bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0390_orf1
Length=163
Score E
Sequences producing significant alignments: (Bits) Value
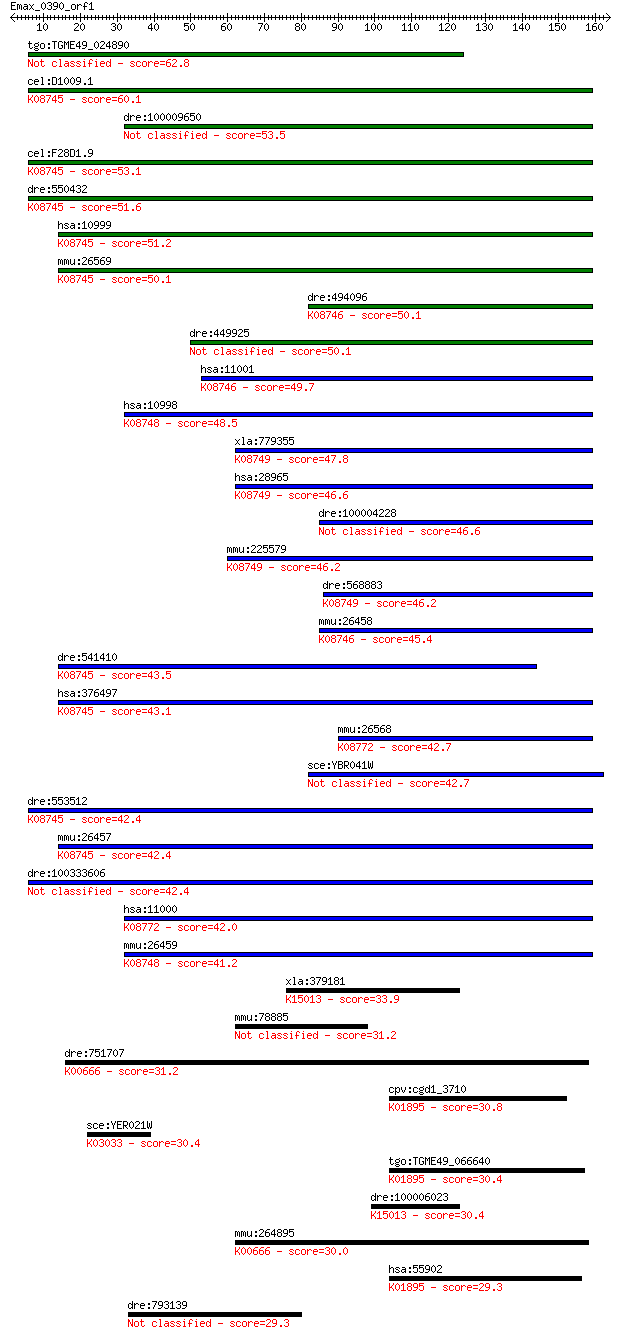
tgo:TGME49_024890 hypothetical protein 62.8 5e-10
cel:D1009.1 hypothetical protein; K08745 solute carrier family... 60.1 3e-09
dre:100009650 zgc:158482 53.5 3e-07
cel:F28D1.9 hypothetical protein; K08745 solute carrier family... 53.1 3e-07
dre:550432 slc27a4, zgc:112138; solute carrier family 27 (fatt... 51.6 1e-06
hsa:10999 SLC27A4, ACSVL4, FATP4, IPS; solute carrier family 2... 51.2 2e-06
mmu:26569 Slc27a4, BB144259, FATP4; solute carrier family 27 (... 50.1 3e-06
dre:494096 zgc:101540 (EC:6.2.1.3); K08746 solute carrier fami... 50.1 3e-06
dre:449925 slc27a2, MGC112376, im:7139614, wu:fb99g05, zgc:112... 50.1 3e-06
hsa:11001 SLC27A2, ACSVL1, FACVL1, FATP2, HsT17226, VLACS, VLC... 49.7 4e-06
hsa:10998 SLC27A5, ACSB, ACSVL6, FACVL3, FATP5, FLJ22987, VLAC... 48.5 9e-06
xla:779355 slc27a6, MGC154930; solute carrier family 27 (fatty... 47.8 2e-05
hsa:28965 SLC27A6, ACSVL2, DKFZp779M0564, FACVL2, FATP6, VLCS-... 46.6 4e-05
dre:100004228 si:dkey-147f23.1 46.6 4e-05
mmu:225579 Slc27a6, 4732438L20Rik, FACVL2, FATP6, VLCS-H1; sol... 46.2 5e-05
dre:568883 solute carrier family 27 (fatty acid transporter), ... 46.2 5e-05
mmu:26458 Slc27a2, ACSVL1, FATP2, VLCS, Vlac, Vlacs; solute ca... 45.4 7e-05
dre:541410 slc27a1a, fc10e12, slc27a1, wu:fc10e12, zgc:101649;... 43.5 3e-04
hsa:376497 SLC27A1, ACSVL5, FATP, FATP1, FLJ00336, MGC71751; s... 43.1 4e-04
mmu:26568 Slc27a3, Acsvl3, FATP3, Vlcs-3; solute carrier famil... 42.7 5e-04
sce:YBR041W FAT1; Fat1p 42.7 6e-04
dre:553512 slc27a1b, Slc27a1, zgc:153860; solute carrier famil... 42.4 7e-04
mmu:26457 Slc27a1, FATP1, Fatp; solute carrier family 27 (fatt... 42.4 7e-04
dre:100333606 solute carrier family 27 (fatty acid transporter... 42.4 7e-04
hsa:11000 SLC27A3, ACSVL3, FATP3, MGC4365, VLCS-3; solute carr... 42.0 9e-04
mmu:26459 Slc27a5, FACVL3, FATP5, MGC143799, VLCS-H2, VLCSH2, ... 41.2 0.001
xla:379181 acsbg2, MGC132130, MGC53673; acyl-CoA synthetase bu... 33.9 0.25
mmu:78885 Coro7, 0610011B16Rik, AW048373, AW556392; coronin 7 31.2
dre:751707 acsf2, MGC152887, im:7145554, wu:fi49b09, zgc:15288... 31.2 1.7
cpv:cgd1_3710 acetyl-coenzyme A synthetase ; K01895 acetyl-CoA... 30.8 2.3
sce:YER021W RPN3, SUN2; Essential, non-ATPase regulatory subun... 30.4 2.6
tgo:TGME49_066640 acetyl-coenzyme A synthetase, putative (EC:6... 30.4 3.0
dre:100006023 long-chain-fatty-acid--CoA ligase ACSBG1-like; K... 30.4 3.0
mmu:264895 Acsf2, MGC25878; acyl-CoA synthetase family member ... 30.0 3.8
hsa:55902 ACSS2, ACAS2, ACECS, ACS, ACSA, DKFZp762G026, dJ1161... 29.3 6.1
dre:793139 evpl, wu:fb17b09; envoplakin 29.3 6.6
> tgo:TGME49_024890 hypothetical protein
Length=1103
Score = 62.8 bits (151), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 34/118 (28%), Positives = 50/118 (42%), Gaps = 0/118 (0%)
Query 6 LFNAWNMPGACAFVPDFAWNSKELERIVEFNEETNEVYRDPTTKRGKEASRDEKTLVQRG 65
L N+ G C F+P+ + R+ F+E E+ R P T +A +RG
