bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0372_orf2
Length=214
Score E
Sequences producing significant alignments: (Bits) Value
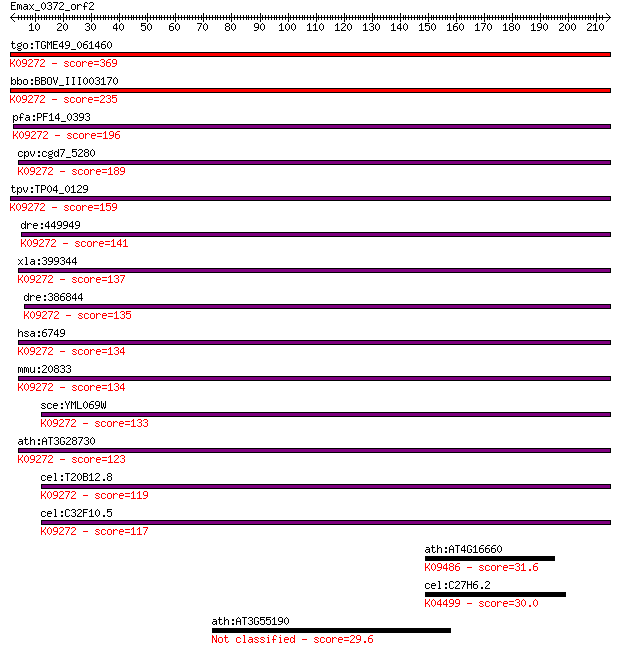
tgo:TGME49_061460 structure specific recognition protein I, pu... 369 5e-102
bbo:BBOV_III003170 17.m07304; structure specific recognition p... 235 1e-61
pfa:PF14_0393 structure specific recognition protein; K09272 s... 196 4e-50
cpv:cgd7_5280 structure-specific recognition protein 1 (SSRP1)... 189 6e-48
tpv:TP04_0129 structure specific recognition protein; K09272 s... 159 6e-39
dre:449949 ssrp1b, im:7140964; structure specific recognition ... 141 2e-33
xla:399344 ssrp1; structure specific recognition protein 1; K0... 137 4e-32
dre:386844 ssrp1a, ssrp1, wu:fa11b03, wu:fd10f06; structure sp... 135 1e-31
hsa:6749 SSRP1, FACT, FACT80, T160; structure specific recogni... 134 3e-31
mmu:20833 Ssrp1, C81323, Hmg1-rs1, Hmgi-rs3, Hmgox, T160; stru... 134 3e-31
sce:YML069W POB3; Pob3p; K09272 structure-specific recognition... 133 4e-31
ath:AT3G28730 ATHMG; ATHMG (ARABIDOPSIS THALIANA HIGH MOBILITY... 123 4e-28
cel:T20B12.8 hmg-4; HMG family member (hmg-4); K09272 structur... 119 6e-27
cel:C32F10.5 hmg-3; HMG family member (hmg-3); K09272 structur... 117 3e-26
ath:AT4G16660 heat shock protein 70, putative / HSP70, putativ... 31.6 2.3
cel:C27H6.2 ruvb-1; RUVB (recombination protein) homolog famil... 30.0 5.8
ath:AT3G55190 esterase/lipase/thioesterase family protein 29.6 9.3
> tgo:TGME49_061460 structure specific recognition protein I,
putative ; K09272 structure-specific recognition protein 1
Length=539
Score = 369 bits (946), Expect = 5e-102, Method: Compositional matrix adjust.
