bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0341_orf1
Length=112
Score E
Sequences producing significant alignments: (Bits) Value
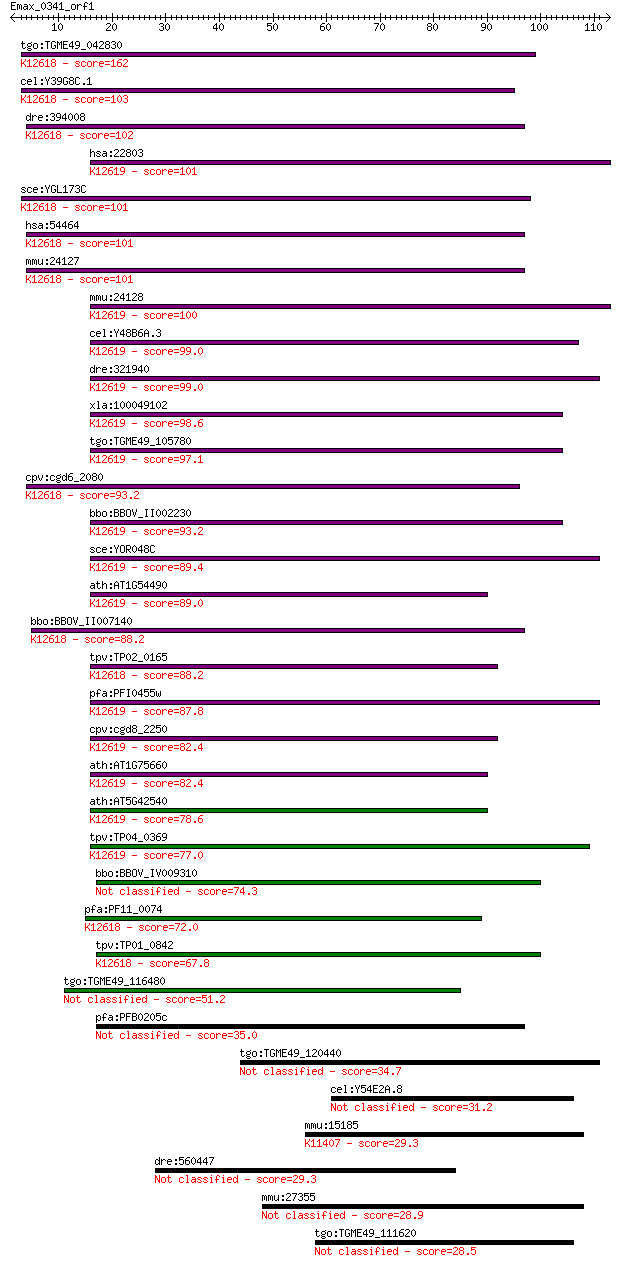
tgo:TGME49_042830 5'-3' exonuclease, putative ; K12618 5'-3' e... 162 3e-40
cel:Y39G8C.1 xrn-1; XRN (mouse/S. cerevisiae) ribonuclease rel... 103 2e-22
dre:394008 xrn1, MGC63635, wu:fk92c07, zgc:63635; 5'-3' exorib... 102 4e-22
hsa:22803 XRN2; 5'-3' exoribonuclease 2 (EC:3.1.13.-); K12619 ... 101 5e-22
sce:YGL173C KEM1, DST2, RAR5, SEP1, SKI1, XRN1; Evolutionarily... 101 5e-22
hsa:54464 XRN1, DKFZp434P0721, DKFZp686B22225, DKFZp686F19113,... 101 6e-22
mmu:24127 Xrn1, Dhm2, exo, mXrn1; 5'-3' exoribonuclease 1; K12... 101 6e-22
mmu:24128 Xrn2; 5'-3' exoribonuclease 2 (EC:3.1.13.-); K12619 ... 100 1e-21
cel:Y48B6A.3 xrn-2; XRN (mouse/S. cerevisiae) ribonuclease rel... 99.0 3e-21
dre:321940 xrn2, wu:fb38g12, wu:fj53d06, zgc:55717; 5'-3' exor... 99.0 3e-21
xla:100049102 xrn2; 5'-3' exoribonuclease 2; K12619 5'-3' exor... 98.6 4e-21
tgo:TGME49_105780 exoribonuclease, putative ; K12619 5'-3' exo... 97.1 1e-20
cpv:cgd6_2080 Kem1p-like 5'-3' exonuclease ; K12618 5'-3' exor... 93.2 2e-19
bbo:BBOV_II002230 18.m06180; XRN 5'-3' exonuclease protein; K1... 93.2 2e-19
sce:YOR048C RAT1, HKE1, TAP1, XRN2; Nuclear 5' to 3' single-st... 89.4 2e-18
ath:AT1G54490 XRN4; XRN4 (EXORIBONUCLEASE 4); 5'-3' exonucleas... 89.0 4e-18
bbo:BBOV_II007140 18.m06591; 5'-3' exoribonuclease (XRN2); K12... 88.2 6e-18
tpv:TP02_0165 hypothetical protein; K12618 5'-3' exoribonuclea... 88.2 6e-18
pfa:PFI0455w exoribonuclease, putative (EC:3.1.11.-); K12619 5... 87.8 7e-18
cpv:cgd8_2250 Rat1 Kar1/Rat1 like 5'-3' exonuclease ; K12619 5... 82.4 3e-16
ath:AT1G75660 XRN3; XRN3; 5'-3' exoribonuclease; K12619 5'-3' ... 82.4 3e-16
ath:AT5G42540 XRN2; XRN2 (EXORIBONUCLEASE 2); 5'-3' exonucleas... 78.6 4e-15
tpv:TP04_0369 exoribonuclease (EC:3.1.11.-); K12619 5'-3' exor... 77.0 1e-14
bbo:BBOV_IV009310 23.m06474; 5'-3' exonuclease (EC:3.1.11.-) 74.3 9e-14
