKQCPRP+++F QCGLP+KI+++L RG +PFP+Q Q IP LMCGRD+IAVAETGSG
Sbjct 963 VRGKQCPRPITTFFQCGLPDKIVKYLTLRGITEPFPIQMQAIPCLMCGRDVIAVAETGSG 1022
> tpv:TP03_0532 ATP-dependent RNA helicase; K12811 ATP-dependent
RNA helicase DDX46/PRP5 [EC:3.6.4.13]
Length=894
Score = 163 bits (412), Expect = 2e-40, Method: Compositional matrix adjust.
Identities = 73/127 (57%), Positives = 95/127 (74%), Gaps = 1/127 (0%)
Query 4 AKEQLANLSYFDLVKRV-GLKKELPLVDHASCKFPPIKKNLYVQVKELTDLKDHEVEAIR 62
K + Y +L+ ++ G KKELP VDH+ + P +KN YVQV +T++ +HEV+A R
Sbjct 340 TKVSMGGPDYMELLPKIRGGKKELPRVDHSKIDYLPFRKNFYVQVSSITNMGEHEVDAFR 399
Query 63 KTNGNIKVRGKQCPRPVSSFHQCGLPEKILRHLEARGFEKPFPVQSQCIPILMCGRDLIA 122
K NGNI+V GK+CPRP+SSF QCGLP+ IL+ LE R +EKPFP+Q QCIP LMCGRD+I
Sbjct 400 KANGNIRVYGKKCPRPISSFSQCGLPDPILKILEKREYEKPFPIQMQCIPALMCGRDVIG 459
Query 123 VAETGSG 129
+AETGSG
Sbjct 460 IAETGSG 466
> bbo:BBOV_I004920 19.m02284; DEAD/DEAH box helicase and helicase
conserved C-terminal domain containing protein (EC:3.6.1.-);
K12811 ATP-dependent RNA helicase DDX46/PRP5 [EC:3.6.4.13]
Length=994
Score = 160 bits (406), Expect = 8e-40, Method: Compositional matrix adjust.
Identities = 70/122 (57%), Positives = 92/122 (75%), Gaps = 1/122 (0%)
Query 9 ANLSYFDLVKRVGLKK-ELPLVDHASCKFPPIKKNLYVQVKELTDLKDHEVEAIRKTNGN 67
+N+ Y +L K + E+P VDH++ + P KKN YVQ+ +T +K+HEVEA RK NGN
Sbjct 316 SNVDYSELFKGTTRSRIEMPKVDHSTIDYQPFKKNFYVQISAITAMKEHEVEAFRKANGN 375
Query 68 IKVRGKQCPRPVSSFHQCGLPEKILRHLEARGFEKPFPVQSQCIPILMCGRDLIAVAETG 127
I+VRGK CPRP+ +F QCGLP+ IL L+ R +EKPFP+Q QCIP LMCGRD++A+AETG
Sbjct 376 IRVRGKYCPRPIYNFSQCGLPDPILSLLQRRNYEKPFPIQMQCIPALMCGRDVLAIAETG 435
Query 128 SG 129
SG
Sbjct 436 SG 437
> pfa:PFE0430w ATP-dependent RNA Helicase, putative; K12811 ATP-dependent
RNA helicase DDX46/PRP5 [EC:3.6.4.13]
Length=1490
Score = 139 bits (350), Expect = 2e-33, Method: Composition-based stats.
Identities = 65/108 (60%), Positives = 83/108 (76%), Gaps = 0/108 (0%)
Query 22 LKKELPLVDHASCKFPPIKKNLYVQVKELTDLKDHEVEAIRKTNGNIKVRGKQCPRPVSS 81
+ K+L V+H + PIKKN+YVQVKE+T++KD +V+ RK NGNI VRGK CPRPV
Sbjct 665 MNKKLLEVNHDEIDYIPIKKNIYVQVKEITNMKDSDVDMFRKNNGNIIVRGKNCPRPVQY 724
Query 82 FHQCGLPEKILRHLEARGFEKPFPVQSQCIPILMCGRDLIAVAETGSG 129
F+QCGLP KIL+ LE + F+K + +Q Q IP LMCGRD+IA+AETGSG
Sbjct 725 FYQCGLPSKILQILEKKNFKKMYNIQMQTIPALMCGRDVIAIAETGSG 772
> mmu:212880 Ddx46, 2200005K02Rik, 8430438J23Rik, AI325430, AI957095,
MGC116676, MGC31579, mKIAA0801; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 46 (EC:3.6.4.13); K12811 ATP-dependent RNA
helicase DDX46/PRP5 [EC:3.6.4.13]
Length=1031
Score = 113 bits (282), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 51/107 (47%), Positives = 69/107 (64%), Gaps = 0/107 (0%)
Query 23 KKELPLVDHASCKFPPIKKNLYVQVKELTDLKDHEVEAIRKTNGNIKVRGKQCPRPVSSF 82
+K L VDH ++ P +KN YV+V EL + EV R I V+GK CP+P+ S+
Sbjct 315 RKLLEPVDHGKIEYEPFRKNFYVEVPELAKMSQEEVNVFRLEMEGITVKGKGCPKPIKSW 374
Query 83 HQCGLPEKILRHLEARGFEKPFPVQSQCIPILMCGRDLIAVAETGSG 129
QCG+ KIL L+ G+EKP P+Q+Q IP +M GRDLI +A+TGSG
Sbjct 375 VQCGISMKILNSLKKHGYEKPTPIQTQAIPAIMSGRDLIGIAKTGSG 421
> hsa:9879 DDX46, FLJ25329, KIAA0801, MGC9936, PRPF5, Prp5; DEAD
(Asp-Glu-Ala-Asp) box polypeptide 46 (EC:3.6.4.13); K12811
ATP-dependent RNA helicase DDX46/PRP5 [EC:3.6.4.13]
Length=1031
Score = 112 bits (281), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 51/107 (47%), Positives = 69/107 (64%), Gaps = 0/107 (0%)
Query 23 KKELPLVDHASCKFPPIKKNLYVQVKELTDLKDHEVEAIRKTNGNIKVRGKQCPRPVSSF 82
+K L VDH ++ P +KN YV+V EL + EV R I V+GK CP+P+ S+
Sbjct 315 RKLLEPVDHGKIEYEPFRKNFYVEVPELAKMSQEEVNVFRLEMEGITVKGKGCPKPIKSW 374
Query 83 HQCGLPEKILRHLEARGFEKPFPVQSQCIPILMCGRDLIAVAETGSG 129
QCG+ KIL L+ G+EKP P+Q+Q IP +M GRDLI +A+TGSG
Sbjct 375 VQCGISMKILNSLKKHGYEKPTPIQTQAIPAIMSGRDLIGIAKTGSG 421
> ath:AT3G09620 DEAD/DEAH box helicase, putative
Length=989
Score = 107 bits (267), Expect = 8e-24, Method: Compositional matrix adjust.