Identities = 172/214 (80%), Positives = 197/214 (92%), Gaps = 0/214 (0%)
Query 1 NAQDIVQVTTPSKNDLAIELVQDDTRDQTEDMLLEIRLFQPAAGDDETDSHLQSLKEKLV 60
+AQ+I QVTTPSKNDLAIEL+QDDTRDQ ED LLE+R +QP AGDD+ + LQ LK+KLV
Sbjct 156 HAQEIAQVTTPSKNDLAIELIQDDTRDQQEDQLLEVRFYQPFAGDDDAEGPLQQLKQKLV 215
Query 61 KKSGVAETKMESIALLNDVPLLVPRGRYEIDIGRRALKFHGKSYDYTILYTNINRMFLVP 120
KKSGVAETKM+S+ALLNDVPLLVPRGRYEIDIGRRALKFHGKSYDYTI Y++INRMFLVP
Sbjct 216 KKSGVAETKMDSVALLNDVPLLVPRGRYEIDIGRRALKFHGKSYDYTIQYSSINRMFLVP 275
Query 121 RPNSPHVNFILSLDNAMRQGQTSYPFVVMQFDSENVTSVEVNLEEAELKEKGLEKQLEGK 180
RPNSPHVNFILSL+NAMRQGQTSYPFVVMQFDSE+V SV+VNLE AEL+++GLEK +EGK
Sbjct 276 RPNSPHVNFILSLENAMRQGQTSYPFVVMQFDSESVHSVDVNLEPAELQQRGLEKLIEGK 335
Query 181 TFDVITRILKSLIGKSVIVPGDFKSVKNQYGITC 214
TF V+TR+ ++L+GKSVIVPGDFKSVK Q+GI C
Sbjct 336 TFHVVTRLFRALVGKSVIVPGDFKSVKQQFGIAC 369
> bbo:BBOV_III003170 17.m07304; structure specific recognition
protein; K09272 structure-specific recognition protein 1
Length=485
Score = 235 bits (599), Expect = 1e-61, Method: Compositional matrix adjust.
Identities = 109/215 (50%), Positives = 155/215 (72%), Gaps = 2/215 (0%)
Query 1 NAQDIVQVTTPSKNDLAIELVQDDTRDQTEDMLLEIRLFQPAAGD-DETDSHLQSLKEKL 59
+A+D+ QVT P+K DLA+E Q+ D L+EIR P D D+ + L+ LK+
Sbjct 137 DAKDVTQVTVPTKTDLAVEFKQNKGYING-DELMEIRFCIPNKPDVDDNELALEDLKQTF 195
Query 60 VKKSGVAETKMESIALLNDVPLLVPRGRYEIDIGRRALKFHGKSYDYTILYTNINRMFLV 119
+ K+G+ E K E++A L DVPL+VPRGR+EI+ R+ +K+HGKSYDYT+ +TNI+RMFLV
Sbjct 196 LLKAGLDELKSETLAFLTDVPLIVPRGRFEIEFSRKHIKYHGKSYDYTMFFTNISRMFLV 255
Query 120 PRPNSPHVNFILSLDNAMRQGQTSYPFVVMQFDSENVTSVEVNLEEAELKEKGLEKQLEG 179
P+PNSPH+NFI+ L MRQGQT YPFVVMQFD+E +E+N+ E +L+ LEK + G
Sbjct 256 PKPNSPHINFIIGLHQPMRQGQTRYPFVVMQFDAEEDIELEINMPEEDLESMKLEKVMTG 315
Query 180 KTFDVITRILKSLIGKSVIVPGDFKSVKNQYGITC 214
KTF+V+T++ +L+ K ++VPG+FKS K + G +C
Sbjct 316 KTFNVVTKLFGTLVNKPIVVPGEFKSEKEEAGFSC 350
> pfa:PF14_0393 structure specific recognition protein; K09272
structure-specific recognition protein 1
Length=506
Score = 196 bits (499), Expect = 4e-50, Method: Compositional matrix adjust.