pfa:PF11_0074 exonuclease, putative; K12618 5'-3' exoribonucle... 72.0 4e-13
tpv:TP01_0842 5'-3' exonuclease; K12618 5'-3' exoribonuclease ... 67.8 9e-12
tgo:TGME49_116480 XRN 5'-3' exonuclease N-terminus domain-cont... 51.2 9e-07
pfa:PFB0205c 5'-3' exonuclease, putative 35.0 0.064
tgo:TGME49_120440 hypothetical protein 34.7 0.074
cel:Y54E2A.8 hypothetical protein 31.2 0.96
mmu:15185 Hdac6, Hd6, Hdac5, Sfc6, mHDA2; histone deacetylase ... 29.3 3.1
dre:560447 deleted in malignant brain tumors 1-like 29.3 3.2
mmu:27355 MGC169319, MMPAL, Pald, mKIAA1274; cDNA sequence X99384 28.9 4.4
tgo:TGME49_111620 WD repeat domain-containing protein (EC:2.7.... 28.5 5.3
> tgo:TGME49_042830 5'-3' exonuclease, putative ; K12618 5'-3'
exoribonuclease 1 [EC:3.1.13.-]
Length=2042
Score = 162 bits (410), Expect = 3e-40, Method: Compositional matrix adjust.
Identities = 73/96 (76%), Positives = 83/96 (86%), Gaps = 0/96 (0%)
Query 3 VINEKIVASSIPTFDNLYLDLSGILHNCSHGNSGGMLHANEDLMWQSVFAALDLVISTIS 62
+INEKI +S+P FDNLYLD++GILH CSHGNSGGMLH +ED MW VFAALDL+IST++
Sbjct 16 LINEKITQTSLPEFDNLYLDMNGILHTCSHGNSGGMLHTSEDAMWVDVFAALDLIISTVN 75
Query 63 PRKLLYLAADGVAPRAKMNQQRARRYRAAKSAKEAA 98
P+K L LAADGVAPRAKMNQQRARRYRAAK A E A
Sbjct 76 PKKFLVLAADGVAPRAKMNQQRARRYRAAKDAAELA 111
> cel:Y39G8C.1 xrn-1; XRN (mouse/S. cerevisiae) ribonuclease related
family member (xrn-1); K12618 5'-3' exoribonuclease 1
[EC:3.1.13.-]
Length=1591
Score = 103 bits (256), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 46/93 (49%), Positives = 68/93 (73%), Gaps = 1/93 (1%)
Query 3 VINEKIVASSIPTFDNLYLDLSGILHNCSHGNSGGM-LHANEDLMWQSVFAALDLVISTI 61
++E I S IP FDNLYLD++GI+HNCSH N + ED ++ ++FA ++ + + I
Sbjct 16 CLSEVINESQIPEFDNLYLDMNGIIHNCSHPNDDDVTFRITEDEIFVNIFAYIENLYNLI 75
Query 62 SPRKLLYLAADGVAPRAKMNQQRARRYRAAKSA 94
P+K+ ++A DGVAPRAKMNQQRARR+ +A++A
Sbjct 76 RPQKVFFMAVDGVAPRAKMNQQRARRFMSARTA 108
> dre:394008 xrn1, MGC63635, wu:fk92c07, zgc:63635; 5'-3' exoribonuclease
1; K12618 5'-3' exoribonuclease 1 [EC:3.1.13.-]
Length=1697
Score = 102 bits (253), Expect = 4e-22, Method: Composition-based stats.