Sbjct 946 LINSLGKMGPCGFIPNTVQGERGTARLARFDEVKQEIVRGPHTGLCLDAELRSTLDQRRG 1005
Query 66 ELVSHIDPEHGLCLFTNYGETEQFLYRDLLEAGDLWCRSGDIMERDAAGFLYLSKRIG 123
E + ++ F + T +YR+ D WCRSGD+M D GF Y G
Sbjct 1006 EAIVNLADRDEWLGFVDDDVTRAMVYRNSFRQNDAWCRSGDLMSVDRLGFFYFLGHTG 1063
> cel:D1009.1 hypothetical protein; K08745 solute carrier family
27 (fatty acid transporter), member 1/4 [EC:6.2.1.-]
Length=655
Score = 60.1 bits (144), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 48/157 (30%), Positives = 71/157 (45%), Gaps = 10/157 (6%)
Query 6 LFNAWNMPGACAFVPDFA-WNSKELERIVEFNEETNEVYRDPTTKRGKEASRDEKTLVQR 64
+ N N GAC F+P + S R+++ + T E+ RD K +
Sbjct 398 IVNVDNHVGACGFMPIYPHIGSLYPVRLIKVDRATGELERD------KNGLCVPCVPGET 451
Query 65 GELVSHIDPEHGLCLFTNY---GETEQFLYRDLLEAGDLWCRSGDIMERDAAGFLYLSKR 121
GE+V I + L F Y G+T + +YRD+ + GD SGDI+ D G+LY R
Sbjct 452 GEMVGVIKEKDILLKFEGYVSEGDTAKKIYRDVFKHGDKVFASGDILHWDDLGYLYFVDR 511
Query 122 IGSSFFIDGEAAPVHELAKSLEHESGVREAHVEGLQV 158
G +F GE E+ L+ V +A V G+ V
Sbjct 512 CGDTFRWKGENVSTTEVEGILQPVMDVEDATVYGVTV 548
> dre:100009650 zgc:158482
Length=619
Score = 53.5 bits (127), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 39/131 (29%), Positives = 62/131 (47%), Gaps = 13/131 (9%)
Query 32 IVEFNEETNEVYRDPTTKRGKEASRDEKTLVQRGELVSHIDPEHGLCLFTNYG----ETE 87
+++F+ E E RD ++ R E + + L+ ++H+ P F Y +TE
Sbjct 395 LIQFDTEREEPVRD-SSGRCVEVPKGQTGLLV--SQITHMAP------FVGYAHDEQQTE 445
Query 88 QFLYRDLLEAGDLWCRSGDIMERDAAGFLYLSKRIGSSFFIDGEAAPVHELAKSLEHESG 147
+ RD+ GD++ SGD+M D FLY R+G +F GE E++ L
Sbjct 446 RKRLRDVFRRGDVYFNSGDLMRMDQDNFLYFIDRVGDTFRWKGENVATTEVSDVLSLLEC 505
Query 148 VREAHVEGLQV 158
V EA V G+ V
Sbjct 506 VAEASVYGVCV 516
> cel:F28D1.9 hypothetical protein; K08745 solute carrier family
27 (fatty acid transporter), member 1/4 [EC:6.2.1.-]
Length=684
Score = 53.1 bits (126), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 44/157 (28%), Positives = 70/157 (44%), Gaps = 10/157 (6%)
Query 6 LFNAWNMPGACAFVPDFAWNSK-ELERIVEFNEETNEVYRDPTTKRGKEASRDEKTLVQR 64
L N GAC F+P K R+++ ++ T E R T G + + +
Sbjct 428 LVNIDGHVGACGFLPISPLTKKMHPVRLIKVDDVTGEAIR---TSDGLCIACNPG---ES 481
Query 65 GELVSHIDPEHGLCLFTNY---GETEQFLYRDLLEAGDLWCRSGDIMERDAAGFLYLSKR 121
G +VS I + L F Y ET + + RD+ GD +GD++ D G++Y R
Sbjct 482 GAMVSTIRKNNPLLQFEGYLNKKETNKKIIRDVFAKGDSCFLTGDLLHWDRLGYVYFKDR 541
Query 122 IGSSFFIDGEAAPVHELAKSLEHESGVREAHVEGLQV 158
G +F GE E+ L +G+ +A V G++V
Sbjct 542 TGDTFRWKGENVSTTEVEAILHPITGLSDATVYGVEV 578
> dre:550432 slc27a4, zgc:112138; solute carrier family 27 (fatty
acid transporter), member 4; K08745 solute carrier family
27 (fatty acid transporter), member 1/4 [EC:6.2.1.-]
Length=643
Score = 51.6 bits (122), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 44/158 (27%), Positives = 69/158 (43%), Gaps = 14/158 (8%)
Query 6 LFNAWNMPGACAFVPDFAWNSKELE-----RIVEFNEETNEVYRDPTTKRGKEASRDEKT 60
L N N GAC F NS+ L R+V+ +EET E+ R P +
Sbjct 393 LGNFDNKTGACGF------NSRILPYVYPIRLVKVDEETMELIRGPDGVCIPCGPGEPGQ 446
Query 61 LVQRGELVSHIDPEHGLCLFTNYGETEQFLYRDLLEAGDLWCRSGDIMERDAAGFLYLSK 120
