bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0405_orf2
Length=83
Score E
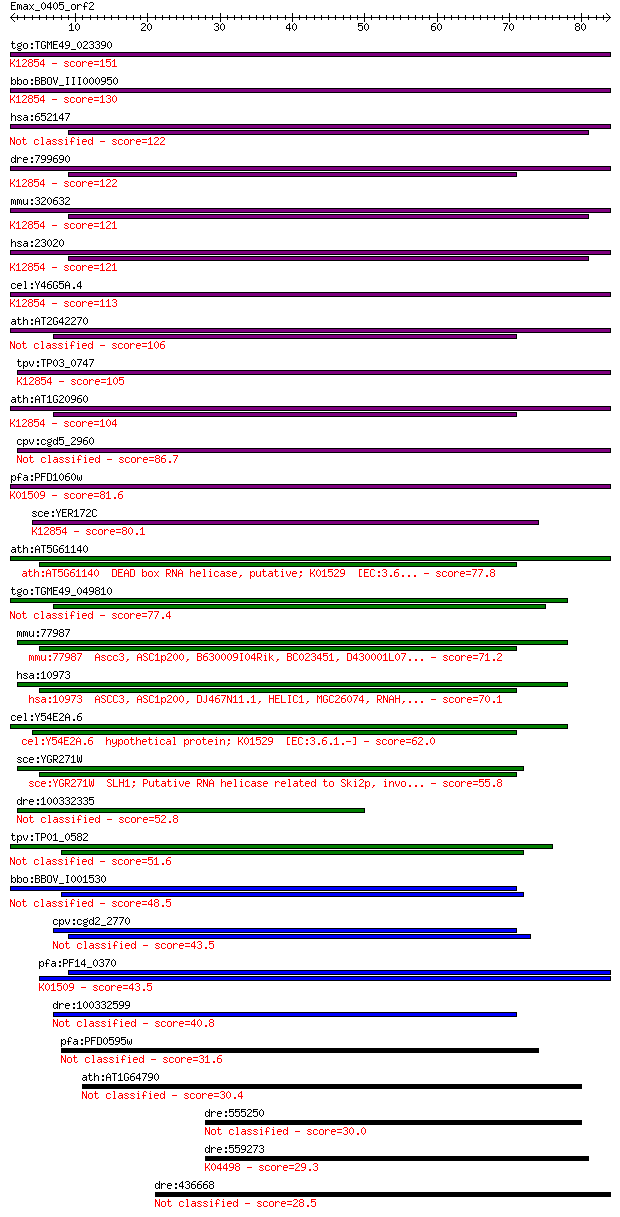
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_023390 sec63 domain-containing DEAD/DEAH box helica... 151 4e-37
bbo:BBOV_III000950 17.m07111; sec63 domain containing protein ... 130 1e-30
hsa:652147 u5 small nuclear ribonucleoprotein 200 kDa helicase... 122 3e-28
dre:799690 fb63a09; wu:fb63a09 (EC:3.6.4.13); K12854 pre-mRNA-... 122 4e-28
mmu:320632 Snrnp200, A330064G03Rik, Ascc3l1, BC011390, HELIC2,... 121 5e-28
hsa:23020 SNRNP200, ASCC3L1, BRR2, FLJ11521, HELIC2, RP33, U5-... 121 5e-28
cel:Y46G5A.4 hypothetical protein; K12854 pre-mRNA-splicing he... 113 2e-25
ath:AT2G42270 U5 small nuclear ribonucleoprotein helicase, put... 106 1e-23
tpv:TP03_0747 ATP-dependent RNA helicase; K12854 pre-mRNA-spli... 105 3e-23
ath:AT1G20960 emb1507 (embryo defective 1507); ATP binding / A... 104 7e-23
cpv:cgd5_2960 U5snrp Brr2 SFII RNA helicase (sec63 and the sec... 86.7 2e-17
pfa:PFD1060w U5 small nuclear ribonucleoprotein-specific prote... 81.6 6e-16
sce:YER172C BRR2, PRP44, RSS1, SLT22, SNU246; Brr2p (EC:3.6.1.... 80.1 1e-15
ath:AT5G61140 DEAD box RNA helicase, putative; K01529 [EC:3.6... 77.8 9e-15
tgo:TGME49_049810 activating signal cointegrator 1 complex sub... 77.4 9e-15
mmu:77987 Ascc3, ASC1p200, B630009I04Rik, BC023451, D430001L07... 71.2 7e-13
hsa:10973 ASCC3, ASC1p200, DJ467N11.1, HELIC1, MGC26074, RNAH,... 70.1 2e-12
cel:Y54E2A.6 hypothetical protein; K01529 [EC:3.6.1.-] 62.0 5e-10
sce:YGR271W SLH1; Putative RNA helicase related to Ski2p, invo... 55.8 4e-08
dre:100332335 mutagen-sensitive 308-like 52.8 2e-07
tpv:TP01_0582 RNA helicase 51.6 6e-07
bbo:BBOV_I001530 19.m02171; helicase 48.5 5e-06
cpv:cgd2_2770 U5 small nuclear ribonucleoprotein 200kDA helica... 43.5 1e-04
pfa:PF14_0370 DEAD/DEAH box helicase, putative; K01509 adenosi... 43.5 2e-04
dre:100332599 mutagen-sensitive 308-like 40.8 0.001
pfa:PFD0595w conserved Apicomplexan protein, unknown function 31.6 0.68
ath:AT1G64790 binding 30.4 1.6
dre:555250 dhx57, fc68d07, si:ch211-250c4.7, wu:fc68d07; DEAH ... 30.0 1.9
dre:559273 ep300a, gb:dq017643, im:7158255; E1A binding protei... 29.3 3.4
dre:436668 zgc:92712 28.5 5.5
> tgo:TGME49_023390 sec63 domain-containing DEAD/DEAH box helicase,
putative ; K12854 pre-mRNA-splicing helicase BRR2 [EC:3.6.4.13]
Length=2198
Score = 151 bits (382), Expect = 4e-37, Method: Composition-based stats.