Identities = 45/94 (47%), Positives = 66/94 (70%), Gaps = 1/94 (1%)
Query 4 INEKIVASSIPTFDNLYLDLSGILHNCSHGNSGGM-LHANEDLMWQSVFAALDLVISTIS 62
++E + IP FDNLYLD++GI+H CSH N + +E+ ++ +F L+++ I
Sbjct 17 LSEVVKEHQIPEFDNLYLDMNGIIHQCSHPNDEDVHFRISEEKIFADIFHYLEVLFRIIK 76
Query 63 PRKLLYLAADGVAPRAKMNQQRARRYRAAKSAKE 96
PRK+ ++A DGVAPRAKMNQQR RR+R+AK A+E
Sbjct 77 PRKVFFMAVDGVAPRAKMNQQRGRRFRSAKEAEE 110
> hsa:22803 XRN2; 5'-3' exoribonuclease 2 (EC:3.1.13.-); K12619
5'-3' exoribonuclease 2 [EC:3.1.13.-]
Length=950
Score = 101 bits (252), Expect = 5e-22, Method: Composition-based stats.
Identities = 50/97 (51%), Positives = 66/97 (68%), Gaps = 4/97 (4%)
Query 16 FDNLYLDLSGILHNCSHGNSGGMLHANEDLMWQSVFAALDLVISTISPRKLLYLAADGVA 75
FDNLYLD++GI+H C+H NED M ++F +D + S + PR+LLY+A DGVA
Sbjct 48 FDNLYLDMNGIIHPCTHPEDKPA-PKNEDEMMVAIFEYIDRLFSIVRPRRLLYMAIDGVA 106
Query 76 PRAKMNQQRARRYRAAKSAKEAAEAQAKQQRAYTEWL 112
PRAKMNQQR+RR+RA +KE EA ++QR E L
Sbjct 107 PRAKMNQQRSRRFRA---SKEGMEAAVEKQRVREEIL 140
> sce:YGL173C KEM1, DST2, RAR5, SEP1, SKI1, XRN1; Evolutionarily-conserved
5'-3' exonuclease component of cytoplasmic processing
(P) bodies involved in mRNA decay; plays a role in microtubule-mediated
processes, filamentous growth, ribosomal
RNA maturation, and telomere maintenance (EC:3.1.11.-); K12618
5'-3' exoribonuclease 1 [EC:3.1.13.-]
Length=1528
Score = 101 bits (252), Expect = 5e-22, Method: Composition-based stats.
Identities = 47/96 (48%), Positives = 66/96 (68%), Gaps = 1/96 (1%)
Query 3 VINEKIVASSIPTFDNLYLDLSGILHNCSHGNSGGML-HANEDLMWQSVFAALDLVISTI 61
+I + I + IP FDNLYLD++ ILHNC+HGN + E+ ++ + +D + TI
Sbjct 16 MILQLIEGTQIPEFDNLYLDMNSILHNCTHGNDDDVTKRLTEEEVFAKICTYIDHLFQTI 75
Query 62 SPRKLLYLAADGVAPRAKMNQQRARRYRAAKSAKEA 97
P+K+ Y+A DGVAPRAKMNQQRARR+R A A++A
Sbjct 76 KPKKIFYMAIDGVAPRAKMNQQRARRFRTAMDAEKA 111
> hsa:54464 XRN1, DKFZp434P0721, DKFZp686B22225, DKFZp686F19113,
FLJ41903, SEP1; 5'-3' exoribonuclease 1; K12618 5'-3' exoribonuclease
1 [EC:3.1.13.-]
Length=1693
Score = 101 bits (251), Expect = 6e-22, Method: Composition-based stats.
Identities = 44/94 (46%), Positives = 66/94 (70%), Gaps = 1/94 (1%)
Query 4 INEKIVASSIPTFDNLYLDLSGILHNCSHGNSGGM-LHANEDLMWQSVFAALDLVISTIS 62
++E + IP FDNLYLD++GI+H CSH N + ++D ++ +F L+++ I
Sbjct 17 LSEVVKEHQIPEFDNLYLDMNGIIHQCSHPNDDDVHFRISDDKIFTDIFHYLEVLFRIIK 76
Query 63 PRKLLYLAADGVAPRAKMNQQRARRYRAAKSAKE 96
PRK+ ++A DGVAPRAKMNQQR RR+R+AK A++
Sbjct 77 PRKVFFMAVDGVAPRAKMNQQRGRRFRSAKEAED 110
> mmu:24127 Xrn1, Dhm2, exo, mXrn1; 5'-3' exoribonuclease 1; K12618
5'-3' exoribonuclease 1 [EC:3.1.13.-]
Length=1723
Score = 101 bits (251), Expect = 6e-22, Method: Composition-based stats.
