bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0384_orf1
Length=189
Score E
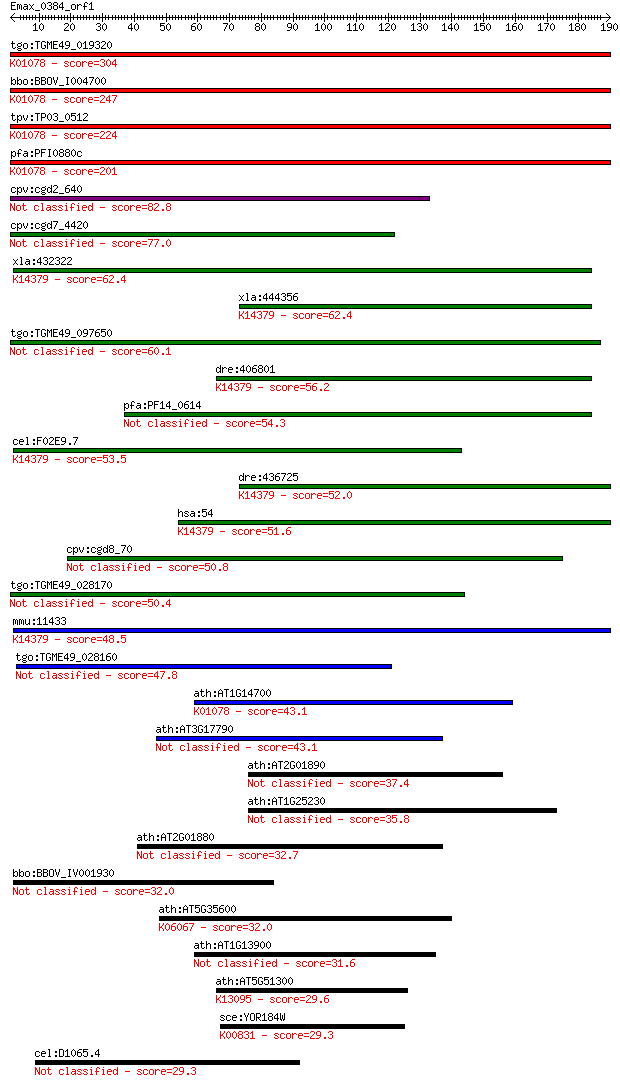
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_019320 acid phosphatase, putative (EC:3.1.3.2); K01... 304 1e-82
bbo:BBOV_I004700 19.m02068; acid phosphatase (EC:3.1.3.2); K01... 247 1e-65
tpv:TP03_0512 acid phosphatase (EC:3.1.3.2); K01078 acid phosp... 224 9e-59
pfa:PFI0880c GAP50; glideosome-associated protein 50 (EC:3.1.3... 201 1e-51
cpv:cgd2_640 acid phosphatase 82.8 7e-16
cpv:cgd7_4420 secreted acid phosphatase (calcineurin family),s... 77.0 4e-14
xla:432322 acp5, MGC78938; acid phosphatase 5, tartrate resist... 62.4 8e-10
xla:444356 MGC82831 protein; K14379 tartrate-resistant acid ph... 62.4 1e-09
tgo:TGME49_097650 serine/threonine protein phosphatase, putati... 60.1 4e-09
dre:406801 acp5a, acp5, sb:cb576, wu:fb19f01, wu:fb30b03, wu:f... 56.2 7e-08
pfa:PF14_0614 phosphatase, putative 54.3 2e-07
cel:F02E9.7 hypothetical protein; K14379 tartrate-resistant ac... 53.5 4e-07
dre:436725 acp5b, zgc:92339; acid phosphatase 5b, tartrate res... 52.0 1e-06
hsa:54 ACP5, MGC117378, TRAP; acid phosphatase 5, tartrate res... 51.6 1e-06
cpv:cgd8_70 hypothetical protein 50.8 3e-06
tgo:TGME49_028170 serine/threonine protein phosphatase, putati... 50.4 4e-06
mmu:11433 Acp5, TRACP, TRAP; acid phosphatase 5, tartrate resi... 48.5 1e-05
tgo:TGME49_028160 serine/threonine protein phosphatase, putati... 47.8 2e-05
ath:AT1G14700 PAP3; PAP3 (PURPLE ACID PHOSPHATASE 3); acid pho... 43.1 5e-04
ath:AT3G17790 PAP17; PAP17; acid phosphatase/ phosphatase/ pro... 43.1 6e-04
ath:AT2G01890 PAP8; PAP8 (PURPLE ACID PHOSPHATASE 8); acid pho... 37.4 0.030
ath:AT1G25230 purple acid phosphatase family protein 35.8 0.092
ath:AT2G01880 PAP7; PAP7 (PURPLE ACID PHOSPHATASE 7); acid pho... 32.7 0.76
bbo:BBOV_IV001930 21.m02940; hypothetical protein 32.0 1.2
ath:AT5G35600 HDA7; HDA7 (histone deacetylase7); histone deace... 32.0 1.4
ath:AT1G13900 calcineurin-like phosphoesterase family protein 31.6 1.7
ath:AT5G51300 splicing factor-related; K13095 splicing factor 1 29.6
sce:YOR184W SER1, ADE9; 3-phosphoserine aminotransferase, cata... 29.3 7.4
cel:D1065.4 srh-210; Serpentine Receptor, class H family membe... 29.3 9.3
> tgo:TGME49_019320 acid phosphatase, putative (EC:3.1.3.2); K01078
acid phosphatase [EC:3.1.3.2]
Length=431
Score = 304 bits (778), Expect = 1e-82, Method: Compositional matrix adjust.