Identities = 92/214 (42%), Positives = 143/214 (66%), Gaps = 3/214 (1%)
Query 2 AQDIVQVTTPSKNDLAIELVQDDTRDQ-TEDMLLEIRLFQPAAGDDETDSHLQSLKEKLV 60
+I Q+ K D+A+E D+ ++ ED L EIR + P D+ + + Q+LK L+
Sbjct 144 TNNINQLNVQIKTDIAMEFKNDENNNKGNEDFLAEIRFYYPHENDE--NQNFQNLKNDLL 201
Query 61 KKSGVAETKMESIALLNDVPLLVPRGRYEIDIGRRALKFHGKSYDYTILYTNINRMFLVP 120
+K + +TK ESIA L+++PLLVPRGRY+I++ K HGKSYD+ I YTNIN+M LVP
Sbjct 202 EKVNIGDTKSESIASLSNIPLLVPRGRYDIEMYSSTFKLHGKSYDFNIQYTNINKMILVP 261
Query 121 RPNSPHVNFILSLDNAMRQGQTSYPFVVMQFDSENVTSVEVNLEEAELKEKGLEKQLEGK 180
+ NS I SL N M+QGQT YPF+++Q ++++ ++++ + + + LEK + GK
Sbjct 262 KSNSNQYVLIFSLSNKMKQGQTEYPFILIQLNNDDDMELDISASDEVMTKYKLEKTISGK 321
Query 181 TFDVITRILKSLIGKSVIVPGDFKSVKNQYGITC 214
DV+T++ +L+ K+VIVPGD+++ KNQ+GITC
Sbjct 322 AHDVVTKLFTALVNKNVIVPGDYRTSKNQHGITC 355
> cpv:cgd7_5280 structure-specific recognition protein 1 (SSRP1)
(recombination signal sequence recognition protein) ; K09272
structure-specific recognition protein 1
Length=523
Score = 189 bits (480), Expect = 6e-48, Method: Compositional matrix adjust.
Identities = 97/218 (44%), Positives = 143/218 (65%), Gaps = 7/218 (3%)
Query 4 DIVQVTTPSKNDLAIELVQDDTRDQTEDMLLEIRLFQPA---AGDDETDSHLQSLKEKLV 60
+I Q+ PSK++L +E + + D L+EIRLF P + D + S + L+ L+
Sbjct 140 NINQIAMPSKSELVLEFNEGVNAGEDCDELMEIRLFVPNQENSLDGNSLSSAEKLRSDLL 199
Query 61 KKSGVAET-KMESIALLNDVPLLVPRGRYEIDIGRRALKFHGKSYDYTILYTNINRMFLV 119
K +G+ + M+ + ND+ LLVPRGRYEI++ LK HGKS+DYTIL+ +I+R+FL+
Sbjct 200 KLTGIGSSGSMDKVCRWNDIHLLVPRGRYEIEVLVNCLKLHGKSFDYTILFQSISRLFLL 259
Query 120 PRPNSPHVNFILSLDNAMRQGQTSYPFVVMQFDSENVTSVE--VNLEEAELKE-KGLEKQ 176
PRP + VN +++L+ MRQG T YPFVVMQFD++ ++E +NL E E++ GL
Sbjct 260 PRPGTSLVNLVVALETPMRQGNTKYPFVVMQFDTQQDENIEMPLNLSEKEIQRFTGLSPI 319
Query 177 LEGKTFDVITRILKSLIGKSVIVPGDFKSVKNQYGITC 214
+ GK +D++TRILKSL G S+IVPGDF+S + I C
Sbjct 320 MTGKFWDIVTRILKSLTGHSIIVPGDFRSASMYHCIRC 357
> tpv:TP04_0129 structure specific recognition protein; K09272
structure-specific recognition protein 1
Length=460
Score = 159 bits (403), Expect = 6e-39, Method: Compositional matrix adjust.