Identities = 44/94 (46%), Positives = 66/94 (70%), Gaps = 1/94 (1%)
Query 4 INEKIVASSIPTFDNLYLDLSGILHNCSHGNSGGM-LHANEDLMWQSVFAALDLVISTIS 62
++E + IP FDNLYLD++GI+H CSH N + ++D ++ +F L+++ I
Sbjct 17 LSEVVKEHQIPEFDNLYLDMNGIIHQCSHPNDDDVHFRISDDKIFTDIFHYLEVLFRIIK 76
Query 63 PRKLLYLAADGVAPRAKMNQQRARRYRAAKSAKE 96
PRK+ ++A DGVAPRAKMNQQR RR+R+AK A++
Sbjct 77 PRKVFFMAVDGVAPRAKMNQQRGRRFRSAKEAED 110
> mmu:24128 Xrn2; 5'-3' exoribonuclease 2 (EC:3.1.13.-); K12619
5'-3' exoribonuclease 2 [EC:3.1.13.-]
Length=951
Score = 100 bits (249), Expect = 1e-21, Method: Composition-based stats.
Identities = 49/97 (50%), Positives = 66/97 (68%), Gaps = 4/97 (4%)
Query 16 FDNLYLDLSGILHNCSHGNSGGMLHANEDLMWQSVFAALDLVISTISPRKLLYLAADGVA 75
FDNLYLD++GI+H C+H NED M ++F +D + + + PR+LLY+A DGVA
Sbjct 48 FDNLYLDMNGIIHPCTHPEDKPA-PKNEDEMMVAIFEYIDRLFNIVRPRRLLYMAIDGVA 106
Query 76 PRAKMNQQRARRYRAAKSAKEAAEAQAKQQRAYTEWL 112
PRAKMNQQR+RR+RA +KE EA ++QR E L
Sbjct 107 PRAKMNQQRSRRFRA---SKEGMEAAVEKQRVREEIL 140
> cel:Y48B6A.3 xrn-2; XRN (mouse/S. cerevisiae) ribonuclease related
family member (xrn-2); K12619 5'-3' exoribonuclease 2
[EC:3.1.13.-]
Length=975
Score = 99.0 bits (245), Expect = 3e-21, Method: Composition-based stats.
Identities = 48/91 (52%), Positives = 64/91 (70%), Gaps = 2/91 (2%)
Query 16 FDNLYLDLSGILHNCSHGNSGGMLHANEDLMWQSVFAALDLVISTISPRKLLYLAADGVA 75
FDNLYLD++GI+H C+H NED M+ +F +D + S + PR+LLY+A DGVA
Sbjct 48 FDNLYLDMNGIIHPCTHPEDRPA-PKNEDEMFALIFEYIDRIYSIVRPRRLLYMAIDGVA 106
Query 76 PRAKMNQQRARRYRAAKSAKEAAEAQAKQQR 106
PRAKMNQQR+RR+RA+K E EA ++QR
Sbjct 107 PRAKMNQQRSRRFRASKEMAE-KEASIEEQR 136
> dre:321940 xrn2, wu:fb38g12, wu:fj53d06, zgc:55717; 5'-3' exoribonuclease
2 (EC:3.1.13.-); K12619 5'-3' exoribonuclease
2 [EC:3.1.13.-]
Length=952
Score = 99.0 bits (245), Expect = 3e-21, Method: Composition-based stats.
Identities = 46/95 (48%), Positives = 64/95 (67%), Gaps = 1/95 (1%)
Query 16 FDNLYLDLSGILHNCSHGNSGGMLHANEDLMWQSVFAALDLVISTISPRKLLYLAADGVA 75
FDNLYLD++GI+H C+H NED M ++F +D + + + PR++LY+A DGVA
Sbjct 48 FDNLYLDMNGIIHPCTHPEDKPA-PKNEDEMMVAIFEYIDRLFNIVRPRRVLYMAIDGVA 106
Query 76 PRAKMNQQRARRYRAAKSAKEAAEAQAKQQRAYTE 110
PRAKMNQQR+RR+RA+K E AE + K + E
Sbjct 107 PRAKMNQQRSRRFRASKEGVELAEEKQKMREEVIE 141
> xla:100049102 xrn2; 5'-3' exoribonuclease 2; K12619 5'-3' exoribonuclease
2 [EC:3.1.13.-]
Length=457
Score = 98.6 bits (244), Expect = 4e-21, Method: Composition-based stats.