Identities = 53/116 (45%), Positives = 75/116 (64%), Gaps = 3/116 (2%)
Query 16 LVKRVGLKK--ELPLVDHASCKFPPIKKNLYVQVKELTDLKDHEVEAIRKTNGNIKVRGK 73
+KRV K +L LVDH+ ++ P +KN Y++VK+++ + V A RK +KV GK
Sbjct 331 FMKRVKKTKAEKLSLVDHSKIEYEPFRKNFYIEVKDISRMTQDAVNAYRK-ELELKVHGK 389
Query 74 QCPRPVSSFHQCGLPEKILRHLEARGFEKPFPVQSQCIPILMCGRDLIAVAETGSG 129
PRP+ +HQ GL KIL L+ +EKP P+Q+Q +PI+M GRD I VA+TGSG
Sbjct 390 DVPRPIQFWHQTGLTSKILDTLKKLNYEKPMPIQAQALPIIMSGRDCIGVAKTGSG 445
> ath:AT1G20920 DEAD box RNA helicase, putative; K12811 ATP-dependent
RNA helicase DDX46/PRP5 [EC:3.6.4.13]
Length=828
Score = 106 bits (265), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 52/116 (44%), Positives = 75/116 (64%), Gaps = 3/116 (2%)
Query 16 LVKRVGLKK--ELPLVDHASCKFPPIKKNLYVQVKELTDLKDHEVEAIRKTNGNIKVRGK 73
+KRV K +L LVDH+ ++ P +KN Y++VK+++ + EV RK +KV GK
Sbjct 126 FMKRVKKTKAEKLSLVDHSKIEYEPFRKNFYIEVKDISRMTQEEVNTYRK-ELELKVHGK 184
Query 74 QCPRPVSSFHQCGLPEKILRHLEARGFEKPFPVQSQCIPILMCGRDLIAVAETGSG 129
PRP+ +HQ GL KIL ++ +EKP P+Q+Q +PI+M GRD I VA+TGSG
Sbjct 185 DVPRPIKFWHQTGLTSKILDTMKKLNYEKPMPIQTQALPIIMSGRDCIGVAKTGSG 240
> dre:321948 ddx46, fb39a03, wu:fb39a03; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 46 (EC:3.6.4.13); K12811 ATP-dependent RNA
helicase DDX46/PRP5 [EC:3.6.4.13]
Length=1018
Score = 105 bits (262), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 48/107 (44%), Positives = 68/107 (63%), Gaps = 0/107 (0%)
Query 23 KKELPLVDHASCKFPPIKKNLYVQVKELTDLKDHEVEAIRKTNGNIKVRGKQCPRPVSSF 82
+K L VDH ++ P +KN YV+V EL + EV R I V+GK CP+P+ ++
Sbjct 283 RKVLEPVDHQKIQYEPFRKNFYVEVPELARMSPEEVSEYRLELEGISVKGKGCPKPIKTW 342
Query 83 HQCGLPEKILRHLEARGFEKPFPVQSQCIPILMCGRDLIAVAETGSG 129
QCG+ K+L L+ +EKP P+Q+Q IP +M GRDLI +A+TGSG
Sbjct 343 VQCGISMKVLNALKKHNYEKPTPIQAQAIPAIMSGRDLIGIAKTGSG 389
> cpv:cgd8_800 Prp5p C terminal KH. eIF4A-1-family RNA SFII helicase
Length=934
Score = 99.4 bits (246), Expect = 2e-21, Method: Composition-based stats.
Identities = 51/121 (42%), Positives = 69/121 (57%), Gaps = 14/121 (11%)
Query 23 KKELPLVDHASCKFPPIKKNLYVQVKELTDLKDHEVEAIRKTNGNIKVRGKQ-------- 74
KK +P ++H +PPI KN Y +V E+ LK HEV+ IR TN I ++ +
Sbjct 151 KKRIPEINHEVINYPPIIKNYYKEVNEIKKLKQHEVDHIRITNNGIHIKKIKNINKTSND 210
Query 75 ------CPRPVSSFHQCGLPEKILRHLEARGFEKPFPVQSQCIPILMCGRDLIAVAETGS 128
+P+ +F QCGLP I +L+ + KPFP+Q Q IPILM G D+I AETGS
Sbjct 211 LNQPYSSIKPILNFSQCGLPLPIHHYLKKKNIIKPFPIQMQSIPILMSGYDMIGNAETGS 270
Query 129 G 129
G
Sbjct 271 G 271
> cel:F53H1.1 hypothetical protein; K12811 ATP-dependent RNA helicase
DDX46/PRP5 [EC:3.6.4.13]
Length=970
Score = 90.5 bits (223), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 39/104 (37%), Positives = 65/104 (62%), Gaps = 0/104 (0%)
Query 26 LPLVDHASCKFPPIKKNLYVQVKELTDLKDHEVEAIRKTNGNIKVRGKQCPRPVSSFHQC 85
L DH+ + KKN Y++ +E+ + EV+A R+ +I V+G CP+P+ ++ QC
Sbjct 250 LAQTDHSKVYYRKFKKNFYIETEEIRRMTKAEVKAYREELDSITVKGIDCPKPIKTWAQC 309
Query 86 GLPEKILRHLEARGFEKPFPVQSQCIPILMCGRDLIAVAETGSG 129
G+ K++ L+ + KP +Q+Q IP +M GRD+I +A+TGSG
Sbjct 310 GVNLKMMNVLKKFEYSKPTSIQAQAIPSIMSGRDVIGIAKTGSG 353
> ath:AT1G55150 DEAD box RNA helicase, putative (RH20); K12823
ATP-dependent RNA helicase DDX5/DBP2 [EC:3.6.4.13]
Length=501
Score = 88.6 bits (218), Expect = 5e-18, Method: Composition-based stats.
Identities = 43/92 (46%), Positives = 56/92 (60%), Gaps = 1/92 (1%)
Query 38 PIKKNLYVQVKELTDLKDHEVEAIRKTNGNIKVRGKQCPRPVSSFHQCGLPEKILRHLEA 97
P +KN YV+ + + D EVE RK I V GK P+PV SF G P+ +L ++
Sbjct 58 PFEKNFYVESPAVAAMTDTEVEEYRKLR-EITVEGKDIPKPVKSFRDVGFPDYVLEEVKK 116
Query 98 RGFEKPFPVQSQCIPILMCGRDLIAVAETGSG 129
GF +P P+QSQ P+ M GRDLI +AETGSG
Sbjct 117 AGFTEPTPIQSQGWPMAMKGRDLIGIAETGSG 148
> mmu:72047 Ddx42, 1810047H21Rik, AW319508, AW556242, B430002H05Rik,
RHELP, RNAHP, SF3b125; DEAD (Asp-Glu-Ala-Asp) box polypeptide
42 (EC:3.6.4.13); K12835 ATP-dependent RNA helicase
DDX42 [EC:3.6.4.13]
Length=929
Score = 85.9 bits (211), Expect = 3e-17, Method: Composition-based stats.