Identities = 69/83 (83%), Positives = 78/83 (93%), Gaps = 0/83 (0%)
Query 1 RVKPEKGLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECAGKNQVLIFV 60
RV PEKGLF FGNHYRPVPLKQTY+GIKDKKAI+RYNTMN+VTYEK++E AGK+QVLIFV
Sbjct 720 RVAPEKGLFFFGNHYRPVPLKQTYIGIKDKKAIKRYNTMNEVTYEKLMESAGKSQVLIFV 779
Query 61 HSRKETVKTAKFICDAALQKDTL 83
HSRKETVKTA+FI D A+Q+DTL
Sbjct 780 HSRKETVKTARFIRDMAMQRDTL 802
> bbo:BBOV_III000950 17.m07111; sec63 domain containing protein
(EC:3.6.1.-); K12854 pre-mRNA-splicing helicase BRR2 [EC:3.6.4.13]
Length=2133
Score = 130 bits (326), Expect = 1e-30, Method: Composition-based stats.
Identities = 57/83 (68%), Positives = 69/83 (83%), Gaps = 0/83 (0%)
Query 1 RVKPEKGLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECAGKNQVLIFV 60
RV PEKGLF FGNHYRPV L+Q Y+GIK+KKA++RYN MN++ YE+V+E AGKNQVL+FV
Sbjct 665 RVDPEKGLFYFGNHYRPVGLEQRYIGIKEKKAVKRYNVMNELVYERVMEDAGKNQVLVFV 724
Query 61 HSRKETVKTAKFICDAALQKDTL 83
HSRKET +TAK I D A + D L
Sbjct 725 HSRKETARTAKLIRDMAFKTDNL 747
> hsa:652147 u5 small nuclear ribonucleoprotein 200 kDa helicase-like
Length=1700
Score = 122 bits (305), Expect = 3e-28, Method: Composition-based stats.
Identities = 56/83 (67%), Positives = 68/83 (81%), Gaps = 0/83 (0%)
Query 1 RVKPEKGLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECAGKNQVLIFV 60
RV P KGLF F N +RPVPL+QTYVGI +KKAI+R+ MN++ YEK++E AGKNQVL+FV
Sbjct 230 RVDPAKGLFYFDNSFRPVPLEQTYVGITEKKAIKRFQIMNEIVYEKIMEHAGKNQVLVFV 289
Query 61 HSRKETVKTAKFICDAALQKDTL 83
HSRKET KTA+ I D L+KDTL
Sbjct 290 HSRKETGKTARAIRDMCLEKDTL 312
Score = 34.3 bits (77), Expect = 0.11, Method: Composition-based stats.
Identities = 24/75 (32%), Positives = 36/75 (48%), Gaps = 3/75 (4%)
Query 9 FVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECAGKNQVLIFVHSRKETVK 68
F F + RPVPL+ G R +M + + + + K V++FV SRK+T
Sbjct 1076 FNFHPNVRPVPLELHIQGFNISHTQTRLLSMAKPVFHAITKHSPKKPVIVFVPSRKQTRL 1135
Query 69 TAKFI---CDAALQK 80
TA I C A +Q+
Sbjct 1136 TAIDILTTCAADIQR 1150
> dre:799690 fb63a09; wu:fb63a09 (EC:3.6.4.13); K12854 pre-mRNA-splicing
helicase BRR2 [EC:3.6.4.13]
Length=2134
Score = 122 bits (305), Expect = 4e-28, Method: Composition-based stats.
Identities = 56/83 (67%), Positives = 68/83 (81%), Gaps = 0/83 (0%)
Query 1 RVKPEKGLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECAGKNQVLIFV 60
RV P KGLF F N +RPVPL+QTYVGI +KKAI+R+ MN++ YEK++E AGKNQVL+FV
Sbjct 663 RVDPSKGLFYFDNSFRPVPLEQTYVGITEKKAIKRFQIMNEIVYEKIMEHAGKNQVLVFV 722
Query 61 HSRKETVKTAKFICDAALQKDTL 83
HSRKET KTA+ I D L+KDTL
Sbjct 723 HSRKETGKTARAIRDMCLEKDTL 745
Score = 33.9 bits (76), Expect = 0.14, Method: Composition-based stats.