Identities = 135/189 (71%), Positives = 164/189 (86%), Gaps = 0/189 (0%)
Query 1 PKWTLPNWWYHYAVHFAATTGSAFISSGHRDMSVGMVFIDTWVLSSAFPFGNVTEKAWKD 60
PKWTLPNWWYHY +HF A TG AFI+SGH+DMSVGM+FIDTWVLSS+FPF NVT +AW D
Sbjct 177 PKWTLPNWWYHYLMHFPANTGGAFINSGHKDMSVGMIFIDTWVLSSSFPFSNVTSRAWAD 236
Query 61 LEKTLEIAPKVFDYIIVVGNRPVYSSGSSKGDSMLQYYLQPLLKKAGVDAYISGYDRDME 120
LEKTLE+APK+ DYIIVV +R VYSSG+SKGDSMLQYYLQPLLKKA VDAYISGYD +E
Sbjct 237 LEKTLELAPKILDYIIVVADRAVYSSGASKGDSMLQYYLQPLLKKANVDAYISGYDFSLE 296
Query 121 VIEEDDIAYINCGTGSLSGGSALVKSSGSKFFSGDRGFCLFEMTADGLITKFISGENGET 180
VI +D+I++++CG GS + GS +VK SGS +++G+ GFCLFE+TA+GL+T+ +SG GET
Sbjct 297 VISDDNISHVSCGAGSKAAGSPIVKHSGSLYYAGETGFCLFELTAEGLVTRLVSGTTGET 356
Query 181 LYEHKQPVK 189
LY HKQP+K
Sbjct 357 LYTHKQPLK 365
> bbo:BBOV_I004700 19.m02068; acid phosphatase (EC:3.1.3.2); K01078
acid phosphatase [EC:3.1.3.2]
Length=395
Score = 247 bits (631), Expect = 1e-65, Method: Compositional matrix adjust.
Identities = 108/189 (57%), Positives = 141/189 (74%), Gaps = 0/189 (0%)
Query 1 PKWTLPNWWYHYAVHFAATTGSAFISSGHRDMSVGMVFIDTWVLSSAFPFGNVTEKAWKD 60
P+WT+PNWWYHY HFA T + + SGH+DMSVG +FIDTW+LS+AFP+ +V+ AW D
Sbjct 140 PRWTMPNWWYHYYTHFATTASMSLLKSGHKDMSVGFIFIDTWILSTAFPYKDVSNAAWAD 199
Query 61 LEKTLEIAPKVFDYIIVVGNRPVYSSGSSKGDSMLQYYLQPLLKKAGVDAYISGYDRDME 120
L+K LEIAPK+ DYIIVVG++P+ SSG SKGD+ L YYL PLL+ A VDAYI+GYD +ME
Sbjct 200 LKKVLEIAPKILDYIIVVGDKPIQSSGPSKGDAQLSYYLLPLLRDAQVDAYIAGYDHNME 259
Query 121 VIEEDDIAYINCGTGSLSGGSALVKSSGSKFFSGDRGFCLFEMTADGLITKFISGENGET 180
VI+ + IA I G G ++K++ S FFS GFC+ E+ ADG+ TKFI+GE G+
Sbjct 260 VIDSNGIAMIVTGNAGTGGRKPIMKTTNSAFFSEKAGFCIHELGADGMETKFINGETGDV 319
Query 181 LYEHKQPVK 189
+Y HKQ +K
Sbjct 320 MYTHKQAIK 328
> tpv:TP03_0512 acid phosphatase (EC:3.1.3.2); K01078 acid phosphatase
[EC:3.1.3.2]
Length=404
Score = 224 bits (572), Expect = 9e-59, Method: Compositional matrix adjust.
Identities = 98/189 (51%), Positives = 137/189 (72%), Gaps = 0/189 (0%)
Query 1 PKWTLPNWWYHYAVHFAATTGSAFISSGHRDMSVGMVFIDTWVLSSAFPFGNVTEKAWKD 60
P+ +PNWWYH+ F+ + + SGH+D+SV VF+DTWVLS+ FP+ +V+ +AW +
Sbjct 152 PRLIMPNWWYHFFTTFSTNASVSLLKSGHKDLSVAFVFVDTWVLSNQFPYKDVSNEAWNE 211
Query 61 LEKTLEIAPKVFDYIIVVGNRPVYSSGSSKGDSMLQYYLQPLLKKAGVDAYISGYDRDME 120
L+KTLEIAPKV DYI+VVG++PV SSG SKGD+ L Y L PLLK+A VDAY++GYD+DME
Sbjct 212 LKKTLEIAPKVVDYIVVVGDKPVLSSGKSKGDTFLSYKLLPLLKQAQVDAYVAGYDQDME 271
Query 121 VIEEDDIAYINCGTGSLSGGSALVKSSGSKFFSGDRGFCLFEMTADGLITKFISGENGET 180
+++ + A + CG+ G +++KS SKF++ GFCL E+ A+G TKF++G GE
Sbjct 272 LLDYEGTALVVCGSSGNKGRKSVIKSPHSKFYTEAPGFCLHELNAEGFTTKFVNGNTGEV 331
Query 181 LYEHKQPVK 189
LY H QP K
Sbjct 332 LYTHVQPKK 340
> pfa:PFI0880c GAP50; glideosome-associated protein 50 (EC:3.1.3.2);
K01078 acid phosphatase [EC:3.1.3.2]
Length=396
Score = 201 bits (511), Expect = 1e-51, Method: Compositional matrix adjust.
