bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0399_orf1
Length=137
Score E
Sequences producing significant alignments: (Bits) Value
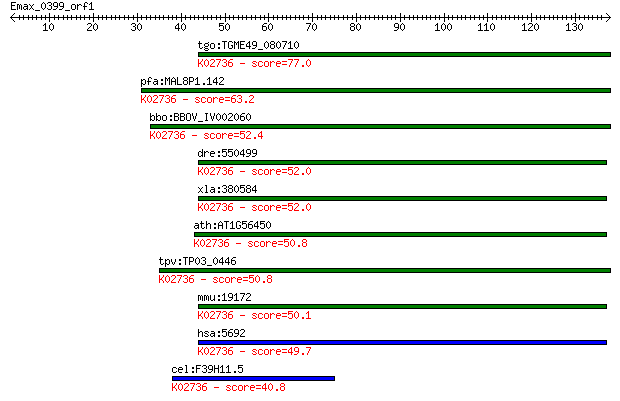
tgo:TGME49_080710 proteasome A-type and B-type domain-containi... 77.0 1e-14
pfa:MAL8P1.142 proteasome beta-subunit; K02736 20S proteasome ... 63.2 2e-10
bbo:BBOV_IV002060 21.m02891; proteasome subunit beta type 4 (E... 52.4 4e-07
dre:550499 psmb4, im:6909492, zgc:110330; proteasome (prosome,... 52.0 5e-07
xla:380584 psmb4, MGC69145; proteasome (prosome, macropain) su... 52.0 6e-07
ath:AT1G56450 PBG1; PBG1; peptidase/ threonine-type endopeptid... 50.8 1e-06
tpv:TP03_0446 20S proteasome subunit beta 7 (EC:3.4.25.1); K02... 50.8 1e-06
mmu:19172 Psmb4, Pros-27; proteasome (prosome, macropain) subu... 50.1 2e-06
hsa:5692 PSMB4, HN3, HsN3, PROS-26, PROS26; proteasome (prosom... 49.7 3e-06
cel:F39H11.5 pbs-7; Proteasome Beta Subunit family member (pbs... 40.8 0.001
> tgo:TGME49_080710 proteasome A-type and B-type domain-containing
protein (EC:3.4.25.1); K02736 20S proteasome subunit beta
7 [EC:3.4.25.1]
Length=266
Score = 77.0 bits (188), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 39/94 (41%), Positives = 56/94 (59%), Gaps = 17/94 (18%)
Query 44 TPKAIASYTARTLYNKRCRFNPYWLSVVVAGYMGPPTGIHQQQQQQQEQQQQEQQQQQGM 103
T A+ +R +Y KR R +P+WLSVVVAGY G ++ E ++ ++Q++
Sbjct 111 TAHQYAALLSRLMYQKRSRMDPWWLSVVVAGYQG----------ERSEAREVSEEQKKPF 160
Query 104 AKAGNEMYLGVADLLGTFFEEQVIATGLGRYFAI 137
LG D+ GTF+EE+VIATGLGRYFA+
Sbjct 161 T-------LGYVDMYGTFYEEEVIATGLGRYFAV 187
> pfa:MAL8P1.142 proteasome beta-subunit; K02736 20S proteasome
subunit beta 7 [EC:3.4.25.1]
Length=265
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 30/113 (26%), Positives = 56/113 (49%), Gaps = 6/113 (5%)
Query 31 NSSSSSSRTEALLTPKAIASYTARTLYNKRCRFNPYWLSVVVAG-----YMGPPTGIHQQ 85
N+ S R E + TP+ SY +R Y ++ R +P + ++++AG Y +
Sbjct 74 NNLSEKKRKEDMYTPQHYHSYVSRVFYVRKNRIDPLFNNIIIAGINSQKYDNNDDNVLLY 133
Query 86 QQQQQEQQQQEQQQQQGMAKA-GNEMYLGVADLLGTFFEEQVIATGLGRYFAI 137
+ + +Q E + + + +++Y+G D+ GT F + I TG RYFA+
Sbjct 134 TNKNNDDEQNEYKNNEEYKEIHKDDLYIGFVDMHGTNFCDDYITTGYARYFAL 186
> bbo:BBOV_IV002060 21.m02891; proteasome subunit beta type 4
(EC:3.4.25.1); K02736 20S proteasome subunit beta 7 [EC:3.4.25.1]
Length=226
Score = 52.4 bits (124), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 28/105 (26%), Positives = 46/105 (43%), Gaps = 30/105 (28%)
Query 33 SSSSSRTEALLTPKAIASYTARTLYNKRCRFNPYWLSVVVAGYMGPPTGIHQQQQQQQEQ 92
+++ L P+A+ +Y R Y +R + NP W S +VAG
Sbjct 73 DNNNDHASCHLGPEALHNYLGRVFYARRSKMNPLWNSAIVAGV----------------- 115
Query 93 QQQEQQQQQGMAKAGNEMYLGVADLLGTFFEEQVIATGLGRYFAI 137
+ N +LG AD+ GT +++ + TG+G+YFAI
Sbjct 116 -------------SQNGPFLGYADMYGTQYKDDFVVTGMGKYFAI 147
> dre:550499 psmb4, im:6909492, zgc:110330; proteasome (prosome,
macropain) subunit, beta type, 4 (EC:3.4.25.1); K02736 20S
proteasome subunit beta 7 [EC:3.4.25.1]
Length=215
Score = 52.0 bits (123), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 29/93 (31%), Positives = 39/93 (41%), Gaps = 30/93 (32%)
Query 44 TPKAIASYTARTLYNKRCRFNPYWLSVVVAGYMGPPTGIHQQQQQQQEQQQQEQQQQQGM 103
TPKAI S+ R +YN+R + NP W +VV+ G+
Sbjct 80 TPKAIHSWLTRVMYNRRSKMNPLWNTVVIGGFY--------------------------- 112
Query 104 AKAGNEMYLGVADLLGTFFEEQVIATGLGRYFA 136
E +LG D LG +E +ATG G Y A
Sbjct 113 ---NGESFLGYVDKLGVAYEAPTVATGFGAYLA 142