Identities = 20/62 (32%), Positives = 30/62 (48%), Gaps = 0/62 (0%)
Query 9 FVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECAGKNQVLIFVHSRKETVK 68
F F + RPVPL+ G R +M Y +++ + VL+FV SR++T
Sbjct 1508 FNFHPNVRPVPLELHIQGFNVSHTQTRLLSMAKPVYHAIMKHSPSKPVLVFVPSRRQTRL 1567
Query 69 TA 70
TA
Sbjct 1568 TA 1569
> mmu:320632 Snrnp200, A330064G03Rik, Ascc3l1, BC011390, HELIC2,
KIAA0788, U5-200-KD, U5-200KD; small nuclear ribonucleoprotein
200 (U5) (EC:3.6.4.13); K12854 pre-mRNA-splicing helicase
BRR2 [EC:3.6.4.13]
Length=2136
Score = 121 bits (304), Expect = 5e-28, Method: Composition-based stats.
Identities = 56/83 (67%), Positives = 68/83 (81%), Gaps = 0/83 (0%)
Query 1 RVKPEKGLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECAGKNQVLIFV 60
RV P KGLF F N +RPVPL+QTYVGI +KKAI+R+ MN++ YEK++E AGKNQVL+FV
Sbjct 666 RVDPAKGLFYFDNSFRPVPLEQTYVGITEKKAIKRFQIMNEIVYEKIMEHAGKNQVLVFV 725
Query 61 HSRKETVKTAKFICDAALQKDTL 83
HSRKET KTA+ I D L+KDTL
Sbjct 726 HSRKETGKTARAIRDMCLEKDTL 748
Score = 35.4 bits (80), Expect = 0.042, Method: Composition-based stats.
Identities = 25/75 (33%), Positives = 36/75 (48%), Gaps = 3/75 (4%)
Query 9 FVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECAGKNQVLIFVHSRKETVK 68
F F + RPVPL+ G R +M Y + + + K V++FV SRK+T
Sbjct 1512 FNFHPNVRPVPLELHIQGFNISHTQTRLLSMAKPVYHAITKHSPKKPVIVFVPSRKQTRL 1571
Query 69 TAKFI---CDAALQK 80
TA I C A +Q+
Sbjct 1572 TAIDILTTCAADIQR 1586
> hsa:23020 SNRNP200, ASCC3L1, BRR2, FLJ11521, HELIC2, RP33, U5-200KD;
small nuclear ribonucleoprotein 200kDa (U5) (EC:3.6.4.13);
K12854 pre-mRNA-splicing helicase BRR2 [EC:3.6.4.13]
Length=2136
Score = 121 bits (304), Expect = 5e-28, Method: Composition-based stats.
Identities = 56/83 (67%), Positives = 68/83 (81%), Gaps = 0/83 (0%)
Query 1 RVKPEKGLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECAGKNQVLIFV 60
RV P KGLF F N +RPVPL+QTYVGI +KKAI+R+ MN++ YEK++E AGKNQVL+FV
Sbjct 666 RVDPAKGLFYFDNSFRPVPLEQTYVGITEKKAIKRFQIMNEIVYEKIMEHAGKNQVLVFV 725
Query 61 HSRKETVKTAKFICDAALQKDTL 83
HSRKET KTA+ I D L+KDTL
Sbjct 726 HSRKETGKTARAIRDMCLEKDTL 748
Score = 35.4 bits (80), Expect = 0.042, Method: Composition-based stats.
Identities = 25/75 (33%), Positives = 36/75 (48%), Gaps = 3/75 (4%)
Query 9 FVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECAGKNQVLIFVHSRKETVK 68
F F + RPVPL+ G R +M Y + + + K V++FV SRK+T
Sbjct 1512 FNFHPNVRPVPLELHIQGFNISHTQTRLLSMAKPVYHAITKHSPKKPVIVFVPSRKQTRL 1571
Query 69 TAKFI---CDAALQK 80
TA I C A +Q+
Sbjct 1572 TAIDILTTCAADIQR 1586
> cel:Y46G5A.4 hypothetical protein; K12854 pre-mRNA-splicing
helicase BRR2 [EC:3.6.4.13]
Length=2145
Score = 113 bits (282), Expect = 2e-25, Method: Composition-based stats.
Identities = 53/83 (63%), Positives = 67/83 (80%), Gaps = 1/83 (1%)
Query 1 RVKPEKGLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECAGKNQVLIFV 60
RVKPE L F N YRPVPL+Q Y+G+ +KKA++R+ MN+V Y+K++E AGK+QVL+FV
Sbjct 660 RVKPEH-LHFFDNSYRPVPLEQQYIGVTEKKALKRFQAMNEVVYDKIMEHAGKSQVLVFV 718
Query 61 HSRKETVKTAKFICDAALQKDTL 83
HSRKET KTAK I DA L+KDTL
Sbjct 719 HSRKETAKTAKAIRDACLEKDTL 741
> ath:AT2G42270 U5 small nuclear ribonucleoprotein helicase, putative
Length=2172
Score = 106 bits (265), Expect = 1e-23, Method: Composition-based stats.