Identities = 90/215 (41%), Positives = 127/215 (59%), Gaps = 38/215 (17%)
Query 1 NAQDIVQVTTPSKNDLAIELVQDDTRDQTEDMLLEIRLFQPAAGDDETDS-HLQSLKEKL 59
+AQ I+Q T PSK DLAIEL +T + + D L+EIR P D E + L+ LK+
Sbjct 156 DAQSIIQATIPSKTDLAIELKNVNTLNNS-DELVEIRFCLPNKLDPEDNEIQLEDLKQTF 214
Query 60 VKKSGVAETKMESIALLNDVPLLVPRGRYEIDIGRRALKFHGKSYDYTILYTNINRMFLV 119
+ KSG+ E K E IALL D+PL+VPRGRYEI+ +R L
Sbjct 215 LVKSGLDEMKSEKIALLMDIPLIVPRGRYEIEFTKRRL---------------------- 252
Query 120 PRPNSPHVNFILSLDNAMRQGQTSYPFVVMQFDSENVTSVEVNLEEAELKEKGLEKQLEG 179
+MRQGQT Y ++VMQF+S++ T V++NL+E +LK+ L+K LEG
Sbjct 253 --------------SQSMRQGQTRYAYIVMQFESDHETKVDLNLQEQDLKQYKLDKVLEG 298
Query 180 KTFDVITRILKSLIGKSVIVPGDFKSVKNQYGITC 214
KT++V++R+ SL+ +S++VPGDFKS K I+C
Sbjct 299 KTYNVVSRLFGSLVNRSIVVPGDFKSEKGDSAISC 333
> dre:449949 ssrp1b, im:7140964; structure specific recognition
protein 1b; K09272 structure-specific recognition protein
1
Length=706
Score = 141 bits (355), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 69/213 (32%), Positives = 125/213 (58%), Gaps = 7/213 (3%)
Query 5 IVQVTTPSKNDLAIELVQDDTRDQTEDMLLEIRLFQPAAGDDETDSHLQSLKEKLVKKSG 64
+ Q TT KN++ +E Q+D TE L+E+R + P DE +++ + ++ K+
Sbjct 136 VSQCTT-GKNEVTVEFHQND---DTEVSLMEVRFYVPPTTGDEGSDPVEAFAQNVLSKAD 191
Query 65 VAETKMESIALLNDVPLLVPRGRYEIDIGRRALKFHGKSYDYTILYTNINRMFLVPRPNS 124
V + +++ + ++ L PRGRY+I I L HGK++DY I YT + R+FL+P +
Sbjct 192 VIQATGDAVCIFRELQCLTPRGRYDIRIYPTFLHLHGKTFDYKIPYTTVLRLFLLPHKDQ 251
Query 125 PHVNFILSLDNAMRQGQTSYPFVVMQFDSENVTSVEVNLEEAELK---EKGLEKQLEGKT 181
+ F++SLD ++QGQT Y F+++ F E S+ +N+ E E++ E L K + G
Sbjct 252 RQMFFVISLDPPIKQGQTRYHFLILLFSKEETISLTLNMNEDEVERRFEGKLNKNMSGSL 311
Query 182 FDVITRILKSLIGKSVIVPGDFKSVKNQYGITC 214
+++++R++K+L+ + + VPG+F+ ITC
Sbjct 312 YEMVSRVMKALVNRKITVPGNFQGHSGAQCITC 344
> xla:399344 ssrp1; structure specific recognition protein 1;
K09272 structure-specific recognition protein 1
Length=693
Score = 137 bits (344), Expect = 4e-32, Method: Compositional matrix adjust.
Identities = 69/214 (32%), Positives = 128/214 (59%), Gaps = 8/214 (3%)
Query 4 DIVQVTTPSKNDLAIELVQDDTRDQTEDMLLEIRLFQPAAGDDETDSHLQSLKEKLVKKS 63
++ Q TT KN++ +E Q+D +E L+EIR + P DD DS +++ + ++ K+
Sbjct 135 NVSQCTT-GKNEVTLEFHQND---DSEVSLMEIRFYVPPTQDDGGDS-VEAFAQNVLSKA 189
Query 64 GVAETKMESIALLNDVPLLVPRGRYEIDIGRRALKFHGKSYDYTILYTNINRMFLVPRPN 123
V + +++ + ++ L PRGRY+I I L HGK++DY I YT + R+FL+P +
Sbjct 190 DVIQATGDAVCIFRELQCLTPRGRYDIRIYPTFLHLHGKTFDYKIPYTTVLRLFLLPHKD 249
Query 124 SPHVNFILSLDNAMRQGQTSYPFVVMQFDSENVTSVEVNLEEAELK---EKGLEKQLEGK 180
+ F++SLD ++QGQT Y F+++ F + ++ +N+ E E++ E L+K + G
Sbjct 250 QRQMFFVISLDPPIKQGQTRYHFLILLFSKDEDMTLTLNMSEEEVERRFEGKLKKSMSGC 309
Query 181 TFDVITRILKSLIGKSVIVPGDFKSVKNQYGITC 214
+++++R++K+L+ + + VPG+F ITC
Sbjct 310 LYEMVSRVMKALVNRKITVPGNFLGHSGSQCITC 343
> dre:386844 ssrp1a, ssrp1, wu:fa11b03, wu:fd10f06; structure
specific recognition protein 1a; K09272 structure-specific recognition
protein 1
Length=518
Score = 135 bits (340), Expect = 1e-31, Method: Compositional matrix adjust.