Identities = 38/104 (36%), Positives = 64/104 (61%), Gaps = 1/104 (0%)
Query 26 LPLVDHASCKFPPIKKNLYVQVKELTDLKDHEVEAIRKTNGNIKVRGKQCPRPVSSFHQC 85
LP +DH+ +PP +KN Y + +E+T+L ++ +R N++V G PRP SSF
Sbjct 200 LPPIDHSEIDYPPFEKNFYNEHEEITNLTPQQLIDLRH-KLNLRVSGAAPPRPGSSFAHF 258
Query 86 GLPEKILRHLEARGFEKPFPVQSQCIPILMCGRDLIAVAETGSG 129
G E+++ + + +P P+Q Q +P+ + GRD+I +A+TGSG
Sbjct 259 GFDEQLMHQIRKSEYTQPTPIQCQGVPVALSGRDMIGIAKTGSG 302
> hsa:11325 DDX42, FLJ43179, RHELP, RNAHP, SF3b125; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 42 (EC:3.6.4.13); K12835 ATP-dependent
RNA helicase DDX42 [EC:3.6.4.13]
Length=938
Score = 85.5 bits (210), Expect = 4e-17, Method: Composition-based stats.
Identities = 38/104 (36%), Positives = 64/104 (61%), Gaps = 1/104 (0%)
Query 26 LPLVDHASCKFPPIKKNLYVQVKELTDLKDHEVEAIRKTNGNIKVRGKQCPRPVSSFHQC 85
LP +DH+ +PP +KN Y + +E+T+L ++ +R N++V G PRP SSF
Sbjct 200 LPPIDHSEIDYPPFEKNFYNEHEEITNLTPQQLIDLRH-KLNLRVSGAAPPRPGSSFAHF 258
Query 86 GLPEKILRHLEARGFEKPFPVQSQCIPILMCGRDLIAVAETGSG 129
G E+++ + + +P P+Q Q +P+ + GRD+I +A+TGSG
Sbjct 259 GFDEQLMHQIRKSEYTQPTPIQCQGVPVALSGRDMIGIAKTGSG 302
> xla:398649 ddx17, MGC80019; DEAD (Asp-Glu-Ala-Asp) box polypeptide
17; K13178 ATP-dependent RNA helicase DDX17 [EC:3.6.4.13]
Length=610
Score = 83.2 bits (204), Expect = 2e-16, Method: Composition-based stats.
Identities = 35/95 (36%), Positives = 59/95 (62%), Gaps = 1/95 (1%)
Query 35 KFPPIKKNLYVQVKELTDLKDHEVEAIRKTNGNIKVRGKQCPRPVSSFHQCGLPEKILRH 94
+ P +KN Y + E+ + H+VE +R+ I +RG CP+P+ +FHQ P+ +L
Sbjct 38 ELPKFEKNFYTEHPEVARMTQHDVEELRRKK-EITIRGVNCPKPLYAFHQANFPQYVLDV 96
Query 95 LEARGFEKPFPVQSQCIPILMCGRDLIAVAETGSG 129
L + F++P P+Q Q P+ + GRD++ +A+TGSG
Sbjct 97 LLDQRFKEPTPIQCQGFPLALSGRDMVGIAQTGSG 131
> sce:YNL112W DBP2; Dbp2p (EC:3.6.1.-); K12823 ATP-dependent RNA
helicase DDX5/DBP2 [EC:3.6.4.13]
Length=546
Score = 83.2 bits (204), Expect = 2e-16, Method: Composition-based stats.
Identities = 37/103 (35%), Positives = 60/103 (58%), Gaps = 1/103 (0%)
Query 27 PLVDHASCKFPPIKKNLYVQVKELTDLKDHEVEAIRKTNGNIKVRGKQCPRPVSSFHQCG 86
P D K P +KN YV+ + + D D E+ RK N + + G P+P+++F + G
Sbjct 61 PNWDEELPKLPTFEKNFYVEHESVRDRSDSEIAQFRKEN-EMTISGHDIPKPITTFDEAG 119
Query 87 LPEKILRHLEARGFEKPFPVQSQCIPILMCGRDLIAVAETGSG 129
P+ +L ++A GF+KP +Q Q P+ + GRD++ +A TGSG
Sbjct 120 FPDYVLNEVKAEGFDKPTGIQCQGWPMALSGRDMVGIAATGSG 162
> dre:503932 ddx42, im:7148194, zgc:111815; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 42 (EC:3.6.1.-); K12835 ATP-dependent RNA
helicase DDX42 [EC:3.6.4.13]
Length=908
Score = 82.8 bits (203), Expect = 2e-16, Method: Composition-based stats.
Identities = 38/104 (36%), Positives = 62/104 (59%), Gaps = 1/104 (0%)
Query 26 LPLVDHASCKFPPIKKNLYVQVKELTDLKDHEVEAIRKTNGNIKVRGKQCPRPVSSFHQC 85
LP +DH+ + P +KN Y + +E++ L EV +R+ N+KV G P+P +SF
Sbjct 202 LPPIDHSEIDYSPFEKNFYNEHEEISSLTGAEVVELRR-KLNLKVSGAAPPKPATSFAHF 260
Query 86 GLPEKILRHLEARGFEKPFPVQSQCIPILMCGRDLIAVAETGSG 129
G E+++ + + +P P+Q Q +PI + GRD I +A+TGSG
Sbjct 261 GFDEQLMHQIRKSEYTQPTPIQCQGVPIALSGRDAIGIAKTGSG 304
> ath:AT2G47330 DEAD/DEAH box helicase, putative; K12835 ATP-dependent
RNA helicase DDX42 [EC:3.6.4.13]
Length=760
Score = 82.8 bits (203), Expect = 3e-16, Method: Composition-based stats.