> xla:380584 psmb4, MGC69145; proteasome (prosome, macropain)
subunit, beta type 4 (EC:3.4.25.1); K02736 20S proteasome subunit
beta 7 [EC:3.4.25.1]
Length=248
Score = 52.0 bits (123), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 29/93 (31%), Positives = 39/93 (41%), Gaps = 30/93 (32%)
Query 44 TPKAIASYTARTLYNKRCRFNPYWLSVVVAGYMGPPTGIHQQQQQQQEQQQQEQQQQQGM 103
+PKAI S+ R +YN+R + NP W +VV+ G+
Sbjct 113 SPKAIHSWLTRVMYNRRSKMNPLWNTVVIGGFY--------------------------- 145
Query 104 AKAGNEMYLGVADLLGTFFEEQVIATGLGRYFA 136
E +LG D LG +E IATG G Y A
Sbjct 146 ---NGESFLGYVDKLGVAYEAPTIATGFGAYLA 175
> ath:AT1G56450 PBG1; PBG1; peptidase/ threonine-type endopeptidase;
K02736 20S proteasome subunit beta 7 [EC:3.4.25.1]
Length=246
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 28/94 (29%), Positives = 41/94 (43%), Gaps = 30/94 (31%)
Query 43 LTPKAIASYTARTLYNKRCRFNPYWLSVVVAGYMGPPTGIHQQQQQQQEQQQQEQQQQQG 102
L PK I +Y R +YN+R +FNP W ++V+ G
Sbjct 106 LGPKEIHNYLTRVMYNRRNKFNPLWNTLVLGG---------------------------- 137
Query 103 MAKAGNEMYLGVADLLGTFFEEQVIATGLGRYFA 136
K G YLG+ ++G FE+ +ATG G + A
Sbjct 138 -VKNGKS-YLGMVSMIGVSFEDDHVATGFGNHLA 169
> tpv:TP03_0446 20S proteasome subunit beta 7 (EC:3.4.25.1); K02736
20S proteasome subunit beta 7 [EC:3.4.25.1]
Length=229
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 30/104 (28%), Positives = 48/104 (46%), Gaps = 32/104 (30%)
Query 35 SSSRTEALLTPKAIASYTARTLYNKRCRFNPYWLSVVVAGYM-GPPTGIHQQQQQQQEQQ 93
++ ++ALL P+ + +YT+R LY++R + +P +S VV G G P
Sbjct 78 NNDASKALLNPEMLLNYTSRLLYHRRTKLDPVLVSAVVGGLSNGKP-------------- 123
Query 94 QQEQQQQQGMAKAGNEMYLGVADLLGTFFEEQVIATGLGRYFAI 137
+LG D GT + + + TGLG+YFAI
Sbjct 124 -----------------FLGYTDYYGTKYTDNFVVTGLGKYFAI 150
> mmu:19172 Psmb4, Pros-27; proteasome (prosome, macropain) subunit,
beta type 4 (EC:3.4.25.1); K02736 20S proteasome subunit
beta 7 [EC:3.4.25.1]
Length=264
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 27/93 (29%), Positives = 41/93 (44%), Gaps = 30/93 (32%)
Query 44 TPKAIASYTARTLYNKRCRFNPYWLSVVVAGYMGPPTGIHQQQQQQQEQQQQEQQQQQGM 103
+P+AI S+ R +Y++R + NP W ++V+ GY
Sbjct 129 SPRAIHSWLTRAMYSRRSKMNPLWNTMVIGGY---------------------------- 160
Query 104 AKAGNEMYLGVADLLGTFFEEQVIATGLGRYFA 136
A E +LG D+LG +E +ATG G Y A
Sbjct 161 --ADGESFLGYVDMLGVAYEAPSLATGYGAYLA 191
> hsa:5692 PSMB4, HN3, HsN3, PROS-26, PROS26; proteasome (prosome,
macropain) subunit, beta type, 4 (EC:3.4.25.1); K02736
20S proteasome subunit beta 7 [EC:3.4.25.1]
Length=264
Score = 49.7 bits (117), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 27/93 (29%), Positives = 41/93 (44%), Gaps = 30/93 (32%)
Query 44 TPKAIASYTARTLYNKRCRFNPYWLSVVVAGYMGPPTGIHQQQQQQQEQQQQEQQQQQGM 103
+P+AI S+ R +Y++R + NP W ++V+ GY
Sbjct 129 SPRAIHSWLTRAMYSRRSKMNPLWNTMVIGGY---------------------------- 160
Query 104 AKAGNEMYLGVADLLGTFFEEQVIATGLGRYFA 136
A E +LG D+LG +E +ATG G Y A
Sbjct 161 --ADGESFLGYVDMLGVAYEAPSLATGYGAYLA 191
> cel:F39H11.5 pbs-7; Proteasome Beta Subunit family member (pbs-7);
K02736 20S proteasome subunit beta 7 [EC:3.4.25.1]
Length=236
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 17/37 (45%), Positives = 23/37 (62%), Gaps = 0/37 (0%)
Query 38 RTEALLTPKAIASYTARTLYNKRCRFNPYWLSVVVAG 74
R + + PKA+ Y LY KRC+ NP W ++VVAG
Sbjct 85 RFDQDIGPKALHGYLTSLLYAKRCKMNPVWNTLVVAG 121
Lambda K H
0.315 0.128 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2428006156
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40