Identities = 101/189 (53%), Positives = 143/189 (75%), Gaps = 0/189 (0%)
Query 1 PKWTLPNWWYHYAVHFAATTGSAFISSGHRDMSVGMVFIDTWVLSSAFPFGNVTEKAWKD 60
PKW +PN+WYHY HF ++G + + +GH+D++ +FIDTWVLSS FP+ + EKAW D
Sbjct 142 PKWIMPNYWYHYFTHFTVSSGPSIVKTGHKDLAAAFIFIDTWVLSSNFPYKKIHEKAWND 201
Query 61 LEKTLEIAPKVFDYIIVVGNRPVYSSGSSKGDSMLQYYLQPLLKKAGVDAYISGYDRDME 120
L+ L +A K+ D+IIVVG++P+YSSG S+G S L YYL PLLK A VD YISG+D +ME
Sbjct 202 LKSQLSVAKKIADFIIVVGDQPIYSSGYSRGSSYLAYYLLPLLKDAEVDLYISGHDNNME 261
Query 121 VIEEDDIAYINCGTGSLSGGSALVKSSGSKFFSGDRGFCLFEMTADGLITKFISGENGET 180
VIE++D+A+I CG+GS+S G + +K+S S FFS D GFC+ E++ +G++TKF+S + GE
Sbjct 262 VIEDNDMAHITCGSGSMSQGKSGMKNSKSLFFSSDIGFCVHELSNNGIVTKFVSSKKGEV 321
Query 181 LYEHKQPVK 189
+Y HK +K
Sbjct 322 IYTHKLNIK 330
> cpv:cgd2_640 acid phosphatase
Length=339
Score = 82.8 bits (203), Expect = 7e-16, Method: Compositional matrix adjust.
Identities = 47/132 (35%), Positives = 75/132 (56%), Gaps = 7/132 (5%)
Query 1 PKWTLPNWWYHYAVHFAATTGSAFISSGHRDMSVGMVFIDTWVLSSAFPFGNVTEKAWKD 60
P+++ PN++YHY H+ T+ RD +V VFIDT++LSS+FP V+E+A+++
Sbjct 121 PRFSFPNYFYHYVSHYTDTSNVL----SRRDGTVLFVFIDTFILSSSFPDHKVSEQAFQN 176
Query 61 LEKTLEIAPKVFDYIIVVGNRPVYSSGSSKGDSMLQYYLQPLLKKAGVDAYISGYDRDME 120
L TL K D+ +VVGN+ Y + S DS L+ +Q L+ V+ ISG +R
Sbjct 177 LNATLHYGHKHHDFTVVVGNK--YLASSYSIDSSLK-KVQQLILDTHVELVISGQNRGTY 233
Query 121 VIEEDDIAYINC 132
+ ++NC
Sbjct 234 NSTIEGTTFLNC 245
> cpv:cgd7_4420 secreted acid phosphatase (calcineurin family),signal
peptide
Length=826
Score = 77.0 bits (188), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 49/138 (35%), Positives = 76/138 (55%), Gaps = 19/138 (13%)
Query 1 PKWTLPNWWYHYAVHFAATTGSAF----ISSGHRDMSVGMV-------FIDTWVLSSAFP 49
P+W LPN+WY+ F + S +SS + + MV + D+W++SS P
Sbjct 125 PRWYLPNFWYYTIEEFESPVNSPHPYLNVSSSPTEETEEMVKTKAIFIYTDSWIISS--P 182
Query 50 FG-NVTEKAWKD----LEKTLEIA-PKVFDYIIVVGNRPVYSSGSSKGDSMLQYYLQPLL 103
G ++T + W + +E TL+ A + D+I V+G+ P YSSG +S + L PLL
Sbjct 183 MGTDITPELWNEQMEFIENTLKAAIMRDIDWIFVIGHFPCYSSGEHGDNSDIHKILDPLL 242
Query 104 KKAGVDAYISGYDRDMEV 121
KK VDAYI+G+D +E+
Sbjct 243 KKYKVDAYIAGHDHHLEL 260
> xla:432322 acp5, MGC78938; acid phosphatase 5, tartrate resistant
(EC:3.1.3.2); K14379 tartrate-resistant acid phosphatase
type 5 [EC:3.1.3.2]
Length=326
Score = 62.4 bits (150), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 54/204 (26%), Positives = 94/204 (46%), Gaps = 38/204 (18%)
Query 2 KWTLPNWWYHYAVHFAATTGSAFISSGHRDMSVGMVFIDTWVL---SSAFPFGNV----- 53
+W P+++Y A + +++V ++ +DT L S F G
Sbjct 131 RWNYPDYYYDLAFTIPGS-----------NVTVRLLMLDTVQLCGISDDFHDGQPRGPNN 179
Query 54 -----TEKAWKDLEKTLEIAPKVFDYIIVVGNRPVYSSGSSKGDSMLQYYLQPLLKKAGV 108
T+ W LE+ L+ A + +Y++V G+ PV+S L + ++PLLKK GV
Sbjct 180 LKMAGTQLEW--LEEKLQSAKE--NYLLVAGHYPVWSVAEHGPTQCLIHTVEPLLKKYGV 235