Identities = 44/88 (50%), Positives = 62/88 (70%), Gaps = 1/88 (1%)
Query 16 FDNLYLDLSGILHNCSHGNSGGMLHANEDLMWQSVFAALDLVISTISPRKLLYLAADGVA 75
FDNLYLD++GI+H C+H NED M ++F +D + + + PR+LLY+A DGVA
Sbjct 48 FDNLYLDMNGIIHPCTHPEDKPA-PKNEDEMMVAIFEYIDRLFNIVRPRRLLYMAIDGVA 106
Query 76 PRAKMNQQRARRYRAAKSAKEAAEAQAK 103
PRAKMNQQR+RR+RA+K E+ E + +
Sbjct 107 PRAKMNQQRSRRFRASKEGVESVEEKNR 134
> tgo:TGME49_105780 exoribonuclease, putative ; K12619 5'-3' exoribonuclease
2 [EC:3.1.13.-]
Length=1239
Score = 97.1 bits (240), Expect = 1e-20, Method: Composition-based stats.
Identities = 46/88 (52%), Positives = 64/88 (72%), Gaps = 1/88 (1%)
Query 16 FDNLYLDLSGILHNCSHGNSGGMLHANEDLMWQSVFAALDLVISTISPRKLLYLAADGVA 75
+D LYLD++GI+H C H + G A E+ M+ S+F +D ++ I PR+LLYLA DGVA
Sbjct 70 YDCLYLDMNGIIHPCCHTDDGS-CPATEEEMFLSIFQYVDRIVDIIRPRQLLYLAIDGVA 128
Query 76 PRAKMNQQRARRYRAAKSAKEAAEAQAK 103
PRAKMNQQR+RR++AAK +E +A A+
Sbjct 129 PRAKMNQQRSRRFKAAKDIQEEEKAYAE 156
> cpv:cgd6_2080 Kem1p-like 5'-3' exonuclease ; K12618 5'-3' exoribonuclease
1 [EC:3.1.13.-]
Length=1588
Score = 93.2 bits (230), Expect = 2e-19, Method: Composition-based stats.
Identities = 44/99 (44%), Positives = 63/99 (63%), Gaps = 7/99 (7%)
Query 4 INEKIVASSIPTFDNLYLDLSGILHNC-------SHGNSGGMLHANEDLMWQSVFAALDL 56
INE+I IP FDNLYLD++GI HN + N + +W ++F ++
Sbjct 17 INEEISDGIIPPFDNLYLDVNGIAHNSVNSSEMRTPSNGINLSEKGSPEIWAAIFRYINK 76
Query 57 VISTISPRKLLYLAADGVAPRAKMNQQRARRYRAAKSAK 95
++ PRKLLY+A DGVAPRAKMNQQR+RR+R+A+ ++
Sbjct 77 LVYIAKPRKLLYIAVDGVAPRAKMNQQRSRRFRSARDSE 115
> bbo:BBOV_II002230 18.m06180; XRN 5'-3' exonuclease protein;
K12619 5'-3' exoribonuclease 2 [EC:3.1.13.-]
Length=988
Score = 93.2 bits (230), Expect = 2e-19, Method: Composition-based stats.
Identities = 44/88 (50%), Positives = 63/88 (71%), Gaps = 1/88 (1%)
Query 16 FDNLYLDLSGILHNCSHGNSGGMLHANEDLMWQSVFAALDLVISTISPRKLLYLAADGVA 75
FDNLY+D++G++H C H G +E++M+Q +F LD ++ I PRK+L+LA DGVA
Sbjct 51 FDNLYIDMNGLIHPCCH-PEGLEQPPSEEVMFQCIFDYLDRLMYIIRPRKILFLAIDGVA 109
Query 76 PRAKMNQQRARRYRAAKSAKEAAEAQAK 103
PRAKMNQQR+RR+++A A AE +K
Sbjct 110 PRAKMNQQRSRRFKSAAEADLEAEIYSK 137
> sce:YOR048C RAT1, HKE1, TAP1, XRN2; Nuclear 5' to 3' single-stranded
RNA exonuclease, involved in RNA metabolism, including
rRNA and snRNA processing as well as poly (A+) dependent
and independent mRNA transcription termination (EC:3.1.13.-);
K12619 5'-3' exoribonuclease 2 [EC:3.1.13.-]
Length=1006
Score = 89.4 bits (220), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 46/95 (48%), Positives = 62/95 (65%), Gaps = 1/95 (1%)
Query 16 FDNLYLDLSGILHNCSHGNSGGMLHANEDLMWQSVFAALDLVISTISPRKLLYLAADGVA 75
DNLYLD++GI+H CSH + ED M +VF + V++ PRK+L +A DGVA
Sbjct 47 LDNLYLDMNGIVHPCSHPENKPPPET-EDEMLLAVFEYTNRVLNMARPRKVLVMAVDGVA 105
Query 76 PRAKMNQQRARRYRAAKSAKEAAEAQAKQQRAYTE 110
PRAKMNQQRARR+R+A+ A+ EA+ + R E
Sbjct 106 PRAKMNQQRARRFRSARDAQIENEAREEIMRQREE 140
> ath:AT1G54490 XRN4; XRN4 (EXORIBONUCLEASE 4); 5'-3' exonuclease/
5'-3' exoribonuclease/ nucleic acid binding; K12619 5'-3'
exoribonuclease 2 [EC:3.1.13.-]
Length=947
Score = 89.0 bits (219), Expect = 4e-18, Method: Composition-based stats.
