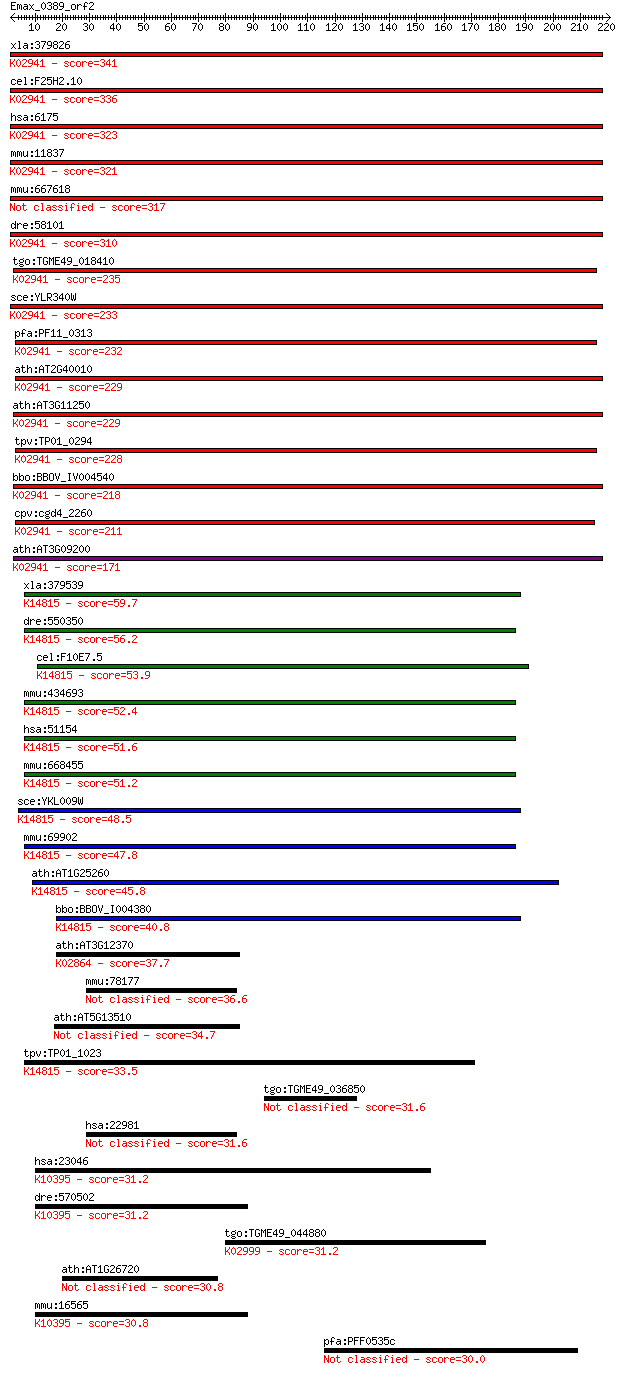
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0389_orf2
Length=220
Score E
Sequences producing significant alignments: (Bits) Value
xla:379826 rplp0, MGC53408, arbp; ribosomal protein, large, P0... 341 1e-93
cel:F25H2.10 rpa-0; Replication Protein A homolog family membe... 336 4e-92
hsa:6175 RPLP0, L10E, MGC111226, MGC88175, P0, PRLP0, RPP0; ri... 323 3e-88
mmu:11837 Rplp0, 36B4, Arbp, MGC107165, MGC107166; ribosomal p... 321 1e-87
mmu:667618 Gm8730, EG667618; predicted pseudogene 8730 317 2e-86
dre:58101 rplp0, arp, fb04a12, wu:fb15a08, wu:fk48c12; ribosom... 310 3e-84
tgo:TGME49_018410 60S acidic ribosomal protein P0 (EC:4.2.99.1... 235 1e-61
sce:YLR340W RPP0, RPL10E; P0; L10E; A0; K02941 large subunit r... 233 3e-61
pfa:PF11_0313 60S ribosomal protein P0; K02941 large subunit r... 232 6e-61
ath:AT2G40010 60S acidic ribosomal protein P0 (RPP0A); K02941 ... 229 5e-60
ath:AT3G11250 60S acidic ribosomal protein P0 (RPP0C); K02941 ... 229 8e-60
tpv:TP01_0294 60S acidic ribosomal protein, P0; K02941 large s... 228 9e-60
bbo:BBOV_IV004540 23.m05776; phosphoriboprotein P0; K02941 lar... 218 1e-56
cpv:cgd4_2260 ribosomal protein PO like protein of the L10 fam... 211 1e-54
ath:AT3G09200 60S acidic ribosomal protein P0 (RPP0B); K02941 ... 171 1e-42
xla:379539 mrto4, MGC68920, mrt4; mRNA turnover 4 homolog; K14... 59.7 8e-09
dre:550350 wu:fb71a02; zgc:110388; K14815 mRNA turnover protein 4 56.2
cel:F10E7.5 hypothetical protein; K14815 mRNA turnover protein 4 53.9
mmu:434693 Gm5633; predicted gene 5633; K14815 mRNA turnover p... 52.4 1e-06
hsa:51154 MRTO4, C1orf33, MRT4, dJ657E11.4; mRNA turnover 4 ho... 51.6 2e-06
mmu:668455 Gm9178, EG668455; predicted gene 9178; K14815 mRNA ... 51.2 2e-06
sce:YKL009W MRT4; Mrt4p; K14815 mRNA turnover protein 4 48.5
mmu:69902 Mrto4, 2610012O22Rik, Mg684, Mrt4; MRT4, mRNA turnov... 47.8 3e-05
ath:AT1G25260 acidic ribosomal protein P0-related; K14815 mRNA... 45.8 1e-04
bbo:BBOV_I004380 19.m02064; hypothetical protein; K14815 mRNA ... 40.8 0.003
ath:AT3G12370 ribosomal protein L10 family protein; K02864 lar... 37.7 0.035
mmu:78177 Ninl, 4930519N13Rik, Gm1004, Gm1634, mKIAA0980; nine... 36.6 0.070
ath:AT5G13510 ribosomal protein L10 family protein 34.7 0.28
tpv:TP01_1023 hypothetical protein; K14815 mRNA turnover prote... 33.5 0.61
tgo:TGME49_036850 hypothetical protein 31.6 2.2
hsa:22981 NINL, FLJ11792, KIAA0980, NLP, dJ691N24.1; ninein-like 31.6 2.4
hsa:23046 KIF21B, FLJ16314; kinesin family member 21B; K10395 ... 31.2 2.6
dre:570502 hCG2027369-like; K10395 kinesin family member 4/7/2... 31.2 2.9
tgo:TGME49_044880 DNA-directed RNA polymerase I largest subuni... 31.2 3.2
ath:AT1G26720 hypothetical protein 30.8 3.7
mmu:16565 Kif21b, 2610511N21Rik, KIAA0449, mKIAA0449; kinesin ... 30.8 3.9
pfa:PFF0535c trancription factor, putative 30.0 7.4
> xla:379826 rplp0, MGC53408, arbp; ribosomal protein, large,
P0; K02941 large subunit ribosomal protein LP0
Length=315
Score = 341 bits (874), Expect = 1e-93, Method: Compositional matrix adjust.