Identities = 36/101 (35%), Positives = 63/101 (62%), Gaps = 1/101 (0%)
Query 29 VDHASCKFPPIKKNLYVQVKELTDLKDHEVEAIRKTNGNIKVRGKQCPRPVSSFHQCGLP 88
+DH+S + PI K+ Y +++ ++ + + E R+ G I+V G RPV +F CG
Sbjct 178 LDHSSIDYEPINKDFYEELESISGMTEQETTDYRQRLG-IRVSGFDVHRPVKTFEDCGFS 236
Query 89 EKILRHLEARGFEKPFPVQSQCIPILMCGRDLIAVAETGSG 129
+I+ ++ + +EKP +Q Q +PI++ GRD+I +A+TGSG
Sbjct 237 SQIMSAIKKQAYEKPTAIQCQALPIVLSGRDVIGIAKTGSG 277
> bbo:BBOV_II004470 18.m06373; p68-like protein; K12823 ATP-dependent
RNA helicase DDX5/DBP2 [EC:3.6.4.13]
Length=529
Score = 82.8 bits (203), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 38/109 (34%), Positives = 58/109 (53%), Gaps = 0/109 (0%)
Query 21 GLKKELPLVDHASCKFPPIKKNLYVQVKELTDLKDHEVEAIRKTNGNIKVRGKQCPRPVS 80
L L +D + +KN Y + E++ + +V+ +RK + G+ P+PV
Sbjct 49 ALGSRLSTIDWSKETLVAFEKNFYKEHSEVSAMSSADVDRVRKEREITIIAGRDVPKPVV 108
Query 81 SFHQCGLPEKILRHLEARGFEKPFPVQSQCIPILMCGRDLIAVAETGSG 129
SF P+ IL+ + A GF P P+Q Q PI + GRD+I +AETGSG
Sbjct 109 SFEHTSFPDYILKAIRAAGFTAPTPIQVQGWPIALSGRDVIGIAETGSG 157
> xla:444634 MGC84147 protein
Length=450
Score = 82.4 bits (202), Expect = 3e-16, Method: Composition-based stats.
Identities = 38/104 (36%), Positives = 61/104 (58%), Gaps = 1/104 (0%)
Query 26 LPLVDHASCKFPPIKKNLYVQVKELTDLKDHEVEAIRKTNGNIKVRGKQCPRPVSSFHQC 85
LP +DH ++PP +KN Y + +E+T ++ +R N++V G PR SSF
Sbjct 198 LPPIDHTEIEYPPFEKNFYEEHEEITSQTPQQITELRH-KLNLRVSGAAAPRLCSSFAHF 256
Query 86 GLPEKILRHLEARGFEKPFPVQSQCIPILMCGRDLIAVAETGSG 129
G E+++ + + KP P+Q Q IP+ + GRD+I +A+TGSG
Sbjct 257 GFDEQLMHQIRKSEYTKPTPIQCQGIPVALSGRDMIGIAKTGSG 300
> xla:380261 ddx42, MGC54025; DEAD (Asp-Glu-Ala-Asp) box polypeptide
42 (EC:3.6.4.13); K12835 ATP-dependent RNA helicase DDX42
[EC:3.6.4.13]
Length=947
Score = 80.9 bits (198), Expect = 8e-16, Method: Composition-based stats.
Identities = 37/104 (35%), Positives = 60/104 (57%), Gaps = 1/104 (0%)
Query 26 LPLVDHASCKFPPIKKNLYVQVKELTDLKDHEVEAIRKTNGNIKVRGKQCPRPVSSFHQC 85
LP +DH ++PP +KN Y + + +T ++ +R N++V G PR SSF
Sbjct 197 LPPIDHTEIEYPPFEKNFYEEHEAITSQTPQQITELRH-KLNLRVSGAAPPRLCSSFAHF 255
Query 86 GLPEKILRHLEARGFEKPFPVQSQCIPILMCGRDLIAVAETGSG 129
G E++L + + +P P+Q Q IP+ + GRD+I +A+TGSG
Sbjct 256 GFDEQLLHQIRKSEYTQPTPIQCQGIPVALSGRDMIGIAKTGSG 299
> tgo:TGME49_036650 DEAD/DEAH box helicase, putative (EC:5.99.1.3);
K12823 ATP-dependent RNA helicase DDX5/DBP2 [EC:3.6.4.13]
Length=550
Score = 79.7 bits (195), Expect = 2e-15, Method: Composition-based stats.
Identities = 41/109 (37%), Positives = 57/109 (52%), Gaps = 0/109 (0%)
Query 21 GLKKELPLVDHASCKFPPIKKNLYVQVKELTDLKDHEVEAIRKTNGNIKVRGKQCPRPVS 80
L +L VD + P +KN YV+ + ++ E E IR+ N V G P+PV
Sbjct 70 ALGSKLQRVDWKTIDLVPFEKNFYVEHPAVANMSAEEAERIRRANEITIVHGHNVPKPVP 129
Query 81 SFHQCGLPEKILRHLEARGFEKPFPVQSQCIPILMCGRDLIAVAETGSG 129
+F P IL + GF+KP +Q Q PI + GRD+I +AETGSG
Sbjct 130 TFEYTSFPSYILDVINQTGFQKPTAIQVQGWPIALSGRDMIGIAETGSG 178
> sce:YBR237W PRP5, RNA5; Prp5p (EC:3.6.1.-); K12811 ATP-dependent
RNA helicase DDX46/PRP5 [EC:3.6.4.13]
Length=849
Score = 79.7 bits (195), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 35/96 (36%), Positives = 61/96 (63%), Gaps = 1/96 (1%)
Query 35 KFPPIKKNLYVQVKELTDLKDHEVEAIRKTNGNIKVRGKQCPRPVSSFHQCGLP-EKILR 93
+ P +KN Y++ + ++ + + EVE +R + NIK++G CP+PV+ + Q GL + ++
Sbjct 210 ELEPFQKNFYIESETVSSMSEMEVEELRLSLDNIKIKGTGCPKPVTKWSQLGLSTDTMVL 269
Query 94 HLEARGFEKPFPVQSQCIPILMCGRDLIAVAETGSG 129
E F P+QSQ +P +M GRD+I +++TGSG
Sbjct 270 ITEKLHFGSLTPIQSQALPAIMSGRDVIGISKTGSG 305
> tpv:TP04_0562 RNA helicase; K12823 ATP-dependent RNA helicase
DDX5/DBP2 [EC:3.6.4.13]
Length=635
Score = 79.3 bits (194), Expect = 3e-15, Method: Composition-based stats.
Identities = 37/90 (41%), Positives = 52/90 (57%), Gaps = 0/90 (0%)
Query 40 KKNLYVQVKELTDLKDHEVEAIRKTNGNIKVRGKQCPRPVSSFHQCGLPEKILRHLEARG 99
+KN YV+ E+ + E + IR+ V G+ P+PV F P IL +EA G
Sbjct 170 EKNFYVEHPEVKAMTQQEADEIRRAKEITVVHGRDVPKPVVKFEYTSFPRYILSSIEAAG 229
Query 100 FEKPFPVQSQCIPILMCGRDLIAVAETGSG 129
F++P P+Q Q PI + GRD+I +AETGSG
Sbjct 230 FKEPTPIQVQSWPIALSGRDMIGIAETGSG 259
> dre:556764 similar to Probable RNA-dependent helicase p72 (DEAD-box
protein p72) (DEAD-box protein 17); K13178 ATP-dependent
RNA helicase DDX17 [EC:3.6.4.13]
Length=671
Score = 78.2 bits (191), Expect = 5e-15, Method: Composition-based stats.