Identities = 67/213 (31%), Positives = 124/213 (58%), Gaps = 8/213 (3%)
Query 6 VQVTTPSKNDLAIELVQDDTRDQTEDMLLEIRLF-QPAAGDDETDSHLQSLKEKLVKKSG 64
V KN++ +E Q+D E L+E+R + P GDD +D +++ + ++ K+
Sbjct 136 VSQCATGKNEVTVEFHQND---DAEVSLMEVRFYVPPNTGDDGSDP-VEAFAQNILSKAD 191
Query 65 VAETKMESIALLNDVPLLVPRGRYEIDIGRRALKFHGKSYDYTILYTNINRMFLVPRPNS 124
V + +++ + ++ L PRGRY+I I L HGK++DY I YT + R+FL+P +
Sbjct 192 VIQATGDAVCIFKELQCLTPRGRYDIRIYPTFLHLHGKTFDYKIPYTTVLRLFLLPHKDQ 251
Query 125 PHVNFILSLDNAMRQGQTSYPFVVMQFDSENVTSVEVNLEEAELK---EKGLEKQLEGKT 181
+ F++SLD ++QGQT Y F+++ F + S+ +N+ E E++ E L K + G
Sbjct 252 RQMFFVISLDPPIKQGQTRYHFLILLFSKDEDISLALNMSEDEVEKRYEGKLSKNMSGPL 311
Query 182 FDVITRILKSLIGKSVIVPGDFKSVKNQYGITC 214
+++++R++K+L+ + + VPG+F+ ITC
Sbjct 312 YEIVSRVMKALVNRKITVPGNFQGHSGSQCITC 344
> hsa:6749 SSRP1, FACT, FACT80, T160; structure specific recognition
protein 1; K09272 structure-specific recognition protein
1
Length=709
Score = 134 bits (337), Expect = 3e-31, Method: Compositional matrix adjust.
Identities = 68/214 (31%), Positives = 126/214 (58%), Gaps = 8/214 (3%)
Query 4 DIVQVTTPSKNDLAIELVQDDTRDQTEDMLLEIRLFQPAAGDDETDSHLQSLKEKLVKKS 63
++ Q TT KN++ +E Q+D E L+E+R + P +D D +++ + ++ K+
Sbjct 135 NVSQCTT-GKNEVTLEFHQND---DAEVSLMEVRFYVPPTQEDGVDP-VEAFAQNVLSKA 189
Query 64 GVAETKMESIALLNDVPLLVPRGRYEIDIGRRALKFHGKSYDYTILYTNINRMFLVPRPN 123
V + ++I + ++ L PRGRY+I I L HGK++DY I YT + R+FL+P +
Sbjct 190 DVIQATGDAICIFRELQCLTPRGRYDIRIYPTFLHLHGKTFDYKIPYTTVLRLFLLPHKD 249
Query 124 SPHVNFILSLDNAMRQGQTSYPFVVMQFDSENVTSVEVNLEEAELK---EKGLEKQLEGK 180
+ F++SLD ++QGQT Y F+++ F + S+ +N+ E E++ E L K + G
Sbjct 250 QRQMFFVISLDPPIKQGQTRYHFLILLFSKDEDISLTLNMNEEEVEKRFEGRLTKNMSGS 309
Query 181 TFDVITRILKSLIGKSVIVPGDFKSVKNQYGITC 214
+++++R++K+L+ + + VPG+F+ ITC
Sbjct 310 LYEMVSRVMKALVNRKITVPGNFQGHSGAQCITC 343
> mmu:20833 Ssrp1, C81323, Hmg1-rs1, Hmgi-rs3, Hmgox, T160; structure
specific recognition protein 1; K09272 structure-specific
recognition protein 1
Length=708
Score = 134 bits (336), Expect = 3e-31, Method: Compositional matrix adjust.