Identities = 161/217 (74%), Positives = 190/217 (87%), Gaps = 0/217 (0%)
Query 1 ASYFTKLVQLLDEYPKCFIVGVDNVGSKQMQQIRHSLREDATLLMGKNTMIRKAIRGILE 60
++YF K++QLLD+YPKCFIVG DNVGSKQMQQIR SLR A +LMGKNTM+RKAIRG LE
Sbjct 11 SNYFLKIIQLLDDYPKCFIVGADNVGSKQMQQIRMSLRGKAVVLMGKNTMMRKAIRGHLE 70
Query 61 KNPALEKLLPYIKSNIGFVFTDKDLSEVRDKILGNKVQAPARAGALAPCDVKIPAQNTGL 120
NPALEKLL +IK N+GFVFT +DL+EVRD +L NKV A ARAGA+APC+V +PAQNTGL
Sbjct 71 NNPALEKLLSHIKGNVGFVFTKEDLTEVRDMLLANKVPASARAGAIAPCEVTVPAQNTGL 130
Query 121 GPEKTSFFQALSIPTKISKGTIEILNEIHLIKKDDRVGASEATLLNMLNISPFTYGLVIR 180
GPEKTSFFQAL I TKIS+GTIEIL+++ LIK D+VGASEATLLNMLNISPF+YGL+I+
Sbjct 131 GPEKTSFFQALGITTKISRGTIEILSDVQLIKTGDKVGASEATLLNMLNISPFSYGLIIQ 190
Query 181 QVYDSGTVFEPEILDITPEDLRARFLEGVRNVAAVFL 217
QVYD+G+++ PE+LDIT E L RFLEGVRNVA+V L
Sbjct 191 QVYDNGSIYSPEVLDITEEALHVRFLEGVRNVASVCL 227
> cel:F25H2.10 rpa-0; Replication Protein A homolog family member
(rpa-0); K02941 large subunit ribosomal protein LP0
Length=312
Score = 336 bits (861), Expect = 4e-92, Method: Compositional matrix adjust.
Identities = 153/217 (70%), Positives = 190/217 (87%), Gaps = 0/217 (0%)
Query 1 ASYFTKLVQLLDEYPKCFIVGVDNVGSKQMQQIRHSLREDATLLMGKNTMIRKAIRGILE 60
A+YFTKLV+L +EYPKC +VGVDNVGSKQMQ+IR ++R A +LMGKNTMIRKA+RG L
Sbjct 11 ANYFTKLVELFEEYPKCLLVGVDNVGSKQMQEIRQAMRGHAEILMGKNTMIRKALRGHLG 70
Query 61 KNPALEKLLPYIKSNIGFVFTDKDLSEVRDKILGNKVQAPARAGALAPCDVKIPAQNTGL 120
KNP+LEKLLP+I N+GFVFT +DL E+R K+L N+ APA+AGA+APCDVK+P QNTG+
Sbjct 71 KNPSLEKLLPHIVENVGFVFTKEDLGEIRSKLLENRKGAPAKAGAIAPCDVKLPPQNTGM 130
Query 121 GPEKTSFFQALSIPTKISKGTIEILNEIHLIKKDDRVGASEATLLNMLNISPFTYGLVIR 180
GPEKTSFFQAL IPTKI++GTIEILN++HLIK+ D+VGASE+ LLNML ++PF+YGLV+R
Sbjct 131 GPEKTSFFQALQIPTKIARGTIEILNDVHLIKEGDKVGASESALLNMLGVTPFSYGLVVR 190
Query 181 QVYDSGTVFEPEILDITPEDLRARFLEGVRNVAAVFL 217
QVYD GT++ PE+LD+T E+LR RFL GVRNVA+V L
Sbjct 191 QVYDDGTLYTPEVLDMTTEELRKRFLSGVRNVASVSL 227
> hsa:6175 RPLP0, L10E, MGC111226, MGC88175, P0, PRLP0, RPP0;
ribosomal protein, large, P0; K02941 large subunit ribosomal
protein LP0
Length=317
Score = 323 bits (828), Expect = 3e-88, Method: Compositional matrix adjust.
Identities = 159/217 (73%), Positives = 192/217 (88%), Gaps = 0/217 (0%)
Query 1 ASYFTKLVQLLDEYPKCFIVGVDNVGSKQMQQIRHSLREDATLLMGKNTMIRKAIRGILE 60
++YF K++QLLD+YPKCFIVG DNVGSKQMQQIR SLR A +LMGKNTM+RKAIRG LE
Sbjct 11 SNYFLKIIQLLDDYPKCFIVGADNVGSKQMQQIRMSLRGKAVVLMGKNTMMRKAIRGHLE 70
Query 61 KNPALEKLLPYIKSNIGFVFTDKDLSEVRDKILGNKVQAPARAGALAPCDVKIPAQNTGL 120
NPALEKLLP+I+ N+GFVFT +DL+E+RD +L NKV A ARAGA+APC+V +PAQNTGL
Sbjct 71 NNPALEKLLPHIRGNVGFVFTKEDLTEIRDMLLANKVPAAARAGAIAPCEVTVPAQNTGL 130
Query 121 GPEKTSFFQALSIPTKISKGTIEILNEIHLIKKDDRVGASEATLLNMLNISPFTYGLVIR 180
GPEKTSFFQAL I TKIS+GTIEIL+++ LIK D+VGASEATLLNMLNISPF++GLVI+
Sbjct 131 GPEKTSFFQALGITTKISRGTIEILSDVQLIKTGDKVGASEATLLNMLNISPFSFGLVIQ 190
Query 181 QVYDSGTVFEPEILDITPEDLRARFLEGVRNVAAVFL 217
QV+D+G+++ PE+LDIT E L +RFLEGVRNVA+V L
Sbjct 191 QVFDNGSIYNPEVLDITEETLHSRFLEGVRNVASVCL 227
> mmu:11837 Rplp0, 36B4, Arbp, MGC107165, MGC107166; ribosomal
protein, large, P0; K02941 large subunit ribosomal protein
LP0
Length=317
Score = 321 bits (822), Expect = 1e-87, Method: Compositional matrix adjust.
Identities = 157/217 (72%), Positives = 192/217 (88%), Gaps = 0/217 (0%)
Query 1 ASYFTKLVQLLDEYPKCFIVGVDNVGSKQMQQIRHSLREDATLLMGKNTMIRKAIRGILE 60
++YF K++QLLD+YPKCFIVG DNVGSKQMQQIR SLR A +LMGKNTM+RKAIRG LE
Sbjct 11 SNYFLKIIQLLDDYPKCFIVGADNVGSKQMQQIRMSLRGKAVVLMGKNTMMRKAIRGHLE 70
Query 61 KNPALEKLLPYIKSNIGFVFTDKDLSEVRDKILGNKVQAPARAGALAPCDVKIPAQNTGL 120
NPALEKLLP+I+ N+GFVFT +DL+E+RD +L NKV A ARAGA+APC+V +PAQNTGL
Sbjct 71 NNPALEKLLPHIRGNVGFVFTKEDLTEIRDMLLANKVPAAARAGAIAPCEVTVPAQNTGL 130
Query 121 GPEKTSFFQALSIPTKISKGTIEILNEIHLIKKDDRVGASEATLLNMLNISPFTYGLVIR 180
GPEKTSFFQAL I TKIS+GTIEIL+++ LIK D+VGASEATLLNMLNISPF++GL+I+
Sbjct 131 GPEKTSFFQALGITTKISRGTIEILSDVQLIKTGDKVGASEATLLNMLNISPFSFGLIIQ 190
Query 181 QVYDSGTVFEPEILDITPEDLRARFLEGVRNVAAVFL 217
QV+D+G+++ PE+LDIT + L +RFLEGVRNVA+V L
Sbjct 191 QVFDNGSIYNPEVLDITEQALHSRFLEGVRNVASVCL 227
> mmu:667618 Gm8730, EG667618; predicted pseudogene 8730
Length=317
Score = 317 bits (812), Expect = 2e-86, Method: Compositional matrix adjust.