Query 109 DAYISGYDRDMEVIEEDD-IAYINCGTGSLSGGSALVKSSGSK----FFSGDR----GFC 159
AY+ G++ +M+ +++D I YI G G+ S + K K FF GD F
Sbjct 236 TAYLCGHEHNMQYLQDDQGIGYILSGAGNFMENSRIHKDDVPKGYLQFFQGDPETMGAFA 295
Query 160 LFEMTADGLITKFISGENGETLYE 183
E+T + ++ NG+ L++
Sbjct 296 YIEITPKEMTVTYVQ-SNGKCLFQ 318
> xla:444356 MGC82831 protein; K14379 tartrate-resistant acid
phosphatase type 5 [EC:3.1.3.2]
Length=325
Score = 62.4 bits (150), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 38/120 (31%), Positives = 63/120 (52%), Gaps = 10/120 (8%)
Query 73 DYIIVVGNRPVYSSGSSKGDSMLQYYLQPLLKKAGVDAYISGYDRDMEVIEEDD-IAYIN 131
DY++V G+ PV+S L + L+PLLKK GV AY+ G++ +M+ +++D I Y+
Sbjct 199 DYLLVAGHYPVWSVAEHGPTHCLLHTLEPLLKKYGVTAYLCGHEHNMQYLQDDQGIGYLL 258
Query 132 CGTGSLSGGSAL----VKSSGSKFFSGDR----GFCLFEMTADGLITKFISGENGETLYE 183
G G+ S + V + KFF GD F E+T + ++ NG+ L++
Sbjct 259 SGAGNFMENSRIHEDDVPTDYLKFFQGDPDTMGAFAYIEITPKEMTITYVQS-NGKCLFQ 317
> tgo:TGME49_097650 serine/threonine protein phosphatase, putative
(EC:3.1.3.2)
Length=679
Score = 60.1 bits (144), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 56/217 (25%), Positives = 96/217 (44%), Gaps = 33/217 (15%)
Query 1 PKWTLPNWWY---HYAVHFAATTGSAFISSG-------------HRDMSVGMVFIDTWVL 44
P+W LPN+WY H + + F++ D++V ++ID+ VL
Sbjct 290 PRWRLPNFWYFTRHVFRNVPRHVKNPFVTGASISDISLVSTEEDKTDVTVVTIYIDSMVL 349
Query 45 -------SSA----FPFGNVTEKAWKDLEKTLEIAPKVFDYIIVVGNRPV--YSSGSSKG 91
SS+ FP + + + LE TL+ A + D+I + G+ PV YS ++K
Sbjct 350 LLEEGSKSSSIRYRFPRDELYYRHLEFLEDTLKAATREADWIFIAGHHPVVNYSVRNAK- 408
Query 92 DSMLQYYLQPLLKKAGVDAYISGYDRDMEVIEEDD--IAYINCGTGSLSGGSALVKSSGS 149
S L L++K VD ++SG++ + +E D +I GTGS + S
Sbjct 409 PSDFAVRLTELMRKYKVDTFLSGHEHALSFFQEPDANTTHIISGTGSKLSARDPIPSKDC 468
Query 150 KFFSGDRGFCLFEMTADGLITKFISGENGETLYEHKQ 186
F + G + + + L F+S +G+ L+ Q
Sbjct 469 LFSVREHGVAVHVLGKEELHHGFVSA-DGKVLFTASQ 504
> dre:406801 acp5a, acp5, sb:cb576, wu:fb19f01, wu:fb30b03, wu:fi14e01,
wu:fj66f03, zgc:63825; acid phosphatase 5a, tartrate
resistant (EC:3.1.3.2); K14379 tartrate-resistant acid phosphatase
type 5 [EC:3.1.3.2]
Length=339
Score = 56.2 bits (134), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 38/126 (30%), Positives = 58/126 (46%), Gaps = 9/126 (7%)
Query 66 EIAPKVFDYIIVVGNRPVYSSGSSKGDSMLQYYLQPLLKKAGVDAYISGYDRDMEVIEED 125
+A DY++V G+ PV+S L L+PLLKK AY+ G+D +++ I+E
Sbjct 200 RLAKSKADYLLVAGHYPVWSISEHGPTDCLLKNLRPLLKKYKATAYLCGHDHNLQYIKES 259
Query 126 DIAYINCGTGSLSGGSALVKSSGS----KFFSGDR----GFCLFEMTADGLITKFISGEN 177
I Y+ G G+ ++ KFF+GD GF E+ + FI
Sbjct 260 GIGYVVSGAGNFMDPDVRHRNRVPKGYLKFFNGDASTLGGFAHIEVDKKQMTVTFIQAR- 318
Query 178 GETLYE 183
G +LY
Sbjct 319 GTSLYR 324
> pfa:PF14_0614 phosphatase, putative
Length=1442
Score = 54.3 bits (129), Expect = 2e-07, Method: Composition-based stats.