Identities = 34/95 (35%), Positives = 59/95 (62%), Gaps = 1/95 (1%)
Query 35 KFPPIKKNLYVQVKELTDLKDHEVEAIRKTNGNIKVRGKQCPRPVSSFHQCGLPEKILRH 94
+ P +KN Y + E+ + ++VE R+ I VRG CP+PV++FHQ P+ ++
Sbjct 51 QLPKFEKNFYNENPEVHHMSQYDVEEYRRKR-EITVRGSGCPKPVTNFHQAQFPQYVMDV 109
Query 95 LEARGFEKPFPVQSQCIPILMCGRDLIAVAETGSG 129
L + F++P +Q+Q P+ + GRD++ +A+TGSG
Sbjct 110 LLQQNFKEPTAIQAQGFPLALSGRDMVGIAQTGSG 144
> mmu:67040 Ddx17, 2610007K22Rik, A430025E01Rik, AI047725, C80929,
Gm926, MGC79147, p72; DEAD (Asp-Glu-Ala-Asp) box polypeptide
17 (EC:3.6.4.13); K13178 ATP-dependent RNA helicase DDX17
[EC:3.6.4.13]
Length=650
Score = 77.4 bits (189), Expect = 1e-14, Method: Composition-based stats.
Identities = 36/96 (37%), Positives = 58/96 (60%), Gaps = 2/96 (2%)
Query 35 KFPPIKKNLYVQVKELTDLKDHEVEAIRKTNGNIKVRGKQ-CPRPVSSFHQCGLPEKILR 93
+ P +KN YV+ E+ L +EV+ +R+ I VRG CP+PV +FH P+ ++
Sbjct 47 ELPKFEKNFYVEHPEVARLTPYEVDELRRKK-EITVRGGDVCPKPVFAFHHANFPQYVMD 105
Query 94 HLEARGFEKPFPVQSQCIPILMCGRDLIAVAETGSG 129
L + F +P P+Q Q P+ + GRD++ +A+TGSG
Sbjct 106 VLMDQHFTEPTPIQCQGFPLALSGRDMVGIAQTGSG 141
> hsa:10521 DDX17, DKFZp761H2016, P72, RH70; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 17 (EC:3.6.4.13); K13178 ATP-dependent
RNA helicase DDX17 [EC:3.6.4.13]
Length=731
Score = 76.6 bits (187), Expect = 2e-14, Method: Composition-based stats.
Identities = 36/96 (37%), Positives = 58/96 (60%), Gaps = 2/96 (2%)
Query 35 KFPPIKKNLYVQVKELTDLKDHEVEAIRKTNGNIKVRGKQ-CPRPVSSFHQCGLPEKILR 93
+ P +KN YV+ E+ L +EV+ +R+ I VRG CP+PV +FH P+ ++
Sbjct 126 ELPKFEKNFYVEHPEVARLTPYEVDELRRKK-EITVRGGDVCPKPVFAFHHANFPQYVMD 184
Query 94 HLEARGFEKPFPVQSQCIPILMCGRDLIAVAETGSG 129
L + F +P P+Q Q P+ + GRD++ +A+TGSG
Sbjct 185 VLMDQHFTEPTPIQCQGFPLALSGRDMVGIAQTGSG 220
> tgo:TGME49_003220 ATP-dependent RNA helicase, putative
Length=774
Score = 75.9 bits (185), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 38/108 (35%), Positives = 61/108 (56%), Gaps = 3/108 (2%)
Query 24 KELPLVDHASCKFPPIKKNLYVQVKELTDLKDHEVEAIRKTNGNIKVRGKQCPRPVSSFH 83
K+L VDH+ +P ++++Y + ++ L HE + + I++ G PRP++SF
Sbjct 146 KQLASVDHSILVYPEFQRDIYKEAADIGSLS-HEAVGELRASLQIRITGLNAPRPIASFL 204
Query 84 QC--GLPEKILRHLEARGFEKPFPVQSQCIPILMCGRDLIAVAETGSG 129
L + + + RGF P P+QS IP LM GRD++ +AETGSG
Sbjct 205 HLKDSLSKALFTGINKRGFTLPTPIQSAAIPCLMRGRDVLGLAETGSG 252
> cpv:cgd2_1010 hypothetical protein ; K12823 ATP-dependent RNA
helicase DDX5/DBP2 [EC:3.6.4.13]
Length=586
Score = 75.5 bits (184), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 34/101 (33%), Positives = 56/101 (55%), Gaps = 0/101 (0%)
Query 29 VDHASCKFPPIKKNLYVQVKELTDLKDHEVEAIRKTNGNIKVRGKQCPRPVSSFHQCGLP 88
+D S P +KN Y + + ++ L + +V+ IRK + G+ P+P++SF G P
Sbjct 117 LDWGSQNLIPFEKNFYHEHESVSSLSNEQVDQIRKERKITIIAGENVPKPITSFVTSGFP 176
Query 89 EKILRHLEARGFEKPFPVQSQCIPILMCGRDLIAVAETGSG 129
++ L GF +P +Q Q P+ + G D+I +AETGSG
Sbjct 177 NFLVDALYRTGFTEPTAIQVQGWPVALSGHDMIGIAETGSG 217
> pfa:PF14_0437 helicase, putative
Length=527
Score = 75.1 bits (183), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 35/106 (33%), Positives = 59/106 (55%), Gaps = 0/106 (0%)
Query 24 KELPLVDHASCKFPPIKKNLYVQVKELTDLKDHEVEAIRKTNGNIKVRGKQCPRPVSSFH 83
K L +D + P +KN Y + ++++ L EV+ IR + + G+ P+PV S +
Sbjct 57 KNLAPIDWKTINLVPFEKNFYKEHEDISKLSTKEVKEIRDKHKITILEGENVPKPVVSIN 116
Query 84 QCGLPEKILRHLEARGFEKPFPVQSQCIPILMCGRDLIAVAETGSG 129
+ G P+ +++ L+ P P+Q Q PI + G+D+I AETGSG
Sbjct 117 KIGFPDYVIKSLKNNNIVAPTPIQIQGWPIALSGKDMIGKAETGSG 162
> mmu:67997 Ddx59, 1210002B07Rik, 4833411G06Rik; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 59 (EC:3.6.4.13)
Length=619
Score = 73.2 bits (178), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 32/80 (40%), Positives = 53/80 (66%), Gaps = 1/80 (1%)
Query 50 LTDLKDHEVEAIRKTNGNIKVRGKQCPRPVSSFHQCGLPEKILRHLEARGFEKPFPVQSQ 109
+ LK+ ++E +++ G I V+G+ RP+ F CG PE + ++L+ G+E P P+Q Q
Sbjct 174 IVTLKEDQIETLKQQLG-ISVQGQDVARPIIDFEHCGFPETLNQNLKKSGYEVPTPIQMQ 232
Query 110 CIPILMCGRDLIAVAETGSG 129
IP+ + GRD++A A+TGSG
Sbjct 233 MIPVGLLGRDILASADTGSG 252
> ath:AT5G63120 ethylene-responsive DEAD box RNA helicase, putative
(RH30)
Length=484
Score = 71.6 bits (174), Expect = 5e-13, Method: Composition-based stats.