Identities = 50/83 (60%), Positives = 61/83 (73%), Gaps = 0/83 (0%)
Query 1 RVKPEKGLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECAGKNQVLIFV 60
RV + GLF+F YRPVPL Q Y+GI KK +RR+ MND+ Y+KV+ AGK+QVLIFV
Sbjct 691 RVDLKNGLFIFDRSYRPVPLGQQYIGINVKKPLRRFQLMNDICYQKVVAVAGKHQVLIFV 750
Query 61 HSRKETVKTAKFICDAALQKDTL 83
HSRKET KTA+ I D A+ DTL
Sbjct 751 HSRKETAKTARAIRDTAMANDTL 773
Score = 31.6 bits (70), Expect = 0.64, Method: Composition-based stats.
Identities = 21/65 (32%), Positives = 31/65 (47%), Gaps = 1/65 (1%)
Query 7 GLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECA-GKNQVLIFVHSRKE 65
G+F F + RPVPL+ G+ R M TY +++ A K ++FV +RK
Sbjct 1536 GVFNFPPNVRPVPLEIHIHGVDILSFEARMQAMTKPTYTAIVQHAKNKKPAIVFVPTRKH 1595
Query 66 TVKTA 70
TA
Sbjct 1596 VRLTA 1600
> tpv:TP03_0747 ATP-dependent RNA helicase; K12854 pre-mRNA-splicing
helicase BRR2 [EC:3.6.4.13]
Length=2249
Score = 105 bits (262), Expect = 3e-23, Method: Composition-based stats.
Identities = 46/82 (56%), Positives = 66/82 (80%), Gaps = 0/82 (0%)
Query 2 VKPEKGLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECAGKNQVLIFVH 61
++ ++GLF FGNHYRPVPL+Q Y+GIK+KKA+++YN N++TYE V++ G+ QVL+FVH
Sbjct 693 LRVDEGLFYFGNHYRPVPLEQHYIGIKEKKALKQYNITNELTYEHVIKNIGEKQVLVFVH 752
Query 62 SRKETVKTAKFICDAALQKDTL 83
SRKET +T+K I D + +D L
Sbjct 753 SRKETYRTSKMILDKIVSEDKL 774
> ath:AT1G20960 emb1507 (embryo defective 1507); ATP binding /
ATP-dependent helicase/ helicase/ nucleic acid binding / nucleoside-triphosphatase/
nucleotide binding; K12854 pre-mRNA-splicing
helicase BRR2 [EC:3.6.4.13]
Length=2171
Score = 104 bits (259), Expect = 7e-23, Method: Composition-based stats.
Identities = 51/83 (61%), Positives = 61/83 (73%), Gaps = 0/83 (0%)
Query 1 RVKPEKGLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECAGKNQVLIFV 60
RV +KGLF F YRPVPL Q Y+GI KK ++R+ MND+ Y+KVL AGK+QVLIFV
Sbjct 690 RVDLKKGLFKFDRSYRPVPLHQQYIGISVKKPLQRFQLMNDLCYQKVLAGAGKHQVLIFV 749
Query 61 HSRKETVKTAKFICDAALQKDTL 83
HSRKET KTA+ I D A+ DTL
Sbjct 750 HSRKETSKTARAIRDTAMANDTL 772
Score = 33.1 bits (74), Expect = 0.22, Method: Composition-based stats.
Identities = 22/65 (33%), Positives = 30/65 (46%), Gaps = 1/65 (1%)
Query 7 GLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECA-GKNQVLIFVHSRKE 65
GLF F RPVPL+ G+ R M TY +++ A K ++FV +RK
Sbjct 1535 GLFNFPPGVRPVPLEIHIQGVDISSFEARMQAMTKPTYTAIVQHAKNKKPAIVFVPTRKH 1594
Query 66 TVKTA 70
TA
Sbjct 1595 VRLTA 1599
> cpv:cgd5_2960 U5snrp Brr2 SFII RNA helicase (sec63 and the second
part of the RNA
Length=1204
Score = 86.7 bits (213), Expect = 2e-17, Method: Composition-based stats.
Identities = 38/82 (46%), Positives = 57/82 (69%), Gaps = 0/82 (0%)
Query 2 VKPEKGLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECAGKNQVLIFVH 61
V P++GL+ FG +RPVPL QT++GIK KK ++ MN + Y+ V++ +Q+L+FVH
Sbjct 711 VNPKRGLYHFGPEFRPVPLLQTFIGIKAKKGFKKLQLMNSLVYDTVIKDITNHQILVFVH 770
Query 62 SRKETVKTAKFICDAALQKDTL 83
SRK+T+ TAK+I D A + L
Sbjct 771 SRKDTIHTAKYIRDTATENGML 792
> pfa:PFD1060w U5 small nuclear ribonucleoprotein-specific protein,
putative; K01509 adenosinetriphosphatase [EC:3.6.1.3]
Length=2874
Score = 81.6 bits (200), Expect = 6e-16, Method: Composition-based stats.
Identities = 46/83 (55%), Positives = 64/83 (77%), Gaps = 0/83 (0%)
Query 1 RVKPEKGLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECAGKNQVLIFV 60
R E+G+F F + +RPV ++Q Y+GIK+KK I++Y MN++TYEKVLE AGKNQ+LIFV
Sbjct 987 RAHIERGIFYFDHSFRPVQIEQHYIGIKEKKGIKKYALMNELTYEKVLEEAGKNQILIFV 1046
Query 61 HSRKETVKTAKFICDAALQKDTL 83
HSRKET +T+K + D ++ D L
Sbjct 1047 HSRKETYRTSKLLIDRFMKSDNL 1069
> sce:YER172C BRR2, PRP44, RSS1, SLT22, SNU246; Brr2p (EC:3.6.1.-);
K12854 pre-mRNA-splicing helicase BRR2 [EC:3.6.4.13]
Length=2163
Score = 80.1 bits (196), Expect = 1e-15, Method: Composition-based stats.