Identities = 54/191 (28%), Positives = 83/191 (43%), Gaps = 45/191 (23%)
Query 37 VFIDTWVLSSAFPFGNVTEKAWKD----LEKTLEIAPKVFDYIIVVGNRPVYSSGSSKGD 92
+FIDTW L FP +A+++ L KTL + K D+I VVG+ P+ SSG +
Sbjct 852 IFIDTWALMVGFPIIR-NYRAFREQFNWLSKTLYESAKKSDWIFVVGHHPLISSGRRSDN 910
Query 93 SMLQYY-----LQPLLKKAGVDAYISGYDRDMEVIEEDDIAYINCGTGS--LSGGSAL-- 143
+ + ++ L VDAY S +D ME I+ + G+ S L S++
Sbjct 911 YSYEEHSFHDIIRDFLFNYHVDAYFSAHDHLMEYIKFGSVDLFINGSSSRVLFDNSSMGR 970
Query 144 ------------------------VKSSG------SKFFS-GDRGFCLFEMTADGLITKF 172
+K G SK+++ D GF ++T D L+T+F
Sbjct 971 GYFGKIIGKLYPLSCYVLKTIHTGLKPKGCNINRYSKWYNKSDIGFSTHKLTKDELVTQF 1030
Query 173 ISGENGETLYE 183
IS G+ L E
Sbjct 1031 ISSRTGKPLSE 1041
> cel:F02E9.7 hypothetical protein; K14379 tartrate-resistant
acid phosphatase type 5 [EC:3.1.3.2]
Length=419
Score = 53.5 bits (127), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 46/156 (29%), Positives = 70/156 (44%), Gaps = 18/156 (11%)
Query 2 KWTLPNWWYHYAVHFAATT------GSAFISSGHRDMSVGMVFIDTWVLSSAFPFG--NV 53
KW P+ +Y +V F T+ + + +D+ FI+ S P G N+
Sbjct 181 KWYFPSLYYKKSVEFNGTSIDFLMIDTISLCGNTKDIQ-NAGFIEMLRNESHDPRGPVNI 239
Query 54 T--EKAWKDLEKTLEIAPKVFDYIIVVGNRPVYSSGSSKGDSMLQYYLQPLLKKAGVDAY 111
T E+ W LE LE + Y+I+ G+ PV+S S L+ L PLLK+ V+AY
Sbjct 240 TAAEEQWAWLENNLEASSA--QYLIISGHYPVHSMSSHGPTDCLRQRLDPLLKRFNVNAY 297
Query 112 ISGYDRDMEVIE-----EDDIAYINCGTGSLSGGSA 142
SG+D ++ E I Y+ G S + S
Sbjct 298 FSGHDHSLQHFTFPGYGEHIINYVVSGAASRADAST 333
> dre:436725 acp5b, zgc:92339; acid phosphatase 5b, tartrate resistant;
K14379 tartrate-resistant acid phosphatase type 5
[EC:3.1.3.2]
Length=327
Score = 52.0 bits (123), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 37/129 (28%), Positives = 61/129 (47%), Gaps = 13/129 (10%)
Query 73 DYIIVVGNRPVYSSGSSKGDSMLQYYLQPLLKKAGVDAYISGYDRDMEVIEEDD-IAYIN 131
D++IVVG+ P++S G L L+PLLKK V Y+SG+D ++ I EDD +++
Sbjct 199 DFVIVVGHYPIWSIGHHGPTKCLISKLRPLLKKYNVSLYLSGHDHSLQFIREDDGSSFVV 258
Query 132 CGTGSLSGGSALVKSS--------GSKFFSGDRGFCLFEMTADGLITKFISGENGETLYE 183
G G S + S S F FE+ ++ ++ +G+ +Y+
Sbjct 259 SGAGVEEDSSTDHRKSFPSAWQLFSSPVNQTSGSFVYFEVNKSEMLINYLQ-PDGKCVYQ 317
Query 184 ---HKQPVK 189
HK V+
Sbjct 318 TSVHKHKVQ 326
> hsa:54 ACP5, MGC117378, TRAP; acid phosphatase 5, tartrate resistant
(EC:3.1.3.2); K14379 tartrate-resistant acid phosphatase
type 5 [EC:3.1.3.2]
Length=325
Score = 51.6 bits (122), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 39/145 (26%), Positives = 73/145 (50%), Gaps = 14/145 (9%)
Query 54 TEKAWKDLEKTLEIAPKVFDYIIVVGNRPVYSSGSSKGDSMLQYYLQPLLKKAGVDAYIS 113
T+ +W L+K L A + DY++V G+ PV+S L L+PLL GV AY+
Sbjct 183 TQLSW--LKKQLAAARE--DYVLVAGHYPVWSIAEHGPTHCLVKQLRPLLATYGVTAYLC 238
Query 114 GYDRDMEVIE-EDDIAYINCGTGSLSGGSAL----VKSSGSKFFSGDR----GFCLFEMT 164
G+D +++ ++ E+ + Y+ G G+ S V + +F G GF E++
Sbjct 239 GHDHNLQYLQDENGVGYVLSGAGNFMDPSKRHQRKVPNGYLRFHYGTEDSLGGFAYVEIS 298
Query 165 ADGLITKFISGENGETLYEHKQPVK 189
+ + +I +G++L++ + P +
Sbjct 299 SKEMTVTYIEA-SGKSLFKTRLPRR 322
> cpv:cgd8_70 hypothetical protein
Length=424
Score = 50.8 bits (120), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 47/163 (28%), Positives = 75/163 (46%), Gaps = 8/163 (4%)
Query 19 TTGSAFISSGHRDMSVGMVFIDTWVLSSAFPFGNVT----EKAWKDLEKTLEIAP-KVFD 73
T+ S + S + + ++ID+W L+ PF + + +E+TL+ A + D
Sbjct 210 TSISEYFSEKQVNSTAVFIYIDSWTLTQD-PFKKTSISYKYSQLEFIEQTLKAAVFENVD 268
Query 74 YIIVVGNRPVYSSGSSKGDSMLQYYLQPLLKKAGVDAYISGYDRDMEV-IEEDDIAYIN- 131
+II+V + +YSSG + L L PL+KK VD ISG+D E+ + ED +Y
Sbjct 269 WIILVTHYSIYSSGLHGPHTRLASILLPLIKKYRVDFIISGHDHHSEILVPEDFNSYFQI 328
Query 132 CGTGSLSGGSALVKSSGSKFFSGDRGFCLFEMTADGLITKFIS 174
G S S S F S F F + D +++ S
Sbjct 329 VGASSKPRTSFGATDENSIFKSNTCSFASFTFSKDIAVSRIFS 371
> tgo:TGME49_028170 serine/threonine protein phosphatase, putative
(EC:3.1.3.2 3.2.1.3)
Length=1491
Score = 50.4 bits (119), Expect = 4e-06, Method: Composition-based stats.