LV R + DP + N T + + +D+ + GD SGD++ D G++Y
Sbjct 447 LVGR---IIQNDPLRRFDGYVNQTATNKKIAQDVFKKGDSAYLSGDVLVMDDFGYMYFRD 503
Query 121 RIGSSFFIDGEAAPVHELAKSLEHESGVREAHVEGLQV 158
R G +F GE E+ +L +++ V G++V
Sbjct 504 RTGDTFRWKGENVSTTEVEGTLSRLLDMKDVVVYGVEV 541
> hsa:10999 SLC27A4, ACSVL4, FATP4, IPS; solute carrier family
27 (fatty acid transporter), member 4 (EC:6.2.1.-); K08745
solute carrier family 27 (fatty acid transporter), member 1/4
[EC:6.2.1.-]
Length=643
Score = 51.2 bits (121), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 41/150 (27%), Positives = 64/150 (42%), Gaps = 14/150 (9%)
Query 14 GACAFVPDFAWNSKELE-----RIVEFNEETNEVYRDPTTKRGKEASRDEKTLVQRGELV 68
GAC F NS+ L R+V NE+T E+ R P + LV R +
Sbjct 401 GACGF------NSRILSFVYPIRLVRVNEDTMELIRGPDGVCIPCQPGEPGQLVGR---I 451
Query 69 SHIDPEHGLCLFTNYGETEQFLYRDLLEAGDLWCRSGDIMERDAAGFLYLSKRIGSSFFI 128
DP + N G + + +D+ + GD +GD++ D G+LY R G +F
Sbjct 452 IQKDPLRRFDGYLNQGANNKKIAKDVFKKGDQAYLTGDVLVMDELGYLYFRDRTGDTFRW 511
Query 129 DGEAAPVHELAKSLEHESGVREAHVEGLQV 158
GE E+ +L + + V G++V
Sbjct 512 KGENVSTTEVEGTLSRLLDMADVAVYGVEV 541
> mmu:26569 Slc27a4, BB144259, FATP4; solute carrier family 27
(fatty acid transporter), member 4 (EC:6.2.1.-); K08745 solute
carrier family 27 (fatty acid transporter), member 1/4 [EC:6.2.1.-]
Length=643
Score = 50.1 bits (118), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 43/153 (28%), Positives = 65/153 (42%), Gaps = 20/153 (13%)
Query 14 GACAFVPDFAWNSKELE-----RIVEFNEETNEVYRDPTTKRGKEASRDEKTLVQRGELV 68
GAC F NS+ L R+V NE+T E+ R P + Q G+LV
Sbjct 401 GACGF------NSRILSFVYPIRLVRVNEDTMELIRGP------DGVCIPCQPGQPGQLV 448
Query 69 SHI---DPEHGLCLFTNYGETEQFLYRDLLEAGDLWCRSGDIMERDAAGFLYLSKRIGSS 125
I DP + N G + + D+ + GD +GD++ D G+LY R G +
Sbjct 449 GRIIQQDPLRRFDGYLNQGANNKKIANDVFKKGDQAYLTGDVLVMDELGYLYFRDRTGDT 508
Query 126 FFIDGEAAPVHELAKSLEHESGVREAHVEGLQV 158
F GE E+ +L + + V G++V
Sbjct 509 FRWKGENVSTTEVEGTLSRLLHMADVAVYGVEV 541
> dre:494096 zgc:101540 (EC:6.2.1.3); K08746 solute carrier family
27 (fatty acid transporter), member 2 [EC:6.2.1.- 6.2.1.3]
Length=620
Score = 50.1 bits (118), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 26/77 (33%), Positives = 41/77 (53%), Gaps = 0/77 (0%)
Query 82 NYGETEQFLYRDLLEAGDLWCRSGDIMERDAAGFLYLSKRIGSSFFIDGEAAPVHELAKS 141
N +T++ D+ E GDL+ SGD++ D F+Y R+G +F GE E+A
Sbjct 441 NKQQTDKKRLADVFEKGDLYFHSGDLLRIDHQNFVYFQDRVGDTFRWKGENVATTEVADI 500
Query 142 LEHESGVREAHVEGLQV 158
L + EA+V G++V
Sbjct 501 LTMVDCIEEANVYGVKV 517
> dre:449925 slc27a2, MGC112376, im:7139614, wu:fb99g05, zgc:112376;
solute carrier family 27 (fatty acid transporter), member
2 (EC:6.2.1.-)
Length=614
Score = 50.1 bits (118), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 35/113 (30%), Positives = 55/113 (48%), Gaps = 7/113 (6%)
Query 50 RGKEASRDEKTLVQRGELVSHIDPEHGLCLFTNYG----ETEQFLYRDLLEAGDLWCRSG 105
RG + E + G LV+ I H L F Y +TE+ RD+ + GD++ +G
Sbjct 402 RGSDGLCVEAAPGETGLLVAKI---HKLAPFEGYAKNSTQTEKKRLRDVFQRGDMYFNTG 458
Query 106 DIMERDAAGFLYLSKRIGSSFFIDGEAAPVHELAKSLEHESGVREAHVEGLQV 158
D++ D GFL+ RIG +F GE E+++ L + A+V G+ V