Identities = 34/90 (37%), Positives = 54/90 (60%), Gaps = 1/90 (1%)
Query 40 KKNLYVQVKELTDLKDHEVEAIRKTNGNIKVRGKQCPRPVSSFHQCGLPEKILRHLEARG 99
+KN YV+ + + + +V A+ +T +I V G+ P+P+ F P+ IL + G
Sbjct 126 EKNFYVESPTVQAMTEQDV-AMYRTERDISVEGRDVPKPMKMFQDANFPDNILEAIAKLG 184
Query 100 FEKPFPVQSQCIPILMCGRDLIAVAETGSG 129
F +P P+Q+Q P+ + GRDLI +AETGSG
Sbjct 185 FTEPTPIQAQGWPMALKGRDLIGIAETGSG 214
> xla:379390 MGC53795; similar to DEAD/H (Asp-Glu-Ala-Asp/His)
box polypeptide 5 (RNA helicase, 68kDa); K12823 ATP-dependent
RNA helicase DDX5/DBP2 [EC:3.6.4.13]
Length=607
Score = 71.2 bits (173), Expect = 6e-13, Method: Composition-based stats.
Identities = 31/95 (32%), Positives = 54/95 (56%), Gaps = 1/95 (1%)
Query 35 KFPPIKKNLYVQVKELTDLKDHEVEAIRKTNGNIKVRGKQCPRPVSSFHQCGLPEKILRH 94
+ P +KN Y ++ +++ E + R++ I VRG CP+PV +FH+ P ++
Sbjct 46 ELPKFEKNFYQELPDVSRRTPQECDQYRRSK-EITVRGLNCPKPVLNFHEASFPANVMEV 104
Query 95 LEARGFEKPFPVQSQCIPILMCGRDLIAVAETGSG 129
++ F +P P+Q Q P+ + G D++ VA TGSG
Sbjct 105 IKRLNFTEPTPIQGQGWPVALSGLDMVGVAMTGSG 139
> hsa:1655 DDX5, DKFZp434E109, DKFZp686J01190, G17P1, HLR1, HUMP68,
p68; DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 (EC:3.6.4.13);
K12823 ATP-dependent RNA helicase DDX5/DBP2 [EC:3.6.4.13]
Length=614
Score = 71.2 bits (173), Expect = 8e-13, Method: Composition-based stats.
Identities = 32/95 (33%), Positives = 54/95 (56%), Gaps = 1/95 (1%)
Query 35 KFPPIKKNLYVQVKELTDLKDHEVEAIRKTNGNIKVRGKQCPRPVSSFHQCGLPEKILRH 94
+ P +KN Y + +L EVE R++ I VRG CP+PV +F++ P ++
Sbjct 50 ELPKFEKNFYQEHPDLARRTAQEVETYRRSK-EITVRGHNCPKPVLNFYEANFPANVMDV 108
Query 95 LEARGFEKPFPVQSQCIPILMCGRDLIAVAETGSG 129
+ + F +P +Q+Q P+ + G D++ VA+TGSG
Sbjct 109 IARQNFTEPTAIQAQGWPVALSGLDMVGVAQTGSG 143
> mmu:13207 Ddx5, 2600009A06Rik, G17P1, HUMP68, Hlr1, MGC118083,
p68; DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 (EC:3.6.4.13);
K12823 ATP-dependent RNA helicase DDX5/DBP2 [EC:3.6.4.13]
Length=615
Score = 70.1 bits (170), Expect = 2e-12, Method: Composition-based stats.
Identities = 31/95 (32%), Positives = 54/95 (56%), Gaps = 1/95 (1%)
Query 35 KFPPIKKNLYVQVKELTDLKDHEVEAIRKTNGNIKVRGKQCPRPVSSFHQCGLPEKILRH 94
+ P +KN Y + +L EV+ R++ I VRG CP+PV +F++ P ++
Sbjct 50 ELPKFEKNFYQEHPDLARRTAQEVDTYRRSK-EITVRGHNCPKPVLNFYEANFPANVMDV 108
Query 95 LEARGFEKPFPVQSQCIPILMCGRDLIAVAETGSG 129
+ + F +P +Q+Q P+ + G D++ VA+TGSG
Sbjct 109 IARQNFTEPTAIQAQGWPVALSGLDMVGVAQTGSG 143
> mmu:432554 Gm12183, OTTMUSG00000005521; predicted gene 12183;
K12823 ATP-dependent RNA helicase DDX5/DBP2 [EC:3.6.4.13]
Length=670
Score = 69.3 bits (168), Expect = 3e-12, Method: Composition-based stats.
Identities = 31/95 (32%), Positives = 53/95 (55%), Gaps = 1/95 (1%)
Query 35 KFPPIKKNLYVQVKELTDLKDHEVEAIRKTNGNIKVRGKQCPRPVSSFHQCGLPEKILRH 94
+ P +KN Y + +L EV+ R++ I VRG CP+PV F++ P ++
Sbjct 105 ELPKFEKNFYQEHPDLARRTAQEVDTYRRSK-EITVRGHNCPKPVLKFYEANFPANVMDV 163
Query 95 LEARGFEKPFPVQSQCIPILMCGRDLIAVAETGSG 129
+ + F +P +Q+Q P+ + G D++ VA+TGSG
Sbjct 164 IARQNFTEPTAIQAQGWPVALSGLDMVGVAQTGSG 198
> hsa:83479 DDX59, DKFZp564B1023, ZNHIT5; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 59 (EC:3.6.4.13)
Length=619
Score = 69.3 bits (168), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 31/80 (38%), Positives = 54/80 (67%), Gaps = 1/80 (1%)
Query 50 LTDLKDHEVEAIRKTNGNIKVRGKQCPRPVSSFHQCGLPEKILRHLEARGFEKPFPVQSQ 109
+ +L++ ++E +++ G I V+G++ RP+ F C LPE + +L+ G+E P P+Q Q
Sbjct 174 ILNLQEDQIENLKQQLG-ILVQGQEVTRPIIDFEHCSLPEVLNHNLKKSGYEVPTPIQMQ 232
Query 110 CIPILMCGRDLIAVAETGSG 129
IP+ + GRD++A A+TGSG
Sbjct 233 MIPVGLLGRDILASADTGSG 252
> cel:C46F11.4 hypothetical protein; K12835 ATP-dependent RNA
helicase DDX42 [EC:3.6.4.13]
Length=811
Score = 69.3 bits (168), Expect = 3e-12, Method: Composition-based stats.