Identities = 37/163 (22%), Positives = 69/163 (42%), Gaps = 21/163 (12%)
Query 1 PKWTLPNWWYHYAVHFAATTGSAFISSGHRDMSVGMVFIDTWVLSSAFPFGNVTEKAWKD 60
P W +PN Y F+ + A + H + ++ ++TW L P N +
Sbjct 505 PNWYMPNDAYTATFSFSTSMTMANGTIQHEAFNATVINVNTWNLFVGNPIANNMQSMMDR 564
Query 61 L----EKTLEIAPKVFDYIIVVGNRPVYSSGSSKGDSMLQY----------------YLQ 100
L ++ + +++I++G+ P+ S+G LQY +Q
Sbjct 565 LMWLSDQLYTAVNQTTNWLIIMGHLPLVSTGPQGEQGRLQYVDDLYKNGQPRGPEAVLIQ 624
Query 101 PLLKKAGVDAYISGYDRDMEVIEEDDIAYINCGTGSLSGGSAL 143
LL VD Y+S +D ME + +D++ N T ++ G+A+
Sbjct 625 MLLSHYQVDLYVSAHDHFMEYVALEDLSK-NTTTAFITSGAAV 666
> mmu:11433 Acp5, TRACP, TRAP; acid phosphatase 5, tartrate resistant
(EC:3.1.3.2); K14379 tartrate-resistant acid phosphatase
type 5 [EC:3.1.3.2]
Length=327
Score = 48.5 bits (114), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 46/208 (22%), Positives = 91/208 (43%), Gaps = 34/208 (16%)
Query 2 KWTLPNWWYHYAVHFAATTGSAFISSGHRDMSVGMVFIDTWVLS-SAFPFGNVTEKAWKD 60
+W P+ +Y T +++V + +DT +L ++ F + K +D
Sbjct 131 RWNFPSPYYRLRFKIPRT-----------NITVAIFMLDTVMLCGNSDDFASQQPKMPRD 179
Query 61 L----------EKTLEIAPKVFDYIIVVGNRPVYSSGSSKGDSMLQYYLQPLLKKAGVDA 110
L +K L A + DY++V G+ P++S L L+PLL GV A
Sbjct 180 LGVARTQLSWLKKQLAAAKE--DYVLVAGHYPIWSIAEHGPTRCLVKNLRPLLATYGVTA 237
Query 111 YISGYDRDMEVIE-EDDIAYINCGTGSLSGGSAL----VKSSGSKFFSGDR----GFCLF 161
Y+ G+D +++ ++ E+ + Y+ G G+ S V + +F G GF
Sbjct 238 YLCGHDHNLQYLQDENGVGYVLSGAGNFMDPSVRHQRKVPNGYLRFHYGSEDSLGGFTHV 297
Query 162 EMTADGLITKFISGENGETLYEHKQPVK 189
E++ + ++ +G++L++ P +
Sbjct 298 EISPKEMTIIYVEA-SGKSLFKTSLPRR 324
> tgo:TGME49_028160 serine/threonine protein phosphatase, putative
(EC:3.1.3.2)
Length=1632
Score = 47.8 bits (112), Expect = 2e-05, Method: Composition-based stats.