Sbjct 459 DLILADRQGFLFFQDRIGDTFRWKGENVATTEVSEILLMLDFIEAANVYGVTV 511
> hsa:11001 SLC27A2, ACSVL1, FACVL1, FATP2, HsT17226, VLACS, VLCS,
hFACVL1; solute carrier family 27 (fatty acid transporter),
member 2 (EC:6.2.1.3); K08746 solute carrier family 27
(fatty acid transporter), member 2 [EC:6.2.1.- 6.2.1.3]
Length=567
Score = 49.7 bits (117), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 35/113 (30%), Positives = 54/113 (47%), Gaps = 7/113 (6%)
Query 53 EASRDEK---TLVQRGELVSHIDPEHGLCLFTNYG----ETEQFLYRDLLEAGDLWCRSG 105
E RDE V +GE+ + L F Y +TE+ RD+ + GDL+ SG
Sbjct 352 EPVRDENGYCVRVPKGEVGLLVCKITQLTPFNGYAGAKAQTEKKKLRDVFKKGDLYFNSG 411
Query 106 DIMERDAAGFLYLSKRIGSSFFIDGEAAPVHELAKSLEHESGVREAHVEGLQV 158
D++ D F+Y R+G +F GE E+A ++ V+E +V G+ V
Sbjct 412 DLLMVDHENFIYFHDRVGDTFRWKGENVATTEVADTVGLVDFVQEVNVYGVHV 464
> hsa:10998 SLC27A5, ACSB, ACSVL6, FACVL3, FATP5, FLJ22987, VLACSR,
VLCS-H2, VLCSH2; solute carrier family 27 (fatty acid
transporter), member 5 (EC:6.2.1.7); K08748 solute carrier family
27 (fatty acid transporter), member 5 [EC:6.2.1.7]
Length=690
Score = 48.5 bits (114), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 33/128 (25%), Positives = 62/128 (48%), Gaps = 7/128 (5%)
Query 32 IVEFNEETNEVYRDPTTKRGKEASRDEKTLVQRGELVSHIDPEHGLCLFTNYGE-TEQFL 90
+V+F+ E E RD + L + G L++ + + + E +E+ L
Sbjct 466 LVQFDMEAAEPVRD------NQGFCIPVGLGEPGLLLTKVVSQQPFVGYRGPRELSERKL 519
Query 91 YRDLLEAGDLWCRSGDIMERDAAGFLYLSKRIGSSFFIDGEAAPVHELAKSLEHESGVRE 150
R++ ++GD++ +GD++ D GFLY R+G +F GE HE+ L +++
Sbjct 520 VRNVRQSGDVYYNTGDVLAMDREGFLYFRDRLGDTFRWKGENVSTHEVEGVLSQVDFLQQ 579
Query 151 AHVEGLQV 158
+V G+ V
Sbjct 580 VNVYGVCV 587
> xla:779355 slc27a6, MGC154930; solute carrier family 27 (fatty
acid transporter), member 6; K08749 solute carrier family
27 (fatty acid transporter), member 6
Length=621
Score = 47.8 bits (112), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 28/101 (27%), Positives = 53/101 (52%), Gaps = 4/101 (3%)
Query 62 VQRGE---LVSHIDPEHGLCLFT-NYGETEQFLYRDLLEAGDLWCRSGDIMERDAAGFLY 117
V++GE L+S ++ + + N T + L ++ GD++ +GD+M +D FLY
Sbjct 418 VKKGETGLLISKVNQNNPFFGYAGNKNHTTKKLLCNVFRKGDVYFNTGDLMVQDHENFLY 477
Query 118 LSKRIGSSFFIDGEAAPVHELAKSLEHESGVREAHVEGLQV 158
RIG +F GE E+A + + ++EA++ G+ +
Sbjct 478 FRDRIGDTFRWKGENVATTEVADIIGMLNFIQEANIYGVAI 518
> hsa:28965 SLC27A6, ACSVL2, DKFZp779M0564, FACVL2, FATP6, VLCS-H1;
solute carrier family 27 (fatty acid transporter), member
6; K08749 solute carrier family 27 (fatty acid transporter),
member 6
Length=619
Score = 46.6 bits (109), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 28/101 (27%), Positives = 53/101 (52%), Gaps = 4/101 (3%)
Query 62 VQRGE---LVSHIDPEHGLCLFTN-YGETEQFLYRDLLEAGDLWCRSGDIMERDAAGFLY 117
V++GE L+S ++ ++ + Y T+ L D+ + GD++ +GD++ +D FLY
Sbjct 416 VKKGEPGLLISRVNAKNPFFGYAGPYKHTKDKLLCDVFKKGDVYLNTGDLIVQDQDNFLY 475
Query 118 LSKRIGSSFFIDGEAAPVHELAKSLEHESGVREAHVEGLQV 158
R G +F GE E+A + ++EA+V G+ +
Sbjct 476 FWDRTGDTFRWKGENVATTEVADVIGMLDFIQEANVYGVAI 516
> dre:100004228 si:dkey-147f23.1
Length=616