Identities = 156/217 (71%), Positives = 190/217 (87%), Gaps = 0/217 (0%)
Query 1 ASYFTKLVQLLDEYPKCFIVGVDNVGSKQMQQIRHSLREDATLLMGKNTMIRKAIRGILE 60
++YF K++QLLD+YPKCFIVG DNVGSKQMQQIR SLR A +LMGKNTM+RKAIRG LE
Sbjct 11 SNYFLKIIQLLDDYPKCFIVGADNVGSKQMQQIRMSLRGKAVVLMGKNTMMRKAIRGHLE 70
Query 61 KNPALEKLLPYIKSNIGFVFTDKDLSEVRDKILGNKVQAPARAGALAPCDVKIPAQNTGL 120
NPALEKLLP+I+ N+GFVFT +DL+E+RD +L NKV A ARAGA+APC+V +PAQNTGL
Sbjct 71 NNPALEKLLPHIRGNVGFVFTKEDLTEIRDMLLANKVPAAARAGAIAPCEVTVPAQNTGL 130
Query 121 GPEKTSFFQALSIPTKISKGTIEILNEIHLIKKDDRVGASEATLLNMLNISPFTYGLVIR 180
GPEKTSFFQAL I TKIS GTIEIL+++ LIK D+V ASEATLLNMLNISPF++GL+I+
Sbjct 131 GPEKTSFFQALGITTKISSGTIEILSDVQLIKTGDKVRASEATLLNMLNISPFSFGLIIQ 190
Query 181 QVYDSGTVFEPEILDITPEDLRARFLEGVRNVAAVFL 217
QV+D+G+++ PE+LDIT + L +RFLEGVRNVA+V L
Sbjct 191 QVFDNGSIYNPEVLDITEQALHSRFLEGVRNVASVCL 227
> dre:58101 rplp0, arp, fb04a12, wu:fb15a08, wu:fk48c12; ribosomal
protein, large, P0; K02941 large subunit ribosomal protein
LP0
Length=319
Score = 310 bits (794), Expect = 3e-84, Method: Compositional matrix adjust.
Identities = 156/220 (70%), Positives = 190/220 (86%), Gaps = 3/220 (1%)
Query 1 ASYFTKLVQLLDEYPKCFIVGVDNVGSKQMQQIRHSLREDATLLMGKNTMIRKAIRGILE 60
++YF K++QLLD++PKCFIVG DNVGSKQMQ IR SLR A +LMGKNTM+RKAIRG LE
Sbjct 11 SNYFLKIIQLLDDFPKCFIVGADNVGSKQMQTIRLSLRGKAVVLMGKNTMMRKAIRGHLE 70
Query 61 KNPALEKLLPYIKSNIGFVFTDKDLSEVRDKILGNKVQAPARAGALAPCDVKIPAQNTGL 120
NPALE+LLP+I+ N+GFVFT +DL+EVRD +L NKV A ARAGA+APC+V +PAQNTGL
Sbjct 71 NNPALERLLPHIRGNVGFVFTKEDLTEVRDLLLANKVPAAARAGAIAPCEVTVPAQNTGL 130
Query 121 GPEKTSFFQALSIPTKISKGTIEILNEIHLIKKDDRVGASEATL---LNMLNISPFTYGL 177
GPEKTSFFQAL I TKIS+GTIEIL+++ LIK D+VGASEATL LNMLNISPF+YGL
Sbjct 131 GPEKTSFFQALGITTKISRGTIEILSDVQLIKPGDKVGASEATLLNMLNMLNISPFSYGL 190
Query 178 VIRQVYDSGTVFEPEILDITPEDLRARFLEGVRNVAAVFL 217
+I+QVYD+G+V+ PE+LDIT + L RFL+GVRN+A+V L
Sbjct 191 IIQQVYDNGSVYSPEVLDITEDALHKRFLKGVRNIASVCL 230
> tgo:TGME49_018410 60S acidic ribosomal protein P0 (EC:4.2.99.18);
K02941 large subunit ribosomal protein LP0
Length=314
Score = 235 bits (599), Expect = 1e-61, Method: Compositional matrix adjust.
Identities = 118/214 (55%), Positives = 151/214 (70%), Gaps = 0/214 (0%)
Query 2 SYFTKLVQLLDEYPKCFIVGVDNVGSKQMQQIRHSLREDATLLMGKNTMIRKAIRGILEK 61
+YF++L LL++YP+ +V D+VGSKQM IR +LR A +LMGKNTMIR A++ + +
Sbjct 13 TYFSRLFALLEKYPRVLVVEADHVGSKQMADIRLALRGKAVVLMGKNTMIRTALKQKMSE 72
Query 62 NPALEKLLPYIKSNIGFVFTDKDLSEVRDKILGNKVQAPARAGALAPCDVKIPAQNTGLG 121
P LEKLLP ++ N+GF+F +D +EVR + NKV APAR G AP DV IPA TG+
Sbjct 73 MPQLEKLLPLVRLNVGFIFCIEDPAEVRRIVAENKVPAPARQGVFAPIDVFIPAGPTGMD 132
Query 122 PEKTSFFQALSIPTKISKGTIEILNEIHLIKKDDRVGASEATLLNMLNISPFTYGLVIRQ 181
P TSFFQAL I TKI KG IEI NE+HLIK+ D+V AS ATLL LNI PF YGL I+
Sbjct 133 PGSTSFFQALGIATKIVKGQIEIQNEVHLIKEGDKVTASAATLLQKLNIKPFEYGLAIQH 192
Query 182 VYDSGTVFEPEILDITPEDLRARFLEGVRNVAAV 215
VYD G+V++ +LDIT E + +F G NVAA+
Sbjct 193 VYDDGSVYKASVLDITDEVILEKFRAGTMNVAAL 226
> sce:YLR340W RPP0, RPL10E; P0; L10E; A0; K02941 large subunit
ribosomal protein LP0
Length=312
Score = 233 bits (595), Expect = 3e-61, Method: Compositional matrix adjust.
Identities = 120/217 (55%), Positives = 166/217 (76%), Gaps = 0/217 (0%)
Query 1 ASYFTKLVQLLDEYPKCFIVGVDNVGSKQMQQIRHSLREDATLLMGKNTMIRKAIRGILE 60
A YF KL + L+EY F+VGVDNV S+QM ++R LR A +LMGKNTM+R+AIRG L
Sbjct 9 AEYFAKLREYLEEYKSLFVVGVDNVSSQQMHEVRKELRGRAVVLMGKNTMVRRAIRGFLS 68
Query 61 KNPALEKLLPYIKSNIGFVFTDKDLSEVRDKILGNKVQAPARAGALAPCDVKIPAQNTGL 120
P EKLLP++K N+GFVFT++ L+E+++ I+ N+V APARAGA+AP D+ + A NTG+
Sbjct 69 DLPDFEKLLPFVKGNVGFVFTNEPLTEIKNVIVSNRVAAPARAGAVAPEDIWVRAVNTGM 128
Query 121 GPEKTSFFQALSIPTKISKGTIEILNEIHLIKKDDRVGASEATLLNMLNISPFTYGLVIR 180
P KTSFFQAL +PTKI++GTIEI++++ ++ ++VG SEA+LLN+LNISPFT+GL +
Sbjct 129 EPGKTSFFQALGVPTKIARGTIEIVSDVKVVDAGNKVGQSEASLLNLLNISPFTFGLTVV 188
Query 181 QVYDSGTVFEPEILDITPEDLRARFLEGVRNVAAVFL 217
QVYD+G VF ILDIT E+L + F+ V +A++ L
Sbjct 189 QVYDNGQVFPSSILDITDEELVSHFVSAVSTIASISL 225
> pfa:PF11_0313 60S ribosomal protein P0; K02941 large subunit
ribosomal protein LP0
Length=316
Score = 232 bits (592), Expect = 6e-61, Method: Compositional matrix adjust.