Identities = 37/138 (26%), Positives = 56/138 (40%), Gaps = 20/138 (14%)
Query 3 WTLPNWWYHYAVHFAATTGSAFISSGHRDMSVGMVFIDTWVLSSAFPFGN----VTEKAW 58
W +PN +Y F A A + ++ I+TW L P N T K W
Sbjct 488 WHMPNDYYTVNFKFGANLTLANGMFEETAFNATVIAINTWDLFVGNPVANNMASWTNKLW 547
Query 59 KDLEKTLEIAPKVFDYIIVVGNRPVYSSGSSKGDSMLQ----------------YYLQPL 102
E+ + K +IIV+G+ P+ S+G+ + LQ + + L
Sbjct 548 WLSEQLYQATQKNSSWIIVMGHHPMLSTGTEGEQARLQNIDDLSKSRRSRGLESFLINTL 607
Query 103 LKKAGVDAYISGYDRDME 120
VDAY+SG+D ME
Sbjct 608 FMHYQVDAYVSGHDHLME 625
> ath:AT1G14700 PAP3; PAP3 (PURPLE ACID PHOSPHATASE 3); acid phosphatase/
protein serine/threonine phosphatase; K01078 acid
phosphatase [EC:3.1.3.2]
Length=364
Score = 43.1 bits (100), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 30/107 (28%), Positives = 56/107 (52%), Gaps = 7/107 (6%)
Query 59 KDLEKTLEIAPK--VFDYIIVVGNRPVYSSGSSKGDSMLQYYLQPLLKKAGVDAYISGYD 116
+L K L++A + V + IV+G+ + S+G L+ +L P+L+ VD Y++G+D
Sbjct 221 NNLLKELDVALRESVAKWKIVIGHHTIKSAGHHGNTIELEKHLLPILQANEVDLYVNGHD 280
Query 117 RDMEVIE--EDDIAYINCGTGSLS---GGSALVKSSGSKFFSGDRGF 158
+E I + +I ++ G GS + G V+ +F+ +GF
Sbjct 281 HCLEHISSVDSNIQFMTSGGGSKAWKGGDVNYVEPEEMRFYYDGQGF 327
> ath:AT3G17790 PAP17; PAP17; acid phosphatase/ phosphatase/ protein
serine/threonine phosphatase
Length=338
Score = 43.1 bits (100), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 31/93 (33%), Positives = 50/93 (53%), Gaps = 5/93 (5%)
Query 47 AFPFGNVTEKAW-KDLEKTLEIAPKVFDYIIVVGNRPVYSSGSSKGDSMLQYYLQPLLKK 105
A P N KA +DLE +L+ + + IVVG+ + S G L L P+LK+
Sbjct 186 AVPSRNSYVKALLRDLEVSLKSSKA--RWKIVVGHHAMRSIGHHGDTKELNEELLPILKE 243
Query 106 AGVDAYISGYDRDMEVIEEDD--IAYINCGTGS 136
GVD Y++G+D ++ + ++D I ++ G GS
Sbjct 244 NGVDLYMNGHDHCLQHMSDEDSPIQFLTSGAGS 276
> ath:AT2G01890 PAP8; PAP8 (PURPLE ACID PHOSPHATASE 8); acid phosphatase/
protein serine/threonine phosphatase
Length=335
Score = 37.4 bits (85), Expect = 0.030, Method: Compositional matrix adjust.
Identities = 27/80 (33%), Positives = 40/80 (50%), Gaps = 11/80 (13%)
Query 76 IVVGNRPVYSSGSSKGDSMLQYYLQPLLKKAGVDAYISGYDRDMEVIEEDDIAYINCGTG 135
IVVG+ + S+G L+ L P+L+ VD YI+G+D +E I+ IN G
Sbjct 213 IVVGHHTIKSAGHHGNTIELEKQLLPILEANEVDLYINGHDHCLE-----HISSINSGIQ 267
Query 136 SLSGGSALVKSSGSKFFSGD 155
++ G GSK + GD
Sbjct 268 FMTSG------GGSKAWKGD 281
> ath:AT1G25230 purple acid phosphatase family protein
Length=339
Score = 35.8 bits (81), Expect = 0.092, Method: Compositional matrix adjust.
Identities = 28/101 (27%), Positives = 46/101 (45%), Gaps = 4/101 (3%)
Query 76 IVVGNRPVYSSGSSKGDSMLQYYLQPLLKKAGVDAYISGYDRDMEVI--EEDDIAYINCG 133
IVVG+ + S+ L+ L P+L+ VD Y++G+D ++ I + I ++ G
Sbjct 214 IVVGHHAIKSASIHGNTKELESLLLPILEANKVDLYMNGHDHCLQHISTSQSPIQFLTSG 273
Query 134 TGSLS--GGSALVKSSGSKFFSGDRGFCLFEMTADGLITKF 172
GS + G KFF +GF ++T L F
Sbjct 274 GGSKAWRGYYNWTTPEDMKFFYDGQGFMSVKITRSELSVVF 314
> ath:AT2G01880 PAP7; PAP7 (PURPLE ACID PHOSPHATASE 7); acid phosphatase/
protein serine/threonine phosphatase
Length=328
Score = 32.7 bits (73), Expect = 0.76, Method: Compositional matrix adjust.