Identities = 34/71 (47%), Positives = 52/71 (73%), Gaps = 1/71 (1%)
Query 4 PEKGLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECAGK-NQVLIFVHS 62
P++GLF F + +RP PL Q + GIK++ ++++ MND YEKVLE + NQ+++FVHS
Sbjct 687 PKEGLFYFDSSFRPCPLSQQFCGIKERNSLKKLKAMNDACYEKVLESINEGNQIIVFVHS 746
Query 63 RKETVKTAKFI 73
RKET +TA ++
Sbjct 747 RKETSRTATWL 757
> ath:AT5G61140 DEAD box RNA helicase, putative; K01529 [EC:3.6.1.-]
Length=2146
Score = 77.8 bits (190), Expect = 9e-15, Method: Composition-based stats.
Identities = 38/84 (45%), Positives = 55/84 (65%), Gaps = 1/84 (1%)
Query 1 RVKPEKGLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECAGK-NQVLIF 59
RV + GLF F + YRPVPL Q Y+GI + R +N++ Y+KV++ + +Q +IF
Sbjct 692 RVNTDTGLFYFDSSYRPVPLAQQYIGITEHNFAARNELLNEICYKKVVDSIKQGHQAMIF 751
Query 60 VHSRKETVKTAKFICDAALQKDTL 83
VHSRK+T KTA+ + D A Q +TL
Sbjct 752 VHSRKDTSKTAEKLVDLARQYETL 775
Score = 43.9 bits (102), Expect = 1e-04, Method: Composition-based stats.
Identities = 26/66 (39%), Positives = 33/66 (50%), Gaps = 0/66 (0%)
Query 5 EKGLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECAGKNQVLIFVHSRK 64
E GLF F RPVP++ G K R N+MN Y + + VLIFV SR+
Sbjct 1525 EIGLFNFKPSVRPVPIEVHIQGYPGKYYCPRMNSMNKPAYAAICTHSPTKPVLIFVSSRR 1584
Query 65 ETVKTA 70
+T TA
Sbjct 1585 QTRLTA 1590
> tgo:TGME49_049810 activating signal cointegrator 1 complex subunit
3, putative (EC:5.99.1.3)
Length=2539
Score = 77.4 bits (189), Expect = 9e-15, Method: Composition-based stats.
Identities = 42/79 (53%), Positives = 53/79 (67%), Gaps = 4/79 (5%)
Query 1 RVKPEKGLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECAGKN--QVLI 58
RV+P + F FG RP+PL+QT VG + A RR +NDV Y KV+E A KN Q L+
Sbjct 878 RVEPSRAFF-FGADTRPIPLEQTLVGALESDAQRRRQKVNDVCYAKVVE-AVKNGHQALV 935
Query 59 FVHSRKETVKTAKFICDAA 77
FVHSR+ETV TA+F+ AA
Sbjct 936 FVHSRRETVATAEFLVQAA 954
Score = 30.4 bits (67), Expect = 1.6, Method: Composition-based stats.
Identities = 20/68 (29%), Positives = 26/68 (38%), Gaps = 0/68 (0%)
Query 7 GLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECAGKNQVLIFVHSRKET 66
GLF F RPVP G K R N MN +E +L A + +H+ E
Sbjct 1702 GLFNFKPAVRPVPCSVHIQGFPQKHYCPRMNAMNKPVFEALLTHASPDLSAEDMHTPPEI 1761
Query 67 VKTAKFIC 74
+ C
Sbjct 1762 LNGVYSTC 1769
> mmu:77987 Ascc3, ASC1p200, B630009I04Rik, BC023451, D430001L07Rik,
D630041L21, Helic1, RNAH; activating signal cointegrator
1 complex subunit 3; K01529 [EC:3.6.1.-]
Length=2198
Score = 71.2 bits (173), Expect = 7e-13, Method: Composition-based stats.
Identities = 34/77 (44%), Positives = 50/77 (64%), Gaps = 1/77 (1%)
Query 2 VKPEKGLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVL-ECAGKNQVLIFV 60
V P GLF F +RPVPL QT++GIK +++ N M++V YE VL + +QV++FV
Sbjct 664 VNPYIGLFYFDGRFRPVPLGQTFLGIKSTNKMQQLNNMDEVCYESVLKQVKAGHQVMVFV 723
Query 61 HSRKETVKTAKFICDAA 77
H+R TV+TA + + A
Sbjct 724 HARNATVRTAMSLIERA 740
Score = 39.7 bits (91), Expect = 0.002, Method: Composition-based stats.