Identities = 116/213 (54%), Positives = 148/213 (69%), Gaps = 0/213 (0%)
Query 3 YFTKLVQLLDEYPKCFIVGVDNVGSKQMQQIRHSLREDATLLMGKNTMIRKAIRGILEKN 62
Y KL L+ +Y K IV VDNVGS QM +R SLR AT+LMGKNT IR A++ L+
Sbjct 13 YIEKLSSLIQQYSKILIVHVDNVGSNQMASVRKSLRGKATILMGKNTRIRTALKKNLQAV 72
Query 63 PALEKLLPYIKSNIGFVFTDKDLSEVRDKILGNKVQAPARAGALAPCDVKIPAQNTGLGP 122
P +EKLLP +K N+GFVF DLSE+R+ IL NK APAR G +AP DV IP TG+ P
Sbjct 73 PQIEKLLPLVKLNMGFVFCKDDLSEIRNIILDNKSPAPARLGVIAPIDVFIPPGPTGMDP 132
Query 123 EKTSFFQALSIPTKISKGTIEILNEIHLIKKDDRVGASEATLLNMLNISPFTYGLVIRQV 182
TSFFQ+L I TKI KG IEI +HLIK+ ++V AS ATLL N+ PF+YG+ +R V
Sbjct 133 SHTSFFQSLGISTKIVKGQIEIQEHVHLIKQGEKVTASSATLLQKFNMKPFSYGVDVRTV 192
Query 183 YDSGTVFEPEILDITPEDLRARFLEGVRNVAAV 215
YD G +++ ++LDIT ED+ +F +GV NVAA+
Sbjct 193 YDDGVIYDAKVLDITDEDILEKFSKGVSNVAAL 225
> ath:AT2G40010 60S acidic ribosomal protein P0 (RPP0A); K02941
large subunit ribosomal protein LP0
Length=317
Score = 229 bits (584), Expect = 5e-60, Method: Compositional matrix adjust.
Identities = 112/217 (51%), Positives = 153/217 (70%), Gaps = 2/217 (0%)
Query 3 YFTKLVQLLDEYPKCFIVGVDNVGSKQMQQIRHSLREDATLLMGKNTMIRKAIRGILEK- 61
Y +KL QLL+EY + +V DNVGS Q+Q IR LR D+ +LMGKNTM+++++R +K
Sbjct 14 YDSKLCQLLNEYSQILVVAADNVGSTQLQNIRKGLRGDSVVLMGKNTMMKRSVRIHADKT 73
Query 62 -NPALEKLLPYIKSNIGFVFTDKDLSEVRDKILGNKVQAPARAGALAPCDVKIPAQNTGL 120
N A LLP ++ N+G +FT DL EV +++ KV APAR G +AP DV + NTGL
Sbjct 74 GNQAFLSLLPLLQGNVGLIFTKGDLKEVSEEVAKYKVGAPARVGLVAPIDVVVQPGNTGL 133
Query 121 GPEKTSFFQALSIPTKISKGTIEILNEIHLIKKDDRVGASEATLLNMLNISPFTYGLVIR 180
P +TSFFQ L+IPTKI+KGT+EI+ + LIKK D+VG+SEA LL L I PF+YGLV+
Sbjct 134 DPSQTSFFQVLNIPTKINKGTVEIITPVELIKKGDKVGSSEAALLAKLGIRPFSYGLVVE 193
Query 181 QVYDSGTVFEPEILDITPEDLRARFLEGVRNVAAVFL 217
VYD+G+VF PE+L++T +DL +F GV + A+ L
Sbjct 194 SVYDNGSVFNPEVLNLTEDDLVEKFAAGVSMITALSL 230
> ath:AT3G11250 60S acidic ribosomal protein P0 (RPP0C); K02941
large subunit ribosomal protein LP0
Length=323
Score = 229 bits (583), Expect = 8e-60, Method: Compositional matrix adjust.
Identities = 111/218 (50%), Positives = 154/218 (70%), Gaps = 2/218 (0%)
Query 2 SYFTKLVQLLDEYPKCFIVGVDNVGSKQMQQIRHSLREDATLLMGKNTMIRKAIRGILEK 61
+Y TKL QL+DEY + +V DNVGS Q+Q IR LR D+ +LMGKNTM+++++R E
Sbjct 12 AYDTKLCQLIDEYTQILVVAADNVGSTQLQNIRKGLRGDSVVLMGKNTMMKRSVRIHSEN 71
Query 62 --NPALEKLLPYIKSNIGFVFTDKDLSEVRDKILGNKVQAPARAGALAPCDVKIPAQNTG 119
N A+ LLP ++ N+G +FT DL EV +++ KV APAR G +AP DV + NTG
Sbjct 72 SGNTAILNLLPLLQGNVGLIFTKGDLKEVSEEVAKYKVGAPARVGLVAPIDVVVQPGNTG 131
Query 120 LGPEKTSFFQALSIPTKISKGTIEILNEIHLIKKDDRVGASEATLLNMLNISPFTYGLVI 179
L P +TSFFQ L+IPTKI+KGT+EI+ + LIK+ D+VG+SEA LL L I PF+YGLV+
Sbjct 132 LDPSQTSFFQVLNIPTKINKGTVEIITPVELIKQGDKVGSSEAALLAKLGIRPFSYGLVV 191
Query 180 RQVYDSGTVFEPEILDITPEDLRARFLEGVRNVAAVFL 217
+ VYD+G+VF PE+LD+T + L +F G+ V ++ L
Sbjct 192 QSVYDNGSVFSPEVLDLTEDQLVEKFASGISMVTSLAL 229
> tpv:TP01_0294 60S acidic ribosomal protein, P0; K02941 large
subunit ribosomal protein LP0
Length=321
Score = 228 bits (582), Expect = 9e-60, Method: Compositional matrix adjust.
Identities = 115/213 (53%), Positives = 148/213 (69%), Gaps = 0/213 (0%)
Query 3 YFTKLVQLLDEYPKCFIVGVDNVGSKQMQQIRHSLREDATLLMGKNTMIRKAIRGILEKN 62
YF +L L+ Y K IV VD+VGS+QM +RHSLR AT+LMGKNT+IR A++ +
Sbjct 22 YFERLTSLMRTYSKILIVSVDHVGSRQMASVRHSLRGMATILMGKNTVIRTALQKNFPDS 81
Query 63 PALEKLLPYIKSNIGFVFTDKDLSEVRDKILGNKVQAPARAGALAPCDVKIPAQNTGLGP 122
P +EK+ +K N GFVF + D EVR+ IL N+V APA+ G +AP DV IPA +TGL P
Sbjct 82 PDVEKVAQCVKLNTGFVFCEADPMEVREVILNNRVPAPAKQGVIAPSDVFIPAGSTGLDP 141
Query 123 EKTSFFQALSIPTKISKGTIEILNEIHLIKKDDRVGASEATLLNMLNISPFTYGLVIRQV 182
+TSFFQAL I TKI KG IEI NE+HLIKKDD+V AS ATLL LNI PF+YGL + ++
Sbjct 142 SQTSFFQALGISTKIVKGQIEIQNEVHLIKKDDKVTASGATLLQKLNIKPFSYGLKVEKI 201
Query 183 YDSGTVFEPEILDITPEDLRARFLEGVRNVAAV 215
YDSG + + +LD+T ED+ + GV A+
Sbjct 202 YDSGAISDASVLDVTDEDILSVVKLGVSYANAM 234
> bbo:BBOV_IV004540 23.m05776; phosphoriboprotein P0; K02941 large
subunit ribosomal protein LP0
Length=312
Score = 218 bits (555), Expect = 1e-56, Method: Compositional matrix adjust.