Identities = 35/105 (33%), Positives = 61/105 (58%), Gaps = 3/105 (2%)
Query 26 LPLVDHASCKFPPIKKNLYVQVKELTDLKDHEVEAIRKTNG-NIKVRGKQCPRPVSSFHQ 84
LP +DH+ ++ KN Y + +++ L H ++ IR N N++V G + PRPV SF
Sbjct 212 LPDIDHSQIQYQKFNKNFYEEHEDIKRL--HYMDVIRLQNTMNLRVGGLKPPRPVCSFAH 269
Query 85 CGLPEKILRHLEARGFEKPFPVQSQCIPILMCGRDLIAVAETGSG 129
+ ++ + +E+P P+Q+ IP + GRD++ +A+TGSG
Sbjct 270 FSFDKLLMEAIRKSEYEQPTPIQAMAIPSALSGRDVLGIAKTGSG 314
> dre:100136871 ddx43, zgc:174910; DEAD (Asp-Glu-Ala-Asp) box
polypeptide 43
Length=405
Score = 69.3 bits (168), Expect = 3e-12, Method: Composition-based stats.
Identities = 40/105 (38%), Positives = 57/105 (54%), Gaps = 15/105 (14%)
Query 36 FPPIKKNLYVQVKELTDLKDHEVEAIRKTNGNIKVRG------KQCPRPVSSF-----HQ 84
P +KKN Y++ K + EV+ RK N NI V + P PV +F H
Sbjct 227 LPVLKKNFYIEAKSVAARSAEEVKIWRKENNNIFVDDLKDGDKRTIPNPVCTFEEAFAHY 286
Query 85 CGLPEKILRHLEARGFEKPFPVQSQCIPILMCGRDLIAVAETGSG 129
G+ E I+R GF+KP P+QSQ P+++ G DLI +A+TG+G
Sbjct 287 PGIMENIVR----VGFKKPTPIQSQAWPVVLNGIDLIGIAQTGTG 327
> cel:F01F1.7 ddx-23; DEAD boX helicase homolog family member
(ddx-23); K12858 ATP-dependent RNA helicase DDX23/PRP28 [EC:3.6.4.13]
Length=730
Score = 68.2 bits (165), Expect = 5e-12, Method: Composition-based stats.
Identities = 31/84 (36%), Positives = 57/84 (67%), Gaps = 1/84 (1%)
Query 46 QVKELTDLKDHEVEAIRKTNGNIKVRGKQCPRPVSSFHQCGLPEKILRHLEARGFEKPFP 105
++KEL+++ D + R+ + NI ++G + PRP+ ++ + G P+++ + ++ G+ +P P
Sbjct 268 RMKELSEMSDRDWRIFRE-DFNISIKGGRVPRPLRNWEEAGFPDEVYQAVKEIGYLEPTP 326
Query 106 VQSQCIPILMCGRDLIAVAETGSG 129
+Q Q IPI + RD+I VAETGSG
Sbjct 327 IQRQAIPIGLQNRDVIGVAETGSG 350
> mmu:100048658 Ddx43, OTTMUSG00000019690; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 43 (EC:3.6.4.13)
Length=646
Score = 68.2 bits (165), Expect = 7e-12, Method: Composition-based stats.
Identities = 36/102 (35%), Positives = 56/102 (54%), Gaps = 9/102 (8%)
Query 36 FPPIKKNLYVQVKELTDLKDHEVEAIRKTNGNIKV------RGKQCPRPVSSFHQC--GL 87
PPIKKN Y++ + + +++ RK N NI + P P+ F
Sbjct 189 LPPIKKNFYIESATTSSMSQVQIDNWRKENFNITCDDLKDGEKRPIPNPICKFEDAFQSY 248
Query 88 PEKILRHLEARGFEKPFPVQSQCIPILMCGRDLIAVAETGSG 129
PE ++ +++ GF+KP P+QSQ PI++ G DLI VA+TG+G
Sbjct 249 PE-VMENIKRAGFQKPTPIQSQAWPIVLQGIDLIGVAQTGTG 289
> xla:399382 ddx5, MGC81559; DEAD (Asp-Glu-Ala-Asp) box polypeptide
5; K12823 ATP-dependent RNA helicase DDX5/DBP2 [EC:3.6.4.13]
Length=608
Score = 67.8 bits (164), Expect = 8e-12, Method: Composition-based stats.
Identities = 29/95 (30%), Positives = 53/95 (55%), Gaps = 1/95 (1%)
Query 35 KFPPIKKNLYVQVKELTDLKDHEVEAIRKTNGNIKVRGKQCPRPVSSFHQCGLPEKILRH 94
+ P +KN Y + ++ E + R++ I VRG CP+P+ +F++ P ++
Sbjct 48 ELPKFEKNFYQEHPDVVRRTPQECDQYRRSK-EITVRGINCPKPILNFNEASFPANVMEA 106
Query 95 LEARGFEKPFPVQSQCIPILMCGRDLIAVAETGSG 129
++ + F +P P+Q Q P+ + G D++ VA TGSG
Sbjct 107 IKRQNFTEPTPIQGQGWPVALSGLDMVGVAMTGSG 141
> ath:AT3G02065 DEAD/DEAH box helicase family protein
Length=505
Score = 67.4 bits (163), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 38/102 (37%), Positives = 59/102 (57%), Gaps = 5/102 (4%)
Query 29 VDHASCKFPPIKKNLYVQVKELTDLKDHEVEAIR-KTNGNIKVRGKQCPRPVSSFHQCGL 87
VD A FP + YV+ + H+ + +R K + +++ +G P PV +F CGL
Sbjct 62 VDSARV-FPATDECFYVRDPGSSS---HDAQLLRRKLDIHVQGQGSAVPPPVLTFTSCGL 117
Query 88 PEKILRHLEARGFEKPFPVQSQCIPILMCGRDLIAVAETGSG 129
P K+L +LE G++ P P+Q Q IP + G+ L+A A+TGSG
Sbjct 118 PPKLLLNLETAGYDFPTPIQMQAIPAALTGKSLLASADTGSG 159
> tpv:TP02_0286 ATP-dependent RNA Helicase
Length=623
Score = 67.4 bits (163), Expect = 1e-11, Method: Composition-based stats.