Score = 46.6 bits (109), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 22/74 (29%), Positives = 39/74 (52%), Gaps = 0/74 (0%)
Query 85 ETEQFLYRDLLEAGDLWCRSGDIMERDAAGFLYLSKRIGSSFFIDGEAAPVHELAKSLEH 144
+TE+ D+ E GD++ +GD+ D F+Y R+G +F GE +E++ +
Sbjct 440 QTEKKKLHDVFEKGDVYFNTGDLFRTDRENFIYFQDRVGDTFRWKGENVSTNEVSDIMTL 499
Query 145 ESGVREAHVEGLQV 158
+ EA+V G+ V
Sbjct 500 VPCIEEANVYGVTV 513
> mmu:225579 Slc27a6, 4732438L20Rik, FACVL2, FATP6, VLCS-H1; solute
carrier family 27 (fatty acid transporter), member 6;
K08749 solute carrier family 27 (fatty acid transporter), member
6
Length=619
Score = 46.2 bits (108), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 31/106 (29%), Positives = 55/106 (51%), Gaps = 10/106 (9%)
Query 60 TLVQRGE---LVSHIDPEHGLCLFTNYG----ETEQFLYRDLLEAGDLWCRSGDIMERDA 112
+ V++GE L+S ++ ++ F Y T+ L D+ GD++ +GD+M +D
Sbjct 414 SCVRKGEPGLLISRVNKKNP---FFGYAGSDTHTKSKLLFDVFRKGDVYFNTGDLMFQDQ 470
Query 113 AGFLYLSKRIGSSFFIDGEAAPVHELAKSLEHESGVREAHVEGLQV 158
F+Y R+G +F GE E+A L ++EA+V G++V
Sbjct 471 ENFVYFWDRLGDTFRWKGENVATTEVADVLGRLDFIQEANVYGVRV 516
> dre:568883 solute carrier family 27 (fatty acid transporter),
member 6-like; K08749 solute carrier family 27 (fatty acid
transporter), member 6
Length=621
Score = 46.2 bits (108), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 23/73 (31%), Positives = 41/73 (56%), Gaps = 0/73 (0%)
Query 86 TEQFLYRDLLEAGDLWCRSGDIMERDAAGFLYLSKRIGSSFFIDGEAAPVHELAKSLEHE 145
+E+ L RD+ + GD++ +GD+M +D F+Y RIG +F GE E+++ L
Sbjct 446 SEKKLLRDVFKTGDVYFNTGDLMLQDHRDFVYFKDRIGDTFRWKGENVSTTEVSEVLGSL 505
Query 146 SGVREAHVEGLQV 158
+ + +V G+ V
Sbjct 506 DFLLDVNVYGVTV 518
> mmu:26458 Slc27a2, ACSVL1, FATP2, VLCS, Vlac, Vlacs; solute
carrier family 27 (fatty acid transporter), member 2 (EC:6.2.1.3);
K08746 solute carrier family 27 (fatty acid transporter),
member 2 [EC:6.2.1.- 6.2.1.3]
Length=620
Score = 45.4 bits (106), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 24/74 (32%), Positives = 39/74 (52%), Gaps = 0/74 (0%)
Query 85 ETEQFLYRDLLEAGDLWCRSGDIMERDAAGFLYLSKRIGSSFFIDGEAAPVHELAKSLEH 144
+TE+ RD+ + GD++ SGD++ D F+Y R+G +F GE E+A +
Sbjct 444 QTEKKKLRDVFKKGDIYFNSGDLLMIDRENFVYFHDRVGDTFRWKGENVATTEVADIVGL 503
Query 145 ESGVREAHVEGLQV 158
V E +V G+ V
Sbjct 504 VDFVEEVNVYGVPV 517
> dre:541410 slc27a1a, fc10e12, slc27a1, wu:fc10e12, zgc:101649;
solute carrier family 27 (fatty acid transporter), member
1a; K08745 solute carrier family 27 (fatty acid transporter),
member 1/4 [EC:6.2.1.-]
Length=647
Score = 43.5 bits (101), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 39/133 (29%), Positives = 58/133 (43%), Gaps = 10/133 (7%)
Query 14 GACAFVPDFAWNSKELERIVEFNEETNEVYRDPTTKRGKEASRDEKTLVQRGELVSHIDP 73
GAC F N + R+V+ NEET E+ RD K+G S + G LV I+
Sbjct 405 GACGFNSRILPNVYPI-RLVKVNEETMELVRD---KQGLCVSCRPG---EPGLLVGRINQ 457
Query 74 EHGLCLFTNYGE---TEQFLYRDLLEAGDLWCRSGDIMERDAAGFLYLSKRIGSSFFIDG 130
+ L F Y T + + ++ D SGD++ D G++Y R G +F G
Sbjct 458 QDPLRRFDGYASQEATRKKIAYNVFRKNDSAYLSGDVLVMDELGYMYFRDRSGDTFRWKG 517
Query 131 EAAPVHELAKSLE 143
E E+ +L
Sbjct 518 ENVSTTEVEGTLS 530
> hsa:376497 SLC27A1, ACSVL5, FATP, FATP1, FLJ00336, MGC71751;