Identities = 115/216 (53%), Positives = 139/216 (64%), Gaps = 0/216 (0%)
Query 2 SYFTKLVQLLDEYPKCFIVGVDNVGSKQMQQIRHSLREDATLLMGKNTMIRKAIRGILEK 61
+YF +L L+ YP+ IV VD VGS+QM +RHSLR A +L+GKNTMIR +
Sbjct 12 AYFERLTHLVKTYPQILIVSVDYVGSRQMAHVRHSLRGKAEILIGKNTMIRMVLNTSFPN 71
Query 62 NPALEKLLPYIKSNIGFVFTDKDLSEVRDKILGNKVQAPARAGALAPCDVKIPAQNTGLG 121
+ A+ KLL +K N+GFVF D EVR IL NKV APA+ G +APCDV I A TG+
Sbjct 72 SEAISKLLSCVKLNVGFVFCMGDPLEVRRIILDNKVPAPAKQGVIAPCDVFISAGATGMD 131
Query 122 PEKTSFFQALSIPTKISKGTIEILNEIHLIKKDDRVGASEATLLNMLNISPFTYGLVIRQ 181
P +TSFFQAL I TKI KG IEI N++HLIK +DRV AS ATLL LN+ PF YGL I +
Sbjct 132 PSQTSFFQALGISTKIVKGQIEIQNDVHLIKVNDRVTASSATLLQKLNMKPFAYGLKIEK 191
Query 182 VYDSGTVFEPEILDITPEDLRARFLEGVRNVAAVFL 217
YDSG + E LDIT +D V NV A L
Sbjct 192 FYDSGHLVEASALDITEDDSLDSVKTAVTNVNAFAL 227
> cpv:cgd4_2260 ribosomal protein PO like protein of the L10 family
; K02941 large subunit ribosomal protein LP0
Length=318
Score = 211 bits (538), Expect = 1e-54, Method: Compositional matrix adjust.
Identities = 104/212 (49%), Positives = 142/212 (66%), Gaps = 0/212 (0%)
Query 3 YFTKLVQLLDEYPKCFIVGVDNVGSKQMQQIRHSLREDATLLMGKNTMIRKAIRGILEKN 62
YF +L + YP+ + D+VGSKQM IR +LR A +LMGKNTMIR A++ +L +
Sbjct 21 YFERLSEYATSYPRILVANADHVGSKQMADIRLALRGKAAVLMGKNTMIRTALKQMLGSH 80
Query 63 PALEKLLPYIKSNIGFVFTDKDLSEVRDKILGNKVQAPARAGALAPCDVKIPAQNTGLGP 122
P LEKL+ ++ N+G +F + SEVR I +V APAR G +APC+V +PA TGL P
Sbjct 81 PELEKLIELVRLNVGLIFCIDEPSEVRKIIEEYRVPAPARQGVIAPCNVVVPAGATGLDP 140
Query 123 EKTSFFQALSIPTKISKGTIEILNEIHLIKKDDRVGASEATLLNMLNISPFTYGLVIRQV 182
+TSFFQAL I TKI KG +EI ++++LI + +V AS+A LL LNI PF+YGL + +
Sbjct 141 SQTSFFQALGIATKIVKGQVEIQSDVNLIDEGKKVTASQAVLLQKLNIKPFSYGLKVNNI 200
Query 183 YDSGTVFEPEILDITPEDLRARFLEGVRNVAA 214
YD G+V+ +LDIT EDL +R E + VAA
Sbjct 201 YDHGSVYSSSVLDITSEDLISRVAEATKYVAA 232
> ath:AT3G09200 60S acidic ribosomal protein P0 (RPP0B); K02941
large subunit ribosomal protein LP0
Length=287
Score = 171 bits (434), Expect = 1e-42, Method: Compositional matrix adjust.
Identities = 93/216 (43%), Positives = 131/216 (60%), Gaps = 31/216 (14%)
Query 2 SYFTKLVQLLDEYPKCFIVGVDNVGSKQMQQIRHSLREDATLLMGKNTMIRKAIRGILEK 61
+Y TKL QL+DEY + + S+R + +NT
Sbjct 12 AYDTKLCQLIDEYTQILV---------------RSVR-----IHSENT-----------G 40
Query 62 NPALEKLLPYIKSNIGFVFTDKDLSEVRDKILGNKVQAPARAGALAPCDVKIPAQNTGLG 121
N A+ LLP ++ N+G +FT DL EV +++ KV APAR G +AP DV + NTGL
Sbjct 41 NTAILNLLPLLQGNVGLIFTKGDLKEVSEEVAKYKVGAPARVGLVAPIDVVVQPGNTGLD 100
Query 122 PEKTSFFQALSIPTKISKGTIEILNEIHLIKKDDRVGASEATLLNMLNISPFTYGLVIRQ 181
P +TSFFQ L+IPTKI+KGT+EI+ + LIK+ D+VG+SEA LL L I PF+YGLV++
Sbjct 101 PSQTSFFQVLNIPTKINKGTVEIITPVELIKQGDKVGSSEAALLAKLGIRPFSYGLVVQS 160
Query 182 VYDSGTVFEPEILDITPEDLRARFLEGVRNVAAVFL 217
VYD+G+VF PE+LD+T + L +F G+ V ++ L
Sbjct 161 VYDNGSVFSPEVLDLTEDQLVEKFASGISMVTSLAL 196
> xla:379539 mrto4, MGC68920, mrt4; mRNA turnover 4 homolog; K14815
mRNA turnover protein 4
Length=240
Score = 59.7 bits (143), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 46/189 (24%), Positives = 93/189 (49%), Gaps = 12/189 (6%)
Query 6 KLVQLLDEYPKCFIVGVDNVGSKQMQQIRHSLREDATLLMGKNTMIRKAI-RGIL-EKNP 63
+L + +D Y F++ V+N+ + +++ +R++ + + L GKN ++ A+ +G+ E
Sbjct 29 ELRKCVDTYKYIFVLSVENMRNNKLKDVRNAWKH-SRLFFGKNKVMMVAMGKGVSDEYKD 87
Query 64 ALEKLLPYIKSNIGFVFTDKDLSEVRDKILGNKVQAPARAGALAPCDVKIPAQ-----NT 118
L KL +K +G +FT++ EV + K ARAG A V + A
Sbjct 88 NLHKLSKCLKGEVGLLFTNRTEKEVEEWFDQYKETDFARAGNKATYSVVLDAGPLDQFTH 147
Query 119 GLGPEKTSFFQALSIPTKISKGTIEILNEIHLIKKDDRVGASEATLLNMLNISPFTYGLV 178
+ P+ + L +PT + KG I +L++ L K+ D + +A +L + + +
Sbjct 148 SMEPQ----LRQLGLPTALKKGVITLLSDYDLCKEGDVLTPEQARILKLFGFQMAEFKVS 203
Query 179 IRQVYDSGT 187
I+ ++ + T
Sbjct 204 IKSIWTAET 212
> dre:550350 wu:fb71a02; zgc:110388; K14815 mRNA turnover protein
4
Length=242
Score = 56.2 bits (134), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 44/183 (24%), Positives = 86/183 (46%), Gaps = 4/183 (2%)
Query 6 KLVQLLDEYPKCFIVGVDNVGSKQMQQIRHSLREDATLLMGKNTMIRKAI-RG-ILEKNP 63
+L + D Y F+ V+N+ + +++ IR + R + GKN ++ A+ +G E
Sbjct 29 ELRKCADIYRYVFVFSVENMRNNKLKDIRTAWRH-SRFFFGKNKVMMIALGKGPTDEYKD 87
Query 64 ALEKLLPYIKSNIGFVFTDKDLSEVRDKILGNKVQAPARAGALAPCDVKIPAQNTGLGPE 123
L KL +++ +G +FT+K EV++ K ARAG A V + P
Sbjct 88 NLHKLSRFLRGEVGVLFTNKTKDEVQEYFGNFKEVDYARAGNTASMAVTLDEGPLDQFPH 147