Identities = 41/74 (55%), Positives = 55/74 (74%), Gaps = 1/74 (1%)
Query 16 FDNLYLDLSGILHNCSHGNSGGMLHANEDLMWQSVFAALDLVISTISPRKLLYLAADGVA 75
FDNLYLD++GI+H C H G A D +++S+F +D + + + PRK+LYLA DGVA
Sbjct 49 FDNLYLDMNGIIHPCFH-PEGKPAPATYDDVFKSMFEYIDHLFTLVRPRKILYLAIDGVA 107
Query 76 PRAKMNQQRARRYR 89
PRAKMNQQR+RR+R
Sbjct 108 PRAKMNQQRSRRFR 121
> bbo:BBOV_II007140 18.m06591; 5'-3' exoribonuclease (XRN2); K12618
5'-3' exoribonuclease 1 [EC:3.1.13.-]
Length=1388
Score = 88.2 bits (217), Expect = 6e-18, Method: Composition-based stats.
Identities = 44/95 (46%), Positives = 65/95 (68%), Gaps = 4/95 (4%)
Query 5 NEKIVASSIPT-FDNLYLDLSGILHNCSHG--NSGGMLHANEDLMWQSVFAALDLVISTI 61
+++ SS+P FDNLYLD++GI+HNCSH L ED+ + +F ++ ++ +
Sbjct 42 DDQYETSSVPDGFDNLYLDVNGIIHNCSHSIEELSQGLRCEEDI-FVLIFQYINNLVKIV 100
Query 62 SPRKLLYLAADGVAPRAKMNQQRARRYRAAKSAKE 96
PRKLLYLA DGVAPRAK+ QQR RR+R+A+ + +
Sbjct 101 RPRKLLYLAIDGVAPRAKIMQQRDRRFRSARDSNK 135
> tpv:TP02_0165 hypothetical protein; K12618 5'-3' exoribonuclease
1 [EC:3.1.13.-]
Length=1726
Score = 88.2 bits (217), Expect = 6e-18, Method: Composition-based stats.
Identities = 45/79 (56%), Positives = 58/79 (73%), Gaps = 5/79 (6%)
Query 16 FDNLYLDLSGILHNCSHGNSGGMLHAN---EDLMWQSVFAALDLVISTISPRKLLYLAAD 72
FDNLYLD++GI+HNCSH S L N ED ++ S+F + +++ + PRKLLYLA D
Sbjct 104 FDNLYLDVNGIVHNCSH--SVVDLCQNITCEDDIFVSIFQCIIKIVNIVRPRKLLYLAVD 161
Query 73 GVAPRAKMNQQRARRYRAA 91
GVAPRAK+ QQR RR+RAA
Sbjct 162 GVAPRAKIIQQRERRFRAA 180
> pfa:PFI0455w exoribonuclease, putative (EC:3.1.11.-); K12619
5'-3' exoribonuclease 2 [EC:3.1.13.-]
Length=1311
Score = 87.8 bits (216), Expect = 7e-18, Method: Composition-based stats.