Identities = 36/91 (39%), Positives = 54/91 (59%), Gaps = 3/91 (3%)
Query 41 KNLYVQVKELTDLKDHEVEAIRKTNGNIKVRGKQCPRPVSSFHQCG--LPEKILRHLEAR 98
KN+Y+ +E+ + E +K NI+ G + P+P+SSF +P IL +E
Sbjct 93 KNIYIPDEEVESMSLEECVNFKK-RFNIETFGTRVPKPISSFIHLSKSIPTTILNRIEKM 151
Query 99 GFEKPFPVQSQCIPILMCGRDLIAVAETGSG 129
GF +P PVQSQ IP ++ GR+ I ++ETGSG
Sbjct 152 GFYEPTPVQSQVIPCILQGRNTIILSETGSG 182
> ath:AT3G01540 DRH1; DRH1 (DEAD BOX RNA HELICASE 1); ATP-dependent
RNA helicase/ ATPase
Length=619
Score = 67.4 bits (163), Expect = 1e-11, Method: Composition-based stats.
Identities = 30/62 (48%), Positives = 41/62 (66%), Gaps = 0/62 (0%)
Query 68 IKVRGKQCPRPVSSFHQCGLPEKILRHLEARGFEKPFPVQSQCIPILMCGRDLIAVAETG 127
I V G Q P P+ SF G P ++LR + + GF P P+Q+Q PI M GRD++A+A+TG
Sbjct 146 ITVSGGQVPPPLMSFEATGFPPELLREVLSAGFSAPTPIQAQSWPIAMQGRDIVAIAKTG 205
Query 128 SG 129
SG
Sbjct 206 SG 207
> dre:322206 ddx5, wu:fa56a07, wu:fb11e01, wu:fb16c10, wu:fb53b05;
DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 (EC:3.6.1.-);
K12823 ATP-dependent RNA helicase DDX5/DBP2 [EC:3.6.4.13]
Length=518
Score = 67.0 bits (162), Expect = 1e-11, Method: Composition-based stats.
Identities = 28/95 (29%), Positives = 55/95 (57%), Gaps = 1/95 (1%)
Query 35 KFPPIKKNLYVQVKELTDLKDHEVEAIRKTNGNIKVRGKQCPRPVSSFHQCGLPEKILRH 94
+ P +KN Y + ++ EVE R++ I V+G+ P+P+ FH+ P+ ++
Sbjct 52 ELPKFEKNFYQENPDVARRSAQEVEHYRRSK-EITVKGRDGPKPIVKFHEANFPKYVMDV 110
Query 95 LEARGFEKPFPVQSQCIPILMCGRDLIAVAETGSG 129
+ + + P P+Q+Q P+ + G+D++ +A+TGSG
Sbjct 111 ITKQNWTDPTPIQAQGWPVALSGKDMVGIAQTGSG 145
> hsa:55510 DDX43, CT13, DKFZp434H2114, HAGE; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 43 (EC:3.6.4.13)
Length=648
Score = 66.2 bits (160), Expect = 2e-11, Method: Composition-based stats.
Identities = 39/103 (37%), Positives = 57/103 (55%), Gaps = 11/103 (10%)
Query 36 FPPIKKNLYVQVKELTDLKDHEVEAIRKTNGNIKV------RGKQCPRPVSSFH---QCG 86
PPIKKN Y + + + E ++ RK N NI + P P +F QC
Sbjct 191 LPPIKKNFYKESTATSAMSKVEADSWRKENFNITWDDLKDGEKRPIPNPTCTFDDAFQC- 249
Query 87 LPEKILRHLEARGFEKPFPVQSQCIPILMCGRDLIAVAETGSG 129
PE ++ +++ GF+KP P+QSQ PI++ G DLI VA+TG+G
Sbjct 250 YPE-VMENIKKAGFQKPTPIQSQAWPIVLQGIDLIGVAQTGTG 291
> dre:562123 ddx59, si:dkey-39e8.2; DEAD (Asp-Glu-Ala-Asp) box
polypeptide 59 (EC:3.6.4.13)
Length=584
Score = 65.9 bits (159), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 30/80 (37%), Positives = 50/80 (62%), Gaps = 1/80 (1%)
Query 50 LTDLKDHEVEAIRKTNGNIKVRGKQCPRPVSSFHQCGLPEKILRHLEARGFEKPFPVQSQ 109
+++L + ++E ++ G + V + C RPV F C P + ++L+ G+E P PVQ Q
Sbjct 141 ISELTEEQIERVKAELGIVSVGTEVC-RPVIEFQHCRFPTVLEKNLKVAGYEAPTPVQMQ 199
Query 110 CIPILMCGRDLIAVAETGSG 129
+P+ + GRD+IA A+TGSG
Sbjct 200 MVPVGLTGRDVIATADTGSG 219
> cel:F58E10.3 hypothetical protein; K12823 ATP-dependent RNA
helicase DDX5/DBP2 [EC:3.6.4.13]
Length=561
Score = 63.9 bits (154), Expect = 1e-10, Method: Composition-based stats.
Identities = 35/101 (34%), Positives = 59/101 (58%), Gaps = 2/101 (1%)
Query 29 VDHASCKFPPIKKNLYVQVKELTDLKDHEVEAIRKTNGNIKVRGKQCPRPVSSFHQCGLP 88
VD ++ PI+K+ Y + ++ + +E++ N + + G+ PRPV F++ LP
Sbjct 80 VDWSAENLTPIEKDFYHENAAVSRREQYEIDQWVSAN-QVTLEGRGVPRPVFEFNEAPLP 138
Query 89 EKILRHLEARGFEKPFPVQSQCIPILMCGRDLIAVAETGSG 129
+I L + F+KP +QS PI M GRD+I++A+TGSG
Sbjct 139 GQIHELLYGK-FQKPTVIQSISWPIAMSGRDIISIAKTGSG 178
> hsa:9416 DDX23, MGC8416, PRPF28, U5-100K, U5-100KD, prp28; DEAD
(Asp-Glu-Ala-Asp) box polypeptide 23 (EC:3.6.4.13); K12858
ATP-dependent RNA helicase DDX23/PRP28 [EC:3.6.4.13]
Length=820
Score = 63.9 bits (154), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 31/82 (37%), Positives = 50/82 (60%), Gaps = 1/82 (1%)
Query 48 KELTDLKDHEVEAIRKTNGNIKVRGKQCPRPVSSFHQCGLPEKILRHLEARGFEKPFPVQ 107
K+L ++ D + R+ + +I +G + P P+ S+ LP IL ++ G+++P P+Q
Sbjct 360 KKLDEMTDRDWRIFRE-DYSITTKGGKIPNPIRSWKDSSLPPHILEVIDKCGYKEPTPIQ 418
Query 108 SQCIPILMCGRDLIAVAETGSG 129
Q IPI + RD+I VAETGSG
Sbjct 419 RQAIPIGLQNRDIIGVAETGSG 440
Lambda K H
0.320 0.138 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2044474180
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40