Query 124 KTS-FFQALSIPTKISKGTIEILNEIHLIKKDDRVGASEATLLNMLNISPFTYGLVIRQV 182
+ L +PT + KG + ++ + + K+ D + +A +L + + L I+ +
Sbjct 148 SMEPQLRQLGLPTALKKGAVTLIKDFEVCKEGDTLTPEQARILKLFGFEMAEFKLSIKCM 207
Query 183 YDS 185
++S
Sbjct 208 WNS 210
> cel:F10E7.5 hypothetical protein; K14815 mRNA turnover protein
4
Length=220
Score = 53.9 bits (128), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 44/188 (23%), Positives = 80/188 (42%), Gaps = 13/188 (6%)
Query 11 LDEYPKCFIVGVDNVGSKQMQQIRHSLREDATLLMGKNTMIRKAI--RGILEKNPALEKL 68
+D+Y FI + N+ S + IR +E++ GKN +I A+ + E L K
Sbjct 34 VDQYKNLFIFTIANMRSTRFIAIRQKYKENSRFFFGKNNVISIALGKQKSDEYANQLHKA 93
Query 69 LPYIKSNIGFVFTDKDLSEVRDKILGNKVQAPARAGALAPCDVKIPAQNTGLGPEKTSFF 128
+K G +FT+ EV + + AR G +A V +P GP F
Sbjct 94 SAILKGQCGLMFTNMSKKEVEAEFSEASEEDYARVGDVATETVVLPE-----GPISQFAF 148
Query 129 ------QALSIPTKISKGTIEILNEIHLIKKDDRVGASEATLLNMLNISPFTYGLVIRQV 182
+ L +PTK+ KG I + + + K+ + + +A +L + + L+ +
Sbjct 149 SMEPQLRKLGLPTKLDKGVITLYQQFEVCKEGEPLTVEQAKILKHFEVKMSQFRLIFKAH 208
Query 183 YDSGTVFE 190
++ F+
Sbjct 209 WNKKDGFK 216
> mmu:434693 Gm5633; predicted gene 5633; K14815 mRNA turnover
protein 4
Length=239
Score = 52.4 bits (124), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 40/187 (21%), Positives = 87/187 (46%), Gaps = 12/187 (6%)
Query 6 KLVQLLDEYPKCFIVGVDNVGSKQMQQIRHSLREDATLLMGKNTMIRKAIRGILEKNPA- 64
+L + +D Y FI V N+ + +++ IR++ + + + GKN ++ A L ++P+
Sbjct 29 ELRKCVDTYKYLFIFSVANMRNSKLKDIRNAWKH-SRMFFGKNKVMMVA----LGRSPSD 83
Query 65 -----LEKLLPYIKSNIGFVFTDKDLSEVRDKILGNKVQAPARAGALAPCDVKIPAQNTG 119
L ++ ++ +G +FT++ EV + ARAG A V +
Sbjct 84 EYKDDLHQVSKKLRGEVGLLFTNRTKEEVNEWFTKYTEMDFARAGNKATLTVSLDPGPLK 143
Query 120 LGPEKTSF-FQALSIPTKISKGTIEILNEIHLIKKDDRVGASEATLLNMLNISPFTYGLV 178
P + L +PT + KG + +L++ + K+ D + +A +L + + +
Sbjct 144 QFPHSMELQLRQLGLPTALEKGVVTLLSDYEVCKEGDVLTPEQARILKLFGYEMAEFKVT 203
Query 179 IRQVYDS 185
I+ ++D+
Sbjct 204 IKYMWDA 210
> hsa:51154 MRTO4, C1orf33, MRT4, dJ657E11.4; mRNA turnover 4
homolog (S. cerevisiae); K14815 mRNA turnover protein 4
Length=239
Score = 51.6 bits (122), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 40/187 (21%), Positives = 87/187 (46%), Gaps = 12/187 (6%)
Query 6 KLVQLLDEYPKCFIVGVDNVGSKQMQQIRHSLREDATLLMGKNTMIRKAIRGILEKNPA- 64
+L + +D Y FI V N+ + +++ IR++ + + + GKN ++ A L ++P+
Sbjct 29 ELRKCVDTYKYLFIFSVANMRNSKLKDIRNAWKH-SRMFFGKNKVMMVA----LGRSPSD 83
Query 65 -----LEKLLPYIKSNIGFVFTDKDLSEVRDKILGNKVQAPARAGALAPCDVKI-PAQNT 118
L ++ ++ +G +FT++ EV + ARAG A V + P
Sbjct 84 EYKDNLHQVSKRLRGEVGLLFTNRTKEEVNEWFTKYTEMDYARAGNKAAFTVSLDPGPLE 143
Query 119 GLGPEKTSFFQALSIPTKISKGTIEILNEIHLIKKDDRVGASEATLLNMLNISPFTYGLV 178
+ L +PT + +G + +L++ + K+ D + +A +L + + +
Sbjct 144 QFPHSMEPQLRQLGLPTALKRGVVTLLSDYEVCKEGDVLTPEQARVLKLFGYEMAEFKVT 203
Query 179 IRQVYDS 185
I+ ++DS
Sbjct 204 IKYMWDS 210
> mmu:668455 Gm9178, EG668455; predicted gene 9178; K14815 mRNA
turnover protein 4
Length=239
Score = 51.2 bits (121), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 40/187 (21%), Positives = 87/187 (46%), Gaps = 12/187 (6%)
Query 6 KLVQLLDEYPKCFIVGVDNVGSKQMQQIRHSLREDATLLMGKNTMIRKAIRGILEKNPA- 64
+L + +D Y FI V N+ + +++ IR++ + + + GKN ++ A L ++P+
Sbjct 29 ELRKCVDTYKYLFIFSVANMRNSKLKDIRNAWKH-SRMFFGKNKVMMVA----LGRSPSD 83
Query 65 -----LEKLLPYIKSNIGFVFTDKDLSEVRDKILGNKVQAPARAGALAPCDVKIPAQNTG 119
L ++ ++ +G +FT++ EV + ARAG A V +
Sbjct 84 EYKDDLHQVSKKLRGEVGLLFTNRTKEEVNEWFTKYTEMDFARAGNKATLTVSLDPGPLK 143
Query 120 LGPEKTSF-FQALSIPTKISKGTIEILNEIHLIKKDDRVGASEATLLNMLNISPFTYGLV 178
P + L +PT + KG + +L++ + K+ D + +A +L + + +
Sbjct 144 QFPHSMELQLRQLGLPTALEKGVVTLLSDYEVCKEGDVLTPEQARILKLFGDEMAEFKVT 203
Query 179 IRQVYDS 185
I+ ++D+
Sbjct 204 IKYMWDA 210
> sce:YKL009W MRT4; Mrt4p; K14815 mRNA turnover protein 4
Length=236
Score = 48.5 bits (114), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 47/200 (23%), Positives = 88/200 (44%), Gaps = 17/200 (8%)
Query 4 FTKLVQLLDEYPKCFIVGVDNVGSKQMQQIRHSLREDATLLMGKNTMIRKAI--RGILEK 61
F ++ + LD Y +++ +D+V + +Q+IR S + L+MGK +++KA+ + E
Sbjct 27 FDEVREALDTYRYVWVLHLDDVRTPVLQEIRTSW-AGSKLIMGKRKVLQKALGEKREEEY 85
Query 62 NPALEKLLPYIKSNIGFVFTDKDLSEVRDKILGNKVQAPARAGALAPCDVKIPA----QN 117
L +L G +FTD+D++ V++ +R AP IP
Sbjct 86 KENLYQLSKLCSGVTGLLFTDEDVNTVKEYFKSYVRSDYSRPNTKAPLTFTIPEGIVYSR 145
Query 118 TGLGP--EKTSFFQALS--------IPTKISKGTIEILNEIHLIKKDDRVGASEATLLNM 167