Identities = 41/95 (43%), Positives = 66/95 (69%), Gaps = 8/95 (8%)
Query 16 FDNLYLDLSGILHNCSHGNSGGMLHANEDLMWQSVFAALDLVISTISPRKLLYLAADGVA 75
FDN+YLD++GI+H CSH ++ +NE++ + +VF ++ + I P+KLLY+A DGVA
Sbjct 68 FDNMYLDMNGIIHLCSHSDNSKRAKSNEEI-FLNVFLYVERLFDIIEPKKLLYMAIDGVA 126
Query 76 PRAKMNQQRARRYRAAKSAKEAAEAQAKQQRAYTE 110
P+AKMNQQR+RR+++ ++ ++RAY E
Sbjct 127 PKAKMNQQRSRRFKSISCSE-------IEKRAYLE 154
> cpv:cgd8_2250 Rat1 Kar1/Rat1 like 5'-3' exonuclease ; K12619
5'-3' exoribonuclease 2 [EC:3.1.13.-]
Length=1350
Score = 82.4 bits (202), Expect = 3e-16, Method: Composition-based stats.
Identities = 39/76 (51%), Positives = 54/76 (71%), Gaps = 1/76 (1%)
Query 16 FDNLYLDLSGILHNCSHGNSGGMLHANEDLMWQSVFAALDLVISTISPRKLLYLAADGVA 75
FD LYLD++GI+H C + SGG +E M+ V +D + + + PR+L+Y+A DGVA
Sbjct 45 FDCLYLDMNGIIHPCCN-PSGGDKPKDEAEMFTRVCDYIDRLYAMVKPRRLIYMAIDGVA 103
Query 76 PRAKMNQQRARRYRAA 91
PRAKMNQQR+RRY+ A
Sbjct 104 PRAKMNQQRSRRYKTA 119
> ath:AT1G75660 XRN3; XRN3; 5'-3' exoribonuclease; K12619 5'-3'
exoribonuclease 2 [EC:3.1.13.-]
Length=1020
Score = 82.4 bits (202), Expect = 3e-16, Method: Composition-based stats.
Identities = 38/74 (51%), Positives = 52/74 (70%), Gaps = 1/74 (1%)
Query 16 FDNLYLDLSGILHNCSHGNSGGMLHANEDLMWQSVFAALDLVISTISPRKLLYLAADGVA 75
+DNLYLD++GI+H C H E++ +Q +F +D + + PRKLLY+A DGVA
Sbjct 48 YDNLYLDMNGIIHPCFHPEDRPSPTTFEEV-FQCMFDYIDRLFVMVRPRKLLYMAIDGVA 106
Query 76 PRAKMNQQRARRYR 89
PRAKMNQQR+RR+R
Sbjct 107 PRAKMNQQRSRRFR 120
> ath:AT5G42540 XRN2; XRN2 (EXORIBONUCLEASE 2); 5'-3' exonuclease/
5'-3' exoribonuclease/ nucleic acid binding; K12619 5'-3'
exoribonuclease 2 [EC:3.1.13.-]
Length=1012
Score = 78.6 bits (192), Expect = 4e-15, Method: Composition-based stats.
Identities = 37/74 (50%), Positives = 51/74 (68%), Gaps = 1/74 (1%)
Query 16 FDNLYLDLSGILHNCSHGNSGGMLHANEDLMWQSVFAALDLVISTISPRKLLYLAADGVA 75
+DNLYLD++GI+H C H ++ +Q +F +D + + PRKLL++A DGVA
Sbjct 50 YDNLYLDMNGIIHPCFHPEDKPSPTTFTEV-FQCMFDYIDRLFVMVRPRKLLFMAIDGVA 108
Query 76 PRAKMNQQRARRYR 89
PRAKMNQQRARR+R
Sbjct 109 PRAKMNQQRARRFR 122
> tpv:TP04_0369 exoribonuclease (EC:3.1.11.-); K12619 5'-3' exoribonuclease
2 [EC:3.1.13.-]
Length=958
Score = 77.0 bits (188), Expect = 1e-14, Method: Composition-based stats.
Identities = 42/104 (40%), Positives = 59/104 (56%), Gaps = 12/104 (11%)
Query 16 FDNLYLDLSGILHNCSHGNSGGMLHANEDLMWQSVFAALDLVISTISPRKLLYLAA---- 71
FDNLYLD++GI+H C H + + E++M+ + LD + + PRKL+YLA
Sbjct 51 FDNLYLDMNGIIHPCCHPENMEQPQS-EEVMFTCILDYLDRIFFMVRPRKLIYLAIGNIK 109
Query 72 -------DGVAPRAKMNQQRARRYRAAKSAKEAAEAQAKQQRAY 108
DGVAPRAK+NQQR+RR+++A A E R Y
Sbjct 110 NYIMICIDGVAPRAKINQQRSRRFKSAALADLEDETYDTLIRDY 153
> bbo:BBOV_IV009310 23.m06474; 5'-3' exonuclease (EC:3.1.11.-)
Length=509
Score = 74.3 bits (181), Expect = 9e-14, Method: Composition-based stats.