solute carrier family 27 (fatty acid transporter), member 1;
K08745 solute carrier family 27 (fatty acid transporter),
member 1/4 [EC:6.2.1.-]
Length=646
Score = 43.1 bits (100), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 43/156 (27%), Positives = 64/156 (41%), Gaps = 26/156 (16%)
Query 14 GACAFVPDFAWNSKELE-----RIVEFNEETNEVYRDPTTKRGKEASRDEKTLVQRGE-- 66
G+C F NS+ L R+V+ NE+T E+ RD ++ Q GE
Sbjct 404 GSCGF------NSRILPHVYPIRLVKVNEDTMELLRD---------AQGLCIPCQAGEPG 448
Query 67 -LVSHIDPEHGLCLFTNY---GETEQFLYRDLLEAGDLWCRSGDIMERDAAGFLYLSKRI 122
LV I+ + L F Y T + + + GD SGD++ D G++Y R
Sbjct 449 LLVGQINQQDPLRRFDGYVSESATSKKIAHSVFSKGDSAYLSGDVLVMDELGYMYFRDRS 508
Query 123 GSSFFIDGEAAPVHELAKSLEHESGVREAHVEGLQV 158
G +F GE E+ L G + V G+ V
Sbjct 509 GDTFRWRGENVSTTEVEGVLSRLLGQTDVAVYGVAV 544
> mmu:26568 Slc27a3, Acsvl3, FATP3, Vlcs-3; solute carrier family
27 (fatty acid transporter), member 3 (EC:6.2.1.-); K08772
solute carrier family 27 (fatty acid transporter), member
3 [EC:6.2.1.-]
Length=667
Score = 42.7 bits (99), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 22/69 (31%), Positives = 39/69 (56%), Gaps = 0/69 (0%)
Query 90 LYRDLLEAGDLWCRSGDIMERDAAGFLYLSKRIGSSFFIDGEAAPVHELAKSLEHESGVR 149
L +D+ +GD++ +GD++ D GFL+ R G +F GE E+A+ LE ++
Sbjct 496 LLKDVFWSGDVFFNTGDLLVCDEQGFLHFHDRTGDTFRWKGENVATTEVAEVLETLDFLQ 555
Query 150 EAHVEGLQV 158
E ++ G+ V
Sbjct 556 EVNIYGVTV 564
> sce:YBR041W FAT1; Fat1p
Length=669
Score = 42.7 bits (99), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 23/80 (28%), Positives = 37/80 (46%), Gaps = 0/80 (0%)
Query 82 NYGETEQFLYRDLLEAGDLWCRSGDIMERDAAGFLYLSKRIGSSFFIDGEAAPVHELAKS 141
N ET+ + RD+ GD W R GD+++ D G Y R+G +F E E+
Sbjct 484 NAKETKSKVVRDVFRRGDAWYRCGDLLKADEYGLWYFLDRMGDTFRWKSENVSTTEVEDQ 543
Query 142 LEHESGVREAHVEGLQVFLP 161
L + + A V + + +P
Sbjct 544 LTASNKEQYAQVLVVGIKVP 563
> dre:553512 slc27a1b, Slc27a1, zgc:153860; solute carrier family
27 (fatty acid transporter), member 1b; K08745 solute carrier
family 27 (fatty acid transporter), member 1/4 [EC:6.2.1.-]
Length=648
Score = 42.4 bits (98), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 44/159 (27%), Positives = 64/159 (40%), Gaps = 16/159 (10%)
Query 6 LFNAWNMPGACAFVPDFAWNSKELERIVEFNEETNEVYRDPTTKRGKEASRDEKTLVQRG 65
L N N GAC F S R++ +E+T E+ RD SR + G
Sbjct 398 LANMDNKVGACGF-NSVVLPSVYPIRLLRADEDTMELIRD---------SRGLCVPCKPG 447
Query 66 E---LVSHIDPEHGLCLFTNYGE---TEQFLYRDLLEAGDLWCRSGDIMERDAAGFLYLS 119
E +V I+P+ L F Y T + + ++ GD SGD+M D G++Y
Sbjct 448 EPGIIVGRINPQDPLRRFDGYANEEATSKKISHNVFRKGDSAYVSGDLMVMDELGYVYFR 507
Query 120 KRIGSSFFIDGEAAPVHELAKSLEHESGVREAHVEGLQV 158
R G +F GE E+ L + V G+ V
Sbjct 508 DRGGDTFRWRGENVSTTEVEGVLSALLKQTDVAVYGVSV 546
> mmu:26457 Slc27a1, FATP1, Fatp; solute carrier family 27 (fatty
acid transporter), member 1; K08745 solute carrier family
27 (fatty acid transporter), member 1/4 [EC:6.2.1.-]
Length=646
Score = 42.4 bits (98), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 44/156 (28%), Positives = 62/156 (39%), Gaps = 26/156 (16%)