Identities = 23/66 (34%), Positives = 33/66 (50%), Gaps = 0/66 (0%)
Query 5 EKGLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECAGKNQVLIFVHSRK 64
+ GLF F RPVPL+ G + R +MN ++ + + VLIFV SR+
Sbjct 1508 QMGLFNFRPSVRPVPLEVHIQGFPGQHYCPRMASMNKPAFQAIRSHSPAKPVLIFVSSRR 1567
Query 65 ETVKTA 70
+T TA
Sbjct 1568 QTRLTA 1573
> hsa:10973 ASCC3, ASC1p200, DJ467N11.1, HELIC1, MGC26074, RNAH,
dJ121G13.4; activating signal cointegrator 1 complex subunit
3; K01529 [EC:3.6.1.-]
Length=2202
Score = 70.1 bits (170), Expect = 2e-12, Method: Composition-based stats.
Identities = 34/77 (44%), Positives = 50/77 (64%), Gaps = 1/77 (1%)
Query 2 VKPEKGLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVL-ECAGKNQVLIFV 60
V P GLF F +RPVPL QT++GIK +++ N M++V YE VL + +QV++FV
Sbjct 663 VNPYIGLFFFDGRFRPVPLGQTFLGIKCANKMQQLNNMDEVCYENVLKQVKAGHQVMVFV 722
Query 61 HSRKETVKTAKFICDAA 77
H+R TV+TA + + A
Sbjct 723 HARNATVRTAMSLIERA 739
Score = 39.7 bits (91), Expect = 0.002, Method: Composition-based stats.
Identities = 23/66 (34%), Positives = 33/66 (50%), Gaps = 0/66 (0%)
Query 5 EKGLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECAGKNQVLIFVHSRK 64
+ GLF F RPVPL+ G + R +MN ++ + + VLIFV SR+
Sbjct 1507 QMGLFNFRPSVRPVPLEVHIQGFPGQHYCPRMASMNKPAFQAIRSHSPAKPVLIFVSSRR 1566
Query 65 ETVKTA 70
+T TA
Sbjct 1567 QTRLTA 1572
> cel:Y54E2A.6 hypothetical protein; K01529 [EC:3.6.1.-]
Length=1798
Score = 62.0 bits (149), Expect = 5e-10, Method: Composition-based stats.
Identities = 32/79 (40%), Positives = 47/79 (59%), Gaps = 2/79 (2%)
Query 1 RVKPEKGLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNT-MNDVTYEKVLECAGK-NQVLI 58
RV P KGLF F +RPVPL Q ++G + R T M++V Y++V++ + +QVL+
Sbjct 279 RVNPYKGLFFFDGRFRPVPLTQKFIGTRKAGNFRDNLTLMDNVCYDEVVDFVKRGHQVLV 338
Query 59 FVHSRKETVKTAKFICDAA 77
FVH+R T K + C A
Sbjct 339 FVHTRNGTAKLGEAFCARA 357
Score = 40.4 bits (93), Expect = 0.001, Method: Composition-based stats.
Identities = 22/67 (32%), Positives = 33/67 (49%), Gaps = 0/67 (0%)
Query 4 PEKGLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECAGKNQVLIFVHSR 63
P++ + F RPVP+ G + R MN Y+ +L + + VLIFV SR
Sbjct 1143 PDEACYNFRPSVRPVPISVHIQGFPGQHYCPRMGLMNKPAYKAILTYSPRKPVLIFVSSR 1202
Query 64 KETVKTA 70
++T TA
Sbjct 1203 RQTRLTA 1209
> sce:YGR271W SLH1; Putative RNA helicase related to Ski2p, involved
in translation inhibition of non-poly(A) mRNAs; required
for repressing propagation of dsRNA viruses (EC:3.6.1.-);
K01529 [EC:3.6.1.-]
Length=1967
Score = 55.8 bits (133), Expect = 4e-08, Method: Composition-based stats.
Identities = 29/72 (40%), Positives = 45/72 (62%), Gaps = 2/72 (2%)
Query 2 VKPEKGLFVFGNHYRPVPLKQTYVGIKDKKAIRRYN-TMNDVTYEKVLECAGKN-QVLIF 59
V + G+F F +RP PL+Q +G + K R+ ++ V Y+K+ E + QV++F
Sbjct 479 VNRQIGMFYFDQSFRPKPLEQQLLGCRGKAGSRQSKENIDKVAYDKLSEMIQRGYQVMVF 538
Query 60 VHSRKETVKTAK 71
VHSRKETVK+A+
Sbjct 539 VHSRKETVKSAR 550
Score = 37.0 bits (84), Expect = 0.014, Method: Composition-based stats.
Identities = 24/67 (35%), Positives = 33/67 (49%), Gaps = 1/67 (1%)
Query 5 EKGLFVFGNHYRPVPLKQTYVGIKDKKAI-RRYNTMNDVTYEKVLECAGKNQVLIFVHSR 63
+ GL+ F + RPVPLK G D A TMN + + + + LIFV SR
Sbjct 1320 DHGLYNFPSSVRPVPLKMYIDGFPDNLAFCPLMKTMNKPVFMAIKQHSPDKPALIFVASR 1379
Query 64 KETVKTA 70
++T TA
Sbjct 1380 RQTRLTA 1386
> dre:100332335 mutagen-sensitive 308-like
Length=654
Score = 52.8 bits (125), Expect = 2e-07, Method: Composition-based stats.