Identities = 68/214 (31%), Positives = 126/214 (58%), Gaps = 8/214 (3%)
Query 4 DIVQVTTPSKNDLAIELVQDDTRDQTEDMLLEIRLFQPAAGDDETDSHLQSLKEKLVKKS 63
++ Q TT KN++ +E Q+D E L+E+R + P +D D +++ + ++ K+
Sbjct 135 NVSQCTT-GKNEVTLEFHQND---DAEVSLMEVRFYVPPTQEDGVDP-VEAFAQNVLSKA 189
Query 64 GVAETKMESIALLNDVPLLVPRGRYEIDIGRRALKFHGKSYDYTILYTNINRMFLVPRPN 123
V + ++I + ++ L PRGRY+I I L HGK++DY I YT + R+FL+P +
Sbjct 190 DVIQATGDAICIFRELQCLTPRGRYDIRIYPTFLHLHGKTFDYKIPYTTVLRLFLLPHKD 249
Query 124 SPHVNFILSLDNAMRQGQTSYPFVVMQFDSENVTSVEVNLEEAELK---EKGLEKQLEGK 180
+ F++SLD ++QGQT Y F+++ F + S+ +N+ E E++ E L K + G
Sbjct 250 QRQMFFVISLDPPIKQGQTRYHFLILLFSKDEDISLTLNMNEEEVEKRFEGRLTKNMSGS 309
Query 181 TFDVITRILKSLIGKSVIVPGDFKSVKNQYGITC 214
+++++R++K+L+ + + VPG+F+ ITC
Sbjct 310 LYEMVSRVMKALVNRKITVPGNFQGHSGAQCITC 343
> sce:YML069W POB3; Pob3p; K09272 structure-specific recognition
protein 1
Length=552
Score = 133 bits (335), Expect = 4e-31, Method: Compositional matrix adjust.
Identities = 76/240 (31%), Positives = 130/240 (54%), Gaps = 37/240 (15%)
Query 12 SKNDLAIEL-VQDDTRDQTEDMLLEIRLFQP----------------------------A 42
SKN++ IE +QD+ D L+E+R + P A
Sbjct 147 SKNEVGIEFNIQDEEYQPAGDELVEMRFYIPGVIQTNVDENMTKKEESSNEVVPKKEDGA 206
Query 43 AGDD-----ETDSHLQSLKEKLVKKSGVAETKMESIALLNDVPLLVPRGRYEIDIGRRAL 97
G+D E S ++ E+L +K+ + E ++I DV PRGRY+IDI + ++
Sbjct 207 EGEDVQMAVEEKSMAEAFYEELKEKADIGEVAGDAIVSFQDVFFTTPRGRYDIDIYKNSI 266
Query 98 KFHGKSYDYTILYTNINRMFLVPRPNSPHVNFILSLDNAMRQGQTSYPFVVMQFDSENVT 157
+ GK+Y+Y + + I R+ +P+ + H +L+++ +RQGQT+YPF+V+QF + T
Sbjct 267 RLRGKTYEYKLQHRQIQRIVSLPKADDIHHLLVLAIEPPLRQGQTTYPFLVLQFQKDEET 326
Query 158 SVEVNLEEAELKEK---GLEKQLEGKTFDVITRILKSLIGKSVIVPGDFKSVKNQYGITC 214
V++NLE+ + +E L+KQ + KT V++ +LK L + VIVPG++KS +Q ++C
Sbjct 327 EVQLNLEDEDYEENYKDKLKKQYDAKTHIVLSHVLKGLTDRRVIVPGEYKSKYDQCAVSC 386
> ath:AT3G28730 ATHMG; ATHMG (ARABIDOPSIS THALIANA HIGH MOBILITY
GROUP); transcription factor; K09272 structure-specific recognition
protein 1
Length=646
Score = 123 bits (309), Expect = 4e-28, Method: Compositional matrix adjust.