Query 14 GACAFVPDFAWNSKELE-----RIVEFNEETNEVYRDPTTKRGKEASRDEKTLVQRGE-- 66
G+C F NS+ L R+V+ NE+T E RD S Q GE
Sbjct 404 GSCGF------NSRILTHVYPIRLVKVNEDTMEPLRD---------SEGLCIPCQPGEPG 448
Query 67 -LVSHIDPEHGLCLFTNY---GETEQFLYRDLLEAGDLWCRSGDIMERDAAGFLYLSKRI 122
LV I+ + L F Y T + + + GD SGD++ D G++Y R
Sbjct 449 LLVGQINQQDPLRRFDGYVSDSATNKKIAHSVFRKGDSAYLSGDVLVMDELGYMYFRDRS 508
Query 123 GSSFFIDGEAAPVHELAKSLEHESGVREAHVEGLQV 158
G +F GE E+ L G + V G+ V
Sbjct 509 GDTFRWRGENVSTTEVEAVLSRLLGQTDVAVYGVAV 544
> dre:100333606 solute carrier family 27 (fatty acid transporter),
member 1-like
Length=523
Score = 42.4 bits (98), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 44/159 (27%), Positives = 64/159 (40%), Gaps = 16/159 (10%)
Query 6 LFNAWNMPGACAFVPDFAWNSKELERIVEFNEETNEVYRDPTTKRGKEASRDEKTLVQRG 65
L N N GAC F S R++ +E+T E+ RD SR + G
Sbjct 273 LANMDNKVGACGF-NSVVLPSVYPIRLLRADEDTMELIRD---------SRGLCVPCKPG 322
Query 66 E---LVSHIDPEHGLCLFTNYGE---TEQFLYRDLLEAGDLWCRSGDIMERDAAGFLYLS 119
E +V I+P+ L F Y T + + ++ GD SGD+M D G++Y
Sbjct 323 EPGIIVGRINPQDPLRRFDGYANEEATSKKISHNVFRKGDSAYVSGDLMVMDELGYVYFR 382
Query 120 KRIGSSFFIDGEAAPVHELAKSLEHESGVREAHVEGLQV 158
R G +F GE E+ L + V G+ V
Sbjct 383 DRGGDTFRWRGENVSTTEVEGVLSALLKQTDVAVYGVSV 421
> hsa:11000 SLC27A3, ACSVL3, FATP3, MGC4365, VLCS-3; solute carrier
family 27 (fatty acid transporter), member 3 (EC:6.2.1.-);
K08772 solute carrier family 27 (fatty acid transporter),
member 3 [EC:6.2.1.-]
Length=730
Score = 42.0 bits (97), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 32/128 (25%), Positives = 57/128 (44%), Gaps = 7/128 (5%)
Query 32 IVEFNEETNEVYRDPTTKRGKEASRDEKTLVQRGELVSHIDPEHGLCLFTNYGETEQF-L 90
++ ++ T E RDP + + + G LV+ + + + E Q L
Sbjct 506 LIRYDVTTGEPIRDP------QGHCMATSPGEPGLLVAPVSQQSPFLGYAGGPELAQGKL 559
Query 91 YRDLLEAGDLWCRSGDIMERDAAGFLYLSKRIGSSFFIDGEAAPVHELAKSLEHESGVRE 150
+D+ GD++ +GD++ D GFL R G +F GE E+A+ E ++E
Sbjct 560 LKDVFRPGDVFFNTGDLLVCDDQGFLRFHDRTGDTFRWKGENVATTEVAEVFEALDFLQE 619
Query 151 AHVEGLQV 158
+V G+ V
Sbjct 620 VNVYGVTV 627
> mmu:26459 Slc27a5, FACVL3, FATP5, MGC143799, VLCS-H2, VLCSH2,
Vlacsr; solute carrier family 27 (fatty acid transporter),
member 5 (EC:6.2.1.7); K08748 solute carrier family 27 (fatty
acid transporter), member 5 [EC:6.2.1.7]
Length=689
Score = 41.2 bits (95), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 33/128 (25%), Positives = 58/128 (45%), Gaps = 7/128 (5%)
Query 32 IVEFNEETNEVYRDPTTKRGKEASRDEKTLVQRGELVSHIDPEHGLCLFT-NYGETEQFL 90
+V+F+ ET E RD K+G + + G L++ + + + E+ + L
Sbjct 465 LVQFDIETAEPLRD---KQGFCIPVEPG---KPGLLLTKVRKNQPFLGYRGSQAESNRKL 518
Query 91 YRDLLEAGDLWCRSGDIMERDAAGFLYLSKRIGSSFFIDGEAAPVHELAKSLEHESGVRE 150
++ GDL+ +GD++ D GF Y R+G +F GE E+ L + E
Sbjct 519 VANVRRVGDLYFNTGDVLTLDQEGFFYFQDRLGDTFRWKGENVSTGEVECVLSSLDFLEE 578
Query 151 AHVEGLQV 158
+V G+ V
Sbjct 579 VNVYGVPV 586
> xla:379181 acsbg2, MGC132130, MGC53673; acyl-CoA synthetase
bubblegum family member 2 (EC:6.2.1.3); K15013 long-chain-fatty-acid--CoA
ligase ACSBG [EC:6.2.1.3]
Length=739
Score = 33.9 bits (76), Expect = 0.25, Method: Composition-based stats.