Identities = 23/48 (47%), Positives = 33/48 (68%), Gaps = 0/48 (0%)
Query 2 VKPEKGLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLE 49
V P GLF F + +RPVPL Q++VGIK +++ + M +V YEKVL+
Sbjct 600 VNPFIGLFYFDSRFRPVPLGQSFVGIKTTNKVQQLHDMEEVCYEKVLK 647
> tpv:TP01_0582 RNA helicase
Length=1764
Score = 51.6 bits (122), Expect = 6e-07, Method: Composition-based stats.
Identities = 31/77 (40%), Positives = 45/77 (58%), Gaps = 6/77 (7%)
Query 1 RVKPEKGLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLEC--AGKNQVLI 58
RV PE + FG YR VPL+Q + GIK+ + N M + ++ ++E +GK Q ++
Sbjct 314 RVPPEH-TYYFGREYRHVPLQQIFYGIKNDDIYK--NNMLTICFDHIVETLESGK-QCMV 369
Query 59 FVHSRKETVKTAKFICD 75
FVHSR ET TA I +
Sbjct 370 FVHSRNETFTTASRIVE 386
Score = 34.3 bits (77), Expect = 0.096, Method: Composition-based stats.
Identities = 20/64 (31%), Positives = 31/64 (48%), Gaps = 0/64 (0%)
Query 8 LFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECAGKNQVLIFVHSRKETV 67
++ F RPV G K R N+MN ++ ++ + VLIFV SR++T
Sbjct 1175 VYNFSPAVRPVKCNLFIDGFSIKAYCPRMNSMNKPCFDTIIRHDHSSNVLIFVSSRRQTR 1234
Query 68 KTAK 71
TA+
Sbjct 1235 MTAQ 1238
> bbo:BBOV_I001530 19.m02171; helicase
Length=1798
Score = 48.5 bits (114), Expect = 5e-06, Method: Composition-based stats.
Identities = 28/71 (39%), Positives = 39/71 (54%), Gaps = 5/71 (7%)
Query 1 RVKPEKGLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLEC-AGKNQVLIF 59
RV PE + FG YR VPL Q + G+K K TM ++ ++ +++ Q +IF
Sbjct 356 RVAPEHA-YHFGPEYRHVPLSQVFYGVKGKDIT---GTMYEICFDHIIQTLENGKQCIIF 411
Query 60 VHSRKETVKTA 70
VHSR ET TA
Sbjct 412 VHSRNETSMTA 422
Score = 32.3 bits (72), Expect = 0.35, Method: Composition-based stats.
Identities = 20/64 (31%), Positives = 29/64 (45%), Gaps = 0/64 (0%)
Query 8 LFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECAGKNQVLIFVHSRKETV 67
+F F RPV G K R N+MN + ++ VL+FV SR++T
Sbjct 1226 VFNFSPAVRPVKCHLYIDGFPLKAYCPRMNSMNRPAFSTIMRHDISAPVLVFVSSRRQTR 1285
Query 68 KTAK 71
TA+
Sbjct 1286 TTAR 1289
> cpv:cgd2_2770 U5 small nuclear ribonucleoprotein 200kDA helicase,
Pre-mRNA splicing helicase BRR2 2 (RNA helicase plus Sec63
domain)
Length=2184
Score = 43.5 bits (101), Expect = 1e-04, Method: Composition-based stats.
Identities = 25/64 (39%), Positives = 33/64 (51%), Gaps = 0/64 (0%)
Query 7 GLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECAGKNQVLIFVHSRKET 66
G + F RPVP G ++K R TMN Y K+L + K V+IFV SR++T
Sbjct 1457 GYYNFPPEIRPVPCTVYISGFQEKNYCPRMATMNRPIYNKILTHSPKKPVIIFVASRRQT 1516
Query 67 VKTA 70
TA
Sbjct 1517 RITA 1520
Score = 38.9 bits (89), Expect = 0.004, Method: Composition-based stats.
Identities = 29/111 (26%), Positives = 43/111 (38%), Gaps = 47/111 (42%)
Query 9 FVFGNHYRPVPLKQTYVGIKDKKA-------IRRYNT----------------------- 38
F F RP PL++T +G+ +K+ IR+ N
Sbjct 446 FFFSQALRPTPLEKTIIGVNEKRVDIEKKREIRKKNDKTSNQDSSLNKEKQREERQKDKI 505
Query 39 ----------------MNDVTYEKVLECAGKN-QVLIFVHSRKETVKTAKF 72
N + ++ VL+C KN Q L+FVHSR ET+ TA +
Sbjct 506 SKQINSKEDISSISDLYNSIAFKIVLDCLEKNEQALVFVHSRNETLSTALY 556
> pfa:PF14_0370 DEAD/DEAH box helicase, putative; K01509 adenosinetriphosphatase
[EC:3.6.1.3]
Length=2472
Score = 43.5 bits (101), Expect = 2e-04, Method: Composition-based stats.
Identities = 25/76 (32%), Positives = 42/76 (55%), Gaps = 1/76 (1%)
Query 9 FVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECAGKN-QVLIFVHSRKETV 67
+ F YR + L +T GI ++ + Y N TY +++ K+ Q +IFV SR ET
Sbjct 648 YYFNEKYRSIQLDKTLYGIHEENNNKLYIAKNIYTYNEIINSLKKDKQCIIFVCSRNETN 707
Query 68 KTAKFICDAALQKDTL 83
KT +F+ + AL+ + +
Sbjct 708 KTIEFLINHALKNNEI 723
Score = 38.9 bits (89), Expect = 0.004, Method: Composition-based stats.