G P E +L IPTKI G I I + + + +++ +A +L
Sbjct 146 GGQIPAEEDVPMIHSLEPTMRNKFEIPTKIKAGKITIDSPYLVCTEGEKLDVRQALILKQ 205
Query 168 LNISPFTYGLVIRQVYDSGT 187
I+ + + + YD+ +
Sbjct 206 FGIAASEFKVKVSAYYDNDS 225
> mmu:69902 Mrto4, 2610012O22Rik, Mg684, Mrt4; MRT4, mRNA turnover
4, homolog (S. cerevisiae); K14815 mRNA turnover protein
4
Length=238
Score = 47.8 bits (112), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 40/187 (21%), Positives = 88/187 (47%), Gaps = 13/187 (6%)
Query 6 KLVQLLDEYPKCFIVGVDNVGSKQMQQIRHSLREDATLLMGKNTMIRKAIRGILEKNPA- 64
+L + +D Y FI V N+ + +++ IR++ + + + GKN ++ A L ++P+
Sbjct 29 ELRKCVDTYKYLFIFSVANMRNSKLKDIRNAWKH-SRMFFGKNKVMMVA----LGRSPSD 83
Query 65 -----LEKLLPYIKSNIGFVFTDKDLSEVRDKILGNKVQAPARAGALAPCDVKI-PAQNT 118
L ++ ++ +G +FT++ EV + ARAG A V + P
Sbjct 84 EYKDNLHQVSKKLRGEVGLLFTNRTKEEVNEWFTKYTEMDFARAGNKATLTVSLDPGPLK 143
Query 119 GLGPEKTSFFQALSIPTKISKGTIEILNEIHLIKKDDRVGASEATLLNMLNISPFTYGLV 178
+ L +PT + KG + +L++ + K+ D + +A +L + + ++
Sbjct 144 QFPHSMEPQLRQLGLPTALKKGVVTLLSDYEVCKEGDVLTPEQARIL-LFGYEMAEFKVI 202
Query 179 IRQVYDS 185
I+ ++D+
Sbjct 203 IKYMWDA 209
> ath:AT1G25260 acidic ribosomal protein P0-related; K14815 mRNA
turnover protein 4
Length=235
Score = 45.8 bits (107), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 36/203 (17%), Positives = 91/203 (44%), Gaps = 15/203 (7%)
Query 9 QLLDEYPKCFIVGVDNVGSKQMQQIRHSLREDATLLMGKNTMIRKAIRGILEKN--PALE 66
+ +++Y ++ +N+ + + ++ R R + +G N +++ A+ E +
Sbjct 32 EAVEKYSSVYVFSFENMRNIKFKEFRQQFRHNGKFFLGSNKVMQVALGRSAEDELRSGIY 91
Query 67 KLLPYIKSNIGFVFTDKDLSEVRDKILGNKVQAPARAGALAPCDVKIPAQNTGLGP---- 122
K+ ++ + G + TD EV + +R G++A V++ GP
Sbjct 92 KVSKLLRGDTGLLVTDMPKEEVESLFNAYEDSDFSRTGSIAVETVELKE-----GPLEQF 146
Query 123 --EKTSFFQALSIPTKISKGTIEILNEIHLIKKDDRVGASEATLLNML--NISPFTYGLV 178
E + L +P +++KGT+E++ + + ++ ++ A +L +L ++ F L+
Sbjct 147 THEMEPLLRKLEMPVRLNKGTVELVADFVVCEEGKQLSPKSAHILRLLRMKMATFKLNLL 206
Query 179 IRQVYDSGTVFEPEILDITPEDL 201
R ++ ++ ++ EDL
Sbjct 207 CRWSPSDFELYREDLSELYREDL 229
> bbo:BBOV_I004380 19.m02064; hypothetical protein; K14815 mRNA
turnover protein 4
Length=225
Score = 40.8 bits (94), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 39/179 (21%), Positives = 83/179 (46%), Gaps = 14/179 (7%)
Query 18 FIVGVDNVGSKQMQQIRHSLREDATLLMGKNTMIRKAIRGILEKN--PALEKLLPYIKSN 75
+++ ++N + ++++R L+ L GKN +++ A+ E L K+ I
Sbjct 46 YVIALNNQRNSPLKELRTILK-PGRLFYGKNKVMQLALGAKPENELLDNLHKIAECISGE 104
Query 76 IGFVFTDKDLSEVRDKILGNKVQAPARAGALA-------PCDVKIPAQNTGLGPEKTSFF 128
+ +++ VR+K+ KV A+AG +A P D + + P+ F
Sbjct 105 RALLVSNEAPDVVRNKLESYKVNDFAKAGNVATETILLKPGDSTLEVFPGNMEPQ----F 160
Query 129 QALSIPTKISKGTIEILNEIHLIKKDDRVGASEATLLNMLNISPFTYGLVIRQVYDSGT 187
+ L +PT + G IE+L + + ++ + ++A +L +L I + I +++GT
Sbjct 161 RHLGMPTTLKMGKIELLGDYLVCEEGKPLTPTQAKVLKVLGIRMALFECTIHAHWNNGT 219
> ath:AT3G12370 ribosomal protein L10 family protein; K02864 large
subunit ribosomal protein L10
Length=171
Score = 37.7 bits (86), Expect = 0.035, Method: Compositional matrix adjust.
Identities = 24/71 (33%), Positives = 38/71 (53%), Gaps = 9/71 (12%)
Query 18 FIVGVDNVG--SKQMQQIRHSLREDAT--LLMGKNTMIRKAIRGILEKNPALEKLLPYIK 73
FI G++ G KQ+Q++R LRE+ LL+ KNT++ KA+ G E L P +K
Sbjct 23 FITGINYNGLSVKQLQELRGILRENTNTKLLVAKNTLVYKALEGT-----KWESLKPCMK 77
Query 74 SNIGFVFTDKD 84
++F +
Sbjct 78 GMNAWLFVQSE 88
> mmu:78177 Ninl, 4930519N13Rik, Gm1004, Gm1634, mKIAA0980; ninein-like
Length=1394
Score = 36.6 bits (83), Expect = 0.070, Method: Composition-based stats.
Identities = 20/55 (36%), Positives = 34/55 (61%), Gaps = 0/55 (0%)
Query 29 QMQQIRHSLREDATLLMGKNTMIRKAIRGILEKNPALEKLLPYIKSNIGFVFTDK 83
++Q SLRE TL + +N+ ++K I ++EK EKL+ ++S++ FV DK
Sbjct 477 RLQAEETSLREKLTLALKENSRLQKEIIEVVEKLSDSEKLVLRLQSDLQFVLKDK 531
> ath:AT5G13510 ribosomal protein L10 family protein
Length=220
Score = 34.7 bits (78), Expect = 0.28, Method: Compositional matrix adjust.
Identities = 21/71 (29%), Positives = 34/71 (47%), Gaps = 8/71 (11%)
Query 17 CFIVGVDN---VGSKQMQQIRHSLREDATLLMGKNTMIRKAIRGILEKNPALEKLLPYIK 73
C ++ N + KQ Q +R +L + L++ KNT++ KAI G E L P +K
Sbjct 62 CHLLAAINYKGLTVKQFQDLRRTLPDTTKLIVAKNTLVFKAIEGT-----KWEALKPCMK 116
Query 74 SNIGFVFTDKD 84
++F D
Sbjct 117 GMNAWLFVQTD 127
> tpv:TP01_1023 hypothetical protein; K14815 mRNA turnover protein
4
Length=225
Score = 33.5 bits (75), Expect = 0.61, Method: Compositional matrix adjust.