Identities = 19/47 (40%), Positives = 24/47 (51%), Gaps = 2/47 (4%)
Query 76 GLCLFTNYGETEQFLYRDLLEAGDLWCRSGDIMERDAAGFLYLSKRI 122
G +F Y E + L E G W SGDI + D GFLY++ RI
Sbjct 528 GRHVFMGYLNMEDKTHESLDEEG--WLHSGDIGKHDENGFLYITGRI 572
> mmu:78885 Coro7, 0610011B16Rik, AW048373, AW556392; coronin
7
Length=922
Score = 31.2 bits (69), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 16/36 (44%), Positives = 19/36 (52%), Gaps = 0/36 (0%)
Query 62 VQRGELVSHIDPEHGLCLFTNYGETEQFLYRDLLEA 97
V L+ DP+ GL L T G+T FLY L EA
Sbjct 726 VAPSTLLPSYDPDTGLVLLTGKGDTRVFLYEVLPEA 761
> dre:751707 acsf2, MGC152887, im:7145554, wu:fi49b09, zgc:152887;
acyl-CoA synthetase family member 2 (EC:6.2.1.-); K00666
fatty-acyl-CoA synthase [EC:6.2.1.-]
Length=606
Score = 31.2 bits (69), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 34/142 (23%), Positives = 56/142 (39%), Gaps = 16/142 (11%)
Query 16 CAFVPDFAWNSKELERIVEFNEETNEVYRDPTTKRGKEASRDEKTLVQRGELVSHIDPEH 75
C F D A +++E + + T DPTT + L +GEL+
Sbjct 410 CGFPVDSA--ERKIETVGCISPHTEAKVVDPTTG-------EIVPLGAQGELMIR----- 455
Query 76 GLCLFTNYGETEQFLYRDLLEAGDLWCRSGDIMERDAAGFLYLSKRIGSSFFIDGEAAPV 135
G C+ Y + E+ + + D W ++GDI D + + RI GE
Sbjct 456 GYCVMLEYWQDEEKTRECITK--DRWYKTGDIASLDQFAYCKIEGRIKDLIIRGGENIYP 513
Query 136 HELAKSLEHESGVREAHVEGLQ 157
E+ + L + EA V G++
Sbjct 514 AEIEQFLHTHPKILEAQVVGVK 535
> cpv:cgd1_3710 acetyl-coenzyme A synthetase ; K01895 acetyl-CoA
synthetase [EC:6.2.1.1]
Length=695
Score = 30.8 bits (68), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 14/48 (29%), Positives = 26/48 (54%), Gaps = 0/48 (0%)
Query 104 SGDIMERDAAGFLYLSKRIGSSFFIDGEAAPVHELAKSLEHESGVREA 151
+GD + RD G+L+++ RI + + G E+ +L + G+ EA
Sbjct 535 TGDGVLRDQDGYLWITGRIDDTINVSGHRLSSKEIEDALTNHFGIAEA 582
> sce:YER021W RPN3, SUN2; Essential, non-ATPase regulatory subunit
of the 26S proteasome lid, similar to the p58 subunit of
the human 26S proteasome; temperature-sensitive alleles cause
metaphase arrest, suggesting a role for the proteasome in
cell cycle control; K03033 26S proteasome regulatory subunit
N3
Length=523
Score = 30.4 bits (67), Expect = 2.6, Method: Composition-based stats.
Identities = 11/17 (64%), Positives = 15/17 (88%), Gaps = 0/17 (0%)
Query 22 FAWNSKELERIVEFNEE 38
F W+SKELE++VEFN +
Sbjct 145 FLWDSKELEQLVEFNRK 161
> tgo:TGME49_066640 acetyl-coenzyme A synthetase, putative (EC:6.2.1.1);
K01895 acetyl-CoA synthetase [EC:6.2.1.1]
Length=535
Score = 30.4 bits (67), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 15/53 (28%), Positives = 27/53 (50%), Gaps = 0/53 (0%)
Query 104 SGDIMERDAAGFLYLSKRIGSSFFIDGEAAPVHELAKSLEHESGVREAHVEGL 156
+GD + RDA G+ +++ R+ + + G E+ +L V EA V G+
Sbjct 377 TGDGVFRDADGYYWITGRVDDTLNVSGHRLTTAEIEHALVQHDDVAEAAVVGV 429
> dre:100006023 long-chain-fatty-acid--CoA ligase ACSBG1-like;
K15013 long-chain-fatty-acid--CoA ligase ACSBG [EC:6.2.1.3]
Length=674
Score = 30.4 bits (67), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 12/24 (50%), Positives = 17/24 (70%), Gaps = 0/24 (0%)
Query 99 DLWCRSGDIMERDAAGFLYLSKRI 122
D W RSGD+ + D GF+Y++ RI
Sbjct 496 DGWLRSGDLGKVDEDGFIYINGRI 519
> mmu:264895 Acsf2, MGC25878; acyl-CoA synthetase family member
2 (EC:6.2.1.-); K00666 fatty-acyl-CoA synthase [EC:6.2.1.-]
Length=615
Score = 30.0 bits (66), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 28/100 (28%), Positives = 39/100 (39%), Gaps = 6/100 (6%)
Query 62 VQRGELVSHIDPE----HGLCLFTNYGETEQFLYRDLLEAGDLWCRSGDIMERDAAGFLY 117
V+ GEL + P G C+ Y Q + + + D W R+GDI D GF
Sbjct 447 VETGELTNLNVPGELYIRGYCVMQGYWGEPQKTFETVGQ--DKWYRTGDIALMDEQGFCK 504
Query 118 LSKRIGSSFFIDGEAAPVHELAKSLEHESGVREAHVEGLQ 157
+ R GE EL V+EA V G++
Sbjct 505 IVGRSKDMIIRGGENIYPAELEDFFLKHPQVQEAQVVGVK 544
> hsa:55902 ACSS2, ACAS2, ACECS, ACS, ACSA, DKFZp762G026, dJ1161H23.1;
acyl-CoA synthetase short-chain family member 2 (EC:6.2.1.1);
K01895 acetyl-CoA synthetase [EC:6.2.1.1]
Length=714
Score = 29.3 bits (64), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 15/52 (28%), Positives = 24/52 (46%), Gaps = 0/52 (0%)
Query 104 SGDIMERDAAGFLYLSKRIGSSFFIDGEAAPVHELAKSLEHESGVREAHVEG 155
+GD +RD G+ +++ RI + G E+ +L V EA V G
Sbjct 563 TGDGCQRDQDGYYWITGRIDDMLNVSGHLLSTAEVESALVEHEAVAEAAVVG 614
> dre:793139 evpl, wu:fb17b09; envoplakin
Length=1771
Score = 29.3 bits (64), Expect = 6.6, Method: Composition-based stats.
Identities = 18/48 (37%), Positives = 28/48 (58%), Gaps = 3/48 (6%)
Query 33 VEFNEETNEVYR-DPTTKRGKEASRDEKTLVQRGELVSHIDPEHGLCL 79
+EFNE +E+YR DP T+R E R L++ G+ + I+ E L +
Sbjct 946 IEFNEIIHEIYRIDPETER--ELRRLRNELIEIGKRRTSIESEISLIM 991
Lambda K H
0.318 0.137 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3832864436
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40