Identities = 25/97 (25%), Positives = 43/97 (44%), Gaps = 3/97 (3%)
Query 41 TWVLSSAFPFGNVTEKAWKDLEKTLEIAPKVFDYIIVVGNRPVYSSGSSKGDSMLQYYLQ 100
T+ + P DL+ LEI + VVG+ + ++G+ L L
Sbjct 176 TYDWRNVLPRNKYISNLLHDLD--LEIKKSRATWKFVVGHHGIKTAGNHGVTQELVDQLL 233
Query 101 PLLKKAGVDAYISGYDRDMEVI-EEDDIAYINCGTGS 136
P+L++ VD YI+G+D ++ I ++ G GS
Sbjct 234 PILEENKVDLYINGHDHCLQHIGSHGKTQFLTSGGGS 270
> bbo:BBOV_IV001930 21.m02940; hypothetical protein
Length=1006
Score = 32.0 bits (71), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 29/102 (28%), Positives = 46/102 (45%), Gaps = 22/102 (21%)
Query 2 KWTLPNWW-YHYAVHFAATTGS-----AFISSGHR--------DMSVGMVF------IDT 41
K +P WW +H+ V +A +G +F+ G + +M++G+VF ID
Sbjct 620 KMVVPAWWVHHHQVSRSAPSGEYLPHGSFMIRGKKNYVQPQRLEMAIGVVFHIEVPDIDE 679
Query 42 WVLSSAFPFGNVTEKAWKDLEKTLEIAPKVFDYIIVVGNRPV 83
+ + P G TE A +D+E A D +I G PV
Sbjct 680 EEVEA--PAGPDTEDAPQDVESDESDASLTVDDLIGHGEEPV 719
> ath:AT5G35600 HDA7; HDA7 (histone deacetylase7); histone deacetylase;
K06067 histone deacetylase 1/2 [EC:3.5.1.98]
Length=409
Score = 32.0 bits (71), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 29/98 (29%), Positives = 46/98 (46%), Gaps = 16/98 (16%)
Query 48 FPFGNVTEKAWKDLEKTLEIAPKVFDYIIVVGNRPVYSSGSSKGDSMLQYYLQPLLKKAG 107
FP G+ E+A+KD ++ + ++ F + G+ + G KG QYY K G
Sbjct 184 FPHGDEVEEAFKDTDRVMTVS---FHKVGDTGD--ISDYGEGKG----QYYSLNAPLKDG 234
Query 108 VD------AYISGYDRDMEVIEEDDIAYINCGTGSLSG 139
+D +I R ME+ E + I + CG SL+G
Sbjct 235 LDDFSLRGLFIPVIHRAMEIYEPEVIV-LQCGADSLAG 271
> ath:AT1G13900 calcineurin-like phosphoesterase family protein
Length=656
Score = 31.6 bits (70), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 22/80 (27%), Positives = 40/80 (50%), Gaps = 9/80 (11%)
Query 59 KDLEKTLEIAPKVFDYIIVVGNRPVYSSGSSKGDSMLQY----YLQPLLKKAGVDAYISG 114
+DLE + K +++V G+RP+Y++ + D+M++ +L+PL K V + G
Sbjct 426 RDLES---VDRKKTPFVVVQGHRPMYTTSNEVRDTMIRQKMVEHLEPLFVKNNVTLALWG 482
Query 115 YDRDMEVIEEDDIAYINCGT 134
+ E I+ CGT
Sbjct 483 HVHRYERFCP--ISNNTCGT 500
> ath:AT5G51300 splicing factor-related; K13095 splicing factor
1
Length=804
Score = 29.6 bits (65), Expect = 7.1, Method: Composition-based stats.
Identities = 21/66 (31%), Positives = 31/66 (46%), Gaps = 6/66 (9%)
Query 66 EIAPKVFDYIIVVGNRPVYSSGSS------KGDSMLQYYLQPLLKKAGVDAYISGYDRDM 119
+ AP D++ + V +GSS G S L+PLL + GV +SG D+D
Sbjct 24 DTAPLALDHMNPQNSESVALNGSSTPIPDTNGSSAKPELLRPLLSENGVSKTLSGNDKDQ 83
Query 120 EVIEED 125
EE+
Sbjct 84 SGGEEE 89
> sce:YOR184W SER1, ADE9; 3-phosphoserine aminotransferase, catalyzes
the formation of phosphoserine from 3-phosphohydroxypyruvate,
required for serine and glycine biosynthesis; regulated
by the general control of amino acid biosynthesis mediated
by Gcn4p (EC:2.6.1.52); K00831 phosphoserine aminotransferase
[EC:2.6.1.52]
Length=395
Score = 29.3 bits (64), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 17/58 (29%), Positives = 28/58 (48%), Gaps = 0/58 (0%)
Query 67 IAPKVFDYIIVVGNRPVYSSGSSKGDSMLQYYLQPLLKKAGVDAYISGYDRDMEVIEE 124
I P FDY VV N Y++ ++ Q +LKK GV+A + + +++ E
Sbjct 253 ITPIAFDYPTVVKNNSAYNTIPIFTLHVMDLVFQHILKKGGVEAQQAENEEKAKILYE 310
> cel:D1065.4 srh-210; Serpentine Receptor, class H family member
(srh-210)
Length=329
Score = 29.3 bits (64), Expect = 9.3, Method: Compositional matrix adjust.
Identities = 21/89 (23%), Positives = 40/89 (44%), Gaps = 15/89 (16%)
Query 9 WYHYAVHFAATTGSAFISSGHRDMSVGMVFIDT--WVLSSAFPFGNV----TEKAWKDLE 62
WY + VHF + T D+S+ ++ I + ++ + FG +AW+
Sbjct 53 WYMFNVHFWSAT---------LDISLSLLVIPYMFFPFAAGYSFGIFKWLDVNRAWQTTV 103
Query 63 KTLEIAPKVFDYIIVVGNRPVYSSGSSKG 91
+EI + +++ NR + + SSKG
Sbjct 104 IVIEIGMTIISILVLFENRFTFLASSSKG 132
Lambda K H
0.318 0.136 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5364689396
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40