Identities = 35/170 (20%), Positives = 77/170 (45%), Gaps = 6/170 (3%)
Query 6 KLVQLLDEYPKCFIVGVDNVGSKQMQQIRHSLREDATLLMGKNTMIRKAI--RGILEKNP 63
K ++ D +++ ++N + ++ +R S+ + GKN ++R A + E +
Sbjct 35 KFIKNNDNSTFVYLIALNNQRNSPLKTLR-SILLPGRVFYGKNKVMRIAFGTKPEDEIHD 93
Query 64 ALEKLLPYIKSNIGFVFTDKDLSEVRDKILGNKVQAPARAGALAPCDVKIPAQNTGLGPE 123
+ K+ I + T ++ V +K+ G KV+ ++AG +A + +
Sbjct 94 NIHKISNNINGETAVLITSENPEVVVNKVKGYKVRDFSKAGNIATDTIVLKVDGNEFDEI 153
Query 124 KTSF---FQALSIPTKISKGTIEILNEIHLIKKDDRVGASEATLLNMLNI 170
S F+ L +PT ++ G I ++ + L +KD + ++ LL + I
Sbjct 154 PGSMEPQFRQLGLPTALNMGKIILMGDYTLCEKDKPLTPNQTHLLKLFGI 203
> tgo:TGME49_036850 hypothetical protein
Length=2744
Score = 31.6 bits (70), Expect = 2.2, Method: Composition-based stats.
Identities = 16/34 (47%), Positives = 19/34 (55%), Gaps = 0/34 (0%)
Query 94 GNKVQAPARAGALAPCDVKIPAQNTGLGPEKTSF 127
+K QA A GA P VK A+ TG PE+ SF
Sbjct 603 ASKTQADAEDGAHNPATVKEEAEKTGPDPERASF 636
> hsa:22981 NINL, FLJ11792, KIAA0980, NLP, dJ691N24.1; ninein-like
Length=1382
Score = 31.6 bits (70), Expect = 2.4, Method: Composition-based stats.
Identities = 17/55 (30%), Positives = 31/55 (56%), Gaps = 0/55 (0%)
Query 29 QMQQIRHSLREDATLLMGKNTMIRKAIRGILEKNPALEKLLPYIKSNIGFVFTDK 83
++Q LRE TL + +N+ ++K I ++EK E+L ++ ++ FV DK
Sbjct 478 RLQAEEAGLREKLTLALKENSRLQKEIVEVVEKLSDSERLALKLQKDLEFVLKDK 532
> hsa:23046 KIF21B, FLJ16314; kinesin family member 21B; K10395
kinesin family member 4/7/21/27
Length=1624
Score = 31.2 bits (69), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 37/148 (25%), Positives = 66/148 (44%), Gaps = 6/148 (4%)
Query 10 LLDEYPKCFIVGVDNVGSK--QMQQIRHSLREDATLLMGKNTMIRKAIRGILEKNPALEK 67
LLD + K I V K Q++ + LR+ +N ++ A+R E +P L+
Sbjct 1033 LLDNFLKASIDKGLQVAQKEAQIRLLEGRLRQTDMAGSSQNHLLLDALREKAEAHPELQA 1092
Query 68 LLPYIKSNIGFVFTDKDLSEVRDKILGNKVQAPARAGALAPCDVKIPAQNTGLGPEKTSF 127
L+ ++ G+ TD+++SE + G+ Q+ G+ + D K ++ K
Sbjct 1093 LIYNVQQENGYASTDEEISEFSE---GSFSQSFTMKGSTSHDDFKFKSEPKLSAQMKAVS 1149
Query 128 FQALSIPTKIS-KGTIEILNEIHLIKKD 154
+ L P IS K + L + IK+D
Sbjct 1150 AECLGPPLDISTKNITKSLASLVEIKED 1177
> dre:570502 hCG2027369-like; K10395 kinesin family member 4/7/21/27
Length=1651
Score = 31.2 bits (69), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 24/80 (30%), Positives = 39/80 (48%), Gaps = 2/80 (2%)
Query 10 LLDEYPKCFIVGVDNVGSK--QMQQIRHSLREDATLLMGKNTMIRKAIRGILEKNPALEK 67
LLD + K I V K Q++ + LR+ + N MI A+R E P L+
Sbjct 1062 LLDYFLKASIDKGLQVAQKEAQIRLLEGQLRQTDVIGSSHNHMILDALREKAECIPELQA 1121
Query 68 LLPYIKSNIGFVFTDKDLSE 87
L+ ++ G+ TD+++SE
Sbjct 1122 LIHNVQQENGYASTDEEVSE 1141
> tgo:TGME49_044880 DNA-directed RNA polymerase I largest subunit,
putative (EC:2.7.7.49 2.7.7.6); K02999 DNA-directed RNA
polymerase I subunit RPA1 [EC:2.7.7.6]
Length=2768
Score = 31.2 bits (69), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 27/100 (27%), Positives = 44/100 (44%), Gaps = 10/100 (10%)
Query 80 FTDKDLSEVRDKILGNKVQAPARAGALAPCDVKIPAQNTGLGPEKTSFFQALSIPTKISK 139
+ ++ +R K++G +V AR LAP D I G+ F LSIP K++
Sbjct 617 WMERKAGTIRQKLMGKRVNYAART-VLAP-DALIATNEVGV---PLDFAMKLSIPEKVTP 671
Query 140 GTIEILNEIHLIKKDDRVGA-----SEATLLNMLNISPFT 174
+ +L+E+ + GA L N+ +SP T
Sbjct 672 RNVHVLSEMVINGPHKHPGALAIMDDAGNLFNLEFLSPAT 711
> ath:AT1G26720 hypothetical protein
Length=169
Score = 30.8 bits (68), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 21/63 (33%), Positives = 34/63 (53%), Gaps = 6/63 (9%)
Query 20 VGVDNVGSKQMQQIRHSLREDATLLMGK---NTMIRKAIRGILEKN---PALEKLLPYIK 73
+G+ VGS++M I ED++L+ + N+M RK IRG + + P + +P K
Sbjct 65 IGLGEVGSQEMHHIYIETGEDSSLVNKQDNINSMPRKVIRGTVSISFSFPIIRIKVPIPK 124
Query 74 SNI 76
NI
Sbjct 125 PNI 127
> mmu:16565 Kif21b, 2610511N21Rik, KIAA0449, mKIAA0449; kinesin
family member 21B; K10395 kinesin family member 4/7/21/27
Length=1624
Score = 30.8 bits (68), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 22/80 (27%), Positives = 40/80 (50%), Gaps = 2/80 (2%)
Query 10 LLDEYPKCFIVGVDNVGSK--QMQQIRHSLREDATLLMGKNTMIRKAIRGILEKNPALEK 67
LLD + K I V K Q++ + LR+ +N ++ A+R E +P L+
Sbjct 1034 LLDNFLKASIDKGLQVAQKEAQIRLLEGRLRQTDMTGSSQNHLLLDALREKAEAHPELQA 1093
Query 68 LLPYIKSNIGFVFTDKDLSE 87
L+ ++ G+ TD+++SE
Sbjct 1094 LIYNVQHENGYASTDEEVSE 1113
> pfa:PFF0535c trancription factor, putative
Length=1280
Score = 30.0 bits (66), Expect = 7.4, Method: Composition-based stats.
Identities = 26/97 (26%), Positives = 38/97 (39%), Gaps = 21/97 (21%)
Query 116 QNTGLGPEKTSFFQALSIPTKISKGTIEILNEIHLIKKDDRVGASEATLLNMLNISPFTY 175
+N G+ EK + S K N +HL KKD+RV + L N++
Sbjct 592 KNNGMAGEKATLGITKSFINK---------NSLHLFKKDERVKILKGELCNLIG------ 636
Query 176 GLVIRQVYDSGTVFEPEIL----DITPEDLRARFLEG 208
I V D+ P+ L P D+ F+EG
Sbjct 637 --TITAVNDNVLTINPDNLAKQFKFLPSDVTKYFIEG 671
Lambda K H
0.320 0.138 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 7202251840
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40