Identities = 67/220 (30%), Positives = 119/220 (54%), Gaps = 10/220 (4%)
Query 4 DIVQVTTPSKNDLAIELVQDDTRDQTE-DMLLEIRLFQPAAG----DDETDSHLQSLKEK 58
D+ Q KND+ +E DDT E D L+EI P + DE Q +
Sbjct 135 DVSQTQLQGKNDVTLEFHVDDTAGANEKDSLMEISFHIPNSNTQFVGDENRPPSQVFNDT 194
Query 59 LVKKSGVAETKMESIALLNDVPLLVPRGRYEIDIGRRALKFHGKSYDYTILYTNINRMFL 118
+V + V+ +++ + +L PRGRY +++ L+ G++ D+ I Y+++ R+FL
Sbjct 195 IVAMADVSPGVEDAVVTFESIAILTPRGRYNVELHLSFLRLQGQANDFKIQYSSVVRLFL 254
Query 119 VPRPNSPHVNFILSLDNAMRQGQTSYPFVVMQFDSENVTSVEVNLEE----AELKEKGLE 174
+P+ N PH ++SLD +R+GQT YP +VMQF+++ V E+++ + + K+K LE
Sbjct 255 LPKSNQPHTFVVISLDPPIRKGQTMYPHIVMQFETDTVVESELSISDELMNTKFKDK-LE 313
Query 175 KQLEGKTFDVITRILKSLIGKSVIVPGDFKSVKNQYGITC 214
+ +G +V T +L+ L G + PG F+S ++ + +
Sbjct 314 RSYKGLIHEVFTTVLRWLSGAKITKPGKFRSSQDGFAVKS 353
> cel:T20B12.8 hmg-4; HMG family member (hmg-4); K09272 structure-specific
recognition protein 1
Length=697
Score = 119 bits (299), Expect = 6e-27, Method: Compositional matrix adjust.
Identities = 67/207 (32%), Positives = 113/207 (54%), Gaps = 8/207 (3%)
Query 12 SKNDLAIELVQ-DDTRDQTEDMLLEIRLFQPAAGDDETDS-HLQSLKEKLVKKSGVAETK 69
+KN+ +E Q DD++ Q L+E+R P ++E D+ ++ K+ ++ +G+
Sbjct 141 NKNEAVLEFHQNDDSKVQ----LMEMRFHMPIDLENEEDADKVEEFKKAVLAYAGLEAET 196
Query 70 MESIALLNDVPLLVPRGRYEIDIGRRALKFHGKSYDYTILYTNINRMFLVPRPNSPHVNF 129
+ I LL D+ PRGRY+I + ++ HGK+YDY I +INR+FLVP + HV F
Sbjct 197 EQPICLLTDILCTTPRGRYDIKVYPTSIALHGKTYDYKIPIKSINRLFLVPHKDGRHVYF 256
Query 130 ILSLDNAMRQGQTSYPFVVMQFDSENVTSVEVNL--EEAELKEKGLEKQLEGKTFDVITR 187
+LSL+ +RQGQT Y +++ +F + +E+ L E+ E L + + G ++ I+
Sbjct 257 VLSLNPPIRQGQTRYSYLIFEFGKDEEQDLELALTDEQLESSNGNLRRDMTGPIYETISI 316
Query 188 ILKSLIGKSVIVPGDFKSVKNQYGITC 214
+ KS+ + VPG F I C
Sbjct 317 LFKSICNLKITVPGRFLGSSGTPAIQC 343
> cel:C32F10.5 hmg-3; HMG family member (hmg-3); K09272 structure-specific
recognition protein 1
Length=689
Score = 117 bits (293), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 64/206 (31%), Positives = 114/206 (55%), Gaps = 6/206 (2%)
Query 12 SKNDLAIELVQDDTRDQTEDMLLEIRLFQPAAGDDETDS-HLQSLKEKLVKKSGVAETKM 70