Identities = 27/79 (34%), Positives = 36/79 (45%), Gaps = 0/79 (0%)
Query 5 EKGLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECAGKNQVLIFVHSRK 64
E LF F + R VP K +G K R + MN ++ + + A VLIFV SR+
Sbjct 1552 ENYLFNFPSSCRIVPCKTHILGFTQKAYCNRMSVMNKNVFDAINQYAQTKNVLIFVSSRR 1611
Query 65 ETVKTAKFICDAALQKDTL 83
+T TA I L L
Sbjct 1612 QTRLTAYDIISLNLSSHNL 1630
> dre:100332599 mutagen-sensitive 308-like
Length=864
Score = 40.8 bits (94), Expect = 0.001, Method: Composition-based stats.
Identities = 24/64 (37%), Positives = 32/64 (50%), Gaps = 0/64 (0%)
Query 7 GLFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECAGKNQVLIFVHSRKET 66
GLF F RPVPL+ G + R TMN ++ + + VLIFV SR++T
Sbjct 387 GLFNFRPSVRPVPLEVHIQGFPGQHYCPRMATMNKPVFQAIRTHSPAKPVLIFVSSRRQT 446
Query 67 VKTA 70
TA
Sbjct 447 RLTA 450
> pfa:PFD0595w conserved Apicomplexan protein, unknown function
Length=778
Score = 31.6 bits (70), Expect = 0.68, Method: Composition-based stats.
Identities = 20/66 (30%), Positives = 33/66 (50%), Gaps = 0/66 (0%)
Query 8 LFVFGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECAGKNQVLIFVHSRKETV 67
LFVF + + L Y+ K KK I Y N + EK+ + +NQ+L F + +
Sbjct 189 LFVFISFFTIFLLPLLYLTFKQKKYICPYCEKNLTSSEKLFKVKRENQILTFQFPKCAII 248
Query 68 KTAKFI 73
+AK++
Sbjct 249 ISAKYL 254
> ath:AT1G64790 binding
Length=2610
Score = 30.4 bits (67), Expect = 1.6, Method: Composition-based stats.
Identities = 19/69 (27%), Positives = 35/69 (50%), Gaps = 4/69 (5%)
Query 11 FGNHYRPVPLKQTYVGIKDKKAIRRYNTMNDVTYEKVLECAGKNQVLIFVHSRKETVKTA 70
G P+ + G+KD +R + +V+ AG++Q+L F+ T++TA
Sbjct 1898 LGERVLPLIIPILSKGLKDPDVDKRQGVC--IGLNEVMASAGRSQLLSFMDQLIPTIRTA 1955
Query 71 KFICDAALQ 79
+CD+AL+
Sbjct 1956 --LCDSALE 1962
> dre:555250 dhx57, fc68d07, si:ch211-250c4.7, wu:fc68d07; DEAH
(Asp-Glu-Ala-Asp/His) box polypeptide 57
Length=1417
Score = 30.0 bits (66), Expect = 1.9, Method: Composition-based stats.
Identities = 21/62 (33%), Positives = 31/62 (50%), Gaps = 10/62 (16%)
Query 28 KDKKAIRRYNTMNDV--------TYEKVLECAGKNQVLIF--VHSRKETVKTAKFICDAA 77
K K++ RRY +M + E +LEC KNQVL+ + +T + +FI D
Sbjct 551 KRKRSSRRYISMQEQRQKLPAWQKREAILECLVKNQVLVISGMTGCGKTTQIPQFILDNF 610
Query 78 LQ 79
LQ
Sbjct 611 LQ 612
> dre:559273 ep300a, gb:dq017643, im:7158255; E1A binding protein
p300 a; K04498 E1A/CREB-binding protein [EC:2.3.1.48]
Length=2667
Score = 29.3 bits (64), Expect = 3.4, Method: Composition-based stats.
Identities = 13/53 (24%), Positives = 30/53 (56%), Gaps = 2/53 (3%)
Query 28 KDKKAIRRYNTMNDVTYEKVLECAGKNQVLIFVHSRKETVKTAKFICDAALQK 80
KD+ ++ +T++ + + ++C K + +H+ ET+ + F+CD L+K
Sbjct 1237 KDQFEKKKNDTLDPELFVECMDCGRKMHQICVLHN--ETIWPSGFVCDGCLKK 1287
> dre:436668 zgc:92712
Length=180
Score = 28.5 bits (62), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 23/69 (33%), Positives = 34/69 (49%), Gaps = 7/69 (10%)
Query 21 KQTY-VGIKDKKAIRRYNTMNDVTYEKVLECAGKNQVLIFVHSRKETVKTAKFICDAA-- 77
K+T+ VG D KA + +YE LE GK+ + F+ +R + KT D A
Sbjct 52 KETFPVGCTDAKATITIEAITGFSYEYTLEINGKS-LQTFLDNRSKISKTWVMKLDGADY 110
Query 78 ---LQKDTL 83
L+KDT+
Sbjct 111 RIVLEKDTM 119
Lambda K H
0.322 0.137 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2044675024
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40