+KN+ +E Q++ Q++ L+E+R P ++E D+ ++ K+ ++ +G+
Sbjct 141 NKNEAILEFHQNE---QSKVQLMEMRFHMPVDLENEEDTDKVEEFKKAVLAYAGLEAETE 197
Query 71 ESIALLNDVPLLVPRGRYEIDIGRRALKFHGKSYDYTILYTNINRMFLVPRPNSPHVNFI 130
+ I LL D+ PRGRY+I + ++ HGK+YDY I INR+FLVP + V F+
Sbjct 198 QPICLLTDILCTTPRGRYDIKVYPTSIALHGKTYDYKIPVKTINRLFLVPHKDGRQVYFV 257
Query 131 LSLDNAMRQGQTSYPFVVMQFDSENVTSVEVNLEEAELK--EKGLEKQLEGKTFDVITRI 188
LSL+ +RQGQT Y +++ +F + +E++L + +L L++++ G ++ I+ +
Sbjct 258 LSLNPPIRQGQTHYSYLIFEFGKDEEEDLELSLTDEQLDYFNGNLQREMTGPIYETISIL 317
Query 189 LKSLIGKSVIVPGDFKSVKNQYGITC 214
KS+ V VPG F I C
Sbjct 318 FKSICNLKVTVPGRFLGSSGTPAIQC 343
> ath:AT4G16660 heat shock protein 70, putative / HSP70, putative;
K09486 hypoxia up-regulated 1
Length=867
Score = 31.6 bits (70), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 14/46 (30%), Positives = 27/46 (58%), Gaps = 0/46 (0%)
Query 149 MQFDSENVTSVEVNLEEAELKEKGLEKQLEGKTFDVITRILKSLIG 194
+Q D+EN T+ EE + G EK+L+ +TF + ++++ +G
Sbjct 584 LQTDAENSTASNTTAEEPAVASLGTEKKLKKRTFRIPLKVVEKTVG 629
> cel:C27H6.2 ruvb-1; RUVB (recombination protein) homolog family
member (ruvb-1); K04499 RuvB-like protein 1 (pontin 52)
Length=458
Score = 30.0 bits (66), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 16/50 (32%), Positives = 27/50 (54%), Gaps = 6/50 (12%)
Query 149 MQFDSENVTSVEVNLEEAELKEKGLEKQLEGKTFDVITRILKSLIGKSVI 198
M+++ E++ + V+ EAE Q E K FD++TR+ G+ VI
Sbjct 382 MKYNEEDIRKILVHRTEAE------NVQFEEKAFDLLTRLCAQTCGREVI 425
> ath:AT3G55190 esterase/lipase/thioesterase family protein
Length=319
Score = 29.6 bits (65), Expect = 9.3, Method: Compositional matrix adjust.
Identities = 26/88 (29%), Positives = 43/88 (48%), Gaps = 6/88 (6%)
Query 73 IALLNDVPLLVPRGRYEI---DIGRRALKFHGKSYDYTILYTNINRMFLVPRPNSPHVNF 129
I+++N V L+P + I DI A+K K ++ + TN N PR + F
Sbjct 159 ISMINMVTNLIPSWKSIIHGPDILNSAIKLPEKRHE---IRTNPNCYNGWPRMKTMSELF 215
Query 130 ILSLDNAMRQGQTSYPFVVMQFDSENVT 157
+SLD R + + PF+V+ + + VT
Sbjct 216 RISLDLENRLNEVTMPFIVLHGEDDKVT 243
Lambda K H
0.316 0.134 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6816416920
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40