Identities = 29/83 (34%), Positives = 54/83 (65%), Gaps = 0/83 (0%)
Query 17 DNLYLDLSGILHNCSHGNSGGMLHANEDLMWQSVFAALDLVISTISPRKLLYLAADGVAP 76
D+LY+D++ ++H +HGN ++ + + + +A++++ + PRK+LY+A DGV P
Sbjct 26 DHLYMDMNAVIHVATHGNISPVVQMANEQRLRRITSAIEMMFDIVRPRKMLYMAVDGVCP 85
Query 77 RAKMNQQRARRYRAAKSAKEAAE 99
AK+NQQR RR+ +K+ A+
Sbjct 86 TAKINQQRGRRFLTSKNVDPLAD 108
> pfa:PF11_0074 exonuclease, putative; K12618 5'-3' exoribonuclease
1 [EC:3.1.13.-]
Length=734
Score = 72.0 bits (175), Expect = 4e-13, Method: Composition-based stats.
Identities = 28/74 (37%), Positives = 49/74 (66%), Gaps = 0/74 (0%)
Query 15 TFDNLYLDLSGILHNCSHGNSGGMLHANEDLMWQSVFAALDLVISTISPRKLLYLAADGV 74
+ D Y+D++G++H+C+H N + +E ++ ++ L + I P+KL+Y+ DGV
Sbjct 73 SVDIFYIDMNGVIHHCTHANKEKLPIYDEHELFSNILQYLKNLFYLIKPKKLIYIGVDGV 132
Query 75 APRAKMNQQRARRY 88
+P+AKMNQQR RR+
Sbjct 133 SPKAKMNQQRKRRF 146
> tpv:TP01_0842 5'-3' exonuclease; K12618 5'-3' exoribonuclease
1 [EC:3.1.13.-]
Length=534
Score = 67.8 bits (164), Expect = 9e-12, Method: Composition-based stats.
Identities = 28/83 (33%), Positives = 49/83 (59%), Gaps = 0/83 (0%)
Query 17 DNLYLDLSGILHNCSHGNSGGMLHANEDLMWQSVFAALDLVISTISPRKLLYLAADGVAP 76
D Y+D++ ++H+ +HGN +L + + + A+ ++P+K++Y+ DGV P
Sbjct 66 DYFYVDMNAVIHSATHGNLFPVLMMEDQQRMRRIVTAMLNTFKLVNPKKMMYIGVDGVCP 125
Query 77 RAKMNQQRARRYRAAKSAKEAAE 99
AK+NQQR RR+R KS+ E
Sbjct 126 SAKINQQRTRRFRLYKSSSSDNE 148
> tgo:TGME49_116480 XRN 5'-3' exonuclease N-terminus domain-containing
protein (EC:3.2.1.3)
Length=2089
Score = 51.2 bits (121), Expect = 9e-07, Method: Composition-based stats.
Identities = 35/79 (44%), Positives = 48/79 (60%), Gaps = 7/79 (8%)
Query 11 SSIPTFDNLYLDLSGILHNCSHGNSGG-----MLHANEDLMWQSVFAALDLVISTISPRK 65
S +P D L+LD + I+H CSHG+ ++H + L+ Q V A L ++S + PRK
Sbjct 651 SRLP-IDALFLDFNAIIHLCSHGHLPSTLPPPLMHC-QPLLLQRVCAYLHRLVSLVRPRK 708
Query 66 LLYLAADGVAPRAKMNQQR 84
LL L DGV P AK+NQQR
Sbjct 709 LLVLTLDGVPPLAKVNQQR 727
> pfa:PFB0205c 5'-3' exonuclease, putative
Length=1188
Score = 35.0 bits (79), Expect = 0.064, Method: Composition-based stats.
Identities = 22/80 (27%), Positives = 37/80 (46%), Gaps = 6/80 (7%)
Query 17 DNLYLDLSGILHNCSHGNSGGMLHANEDLMWQSVFAALDLVISTISPRKLLYLAADGVAP 76
DNL DL+ +LH + + N D + + + V+ P+K + A DG+ P
Sbjct 144 DNLLFDLNQLLHKAN------VKFINYDNYFLKLTRLIKNVLKKFEPKKNVVFAIDGICP 197
Query 77 RAKMNQQRARRYRAAKSAKE 96
+K+ Q RR ++ KE
Sbjct 198 FSKLKLQIKRRAKSKLKNKE 217
> tgo:TGME49_120440 hypothetical protein
Length=2256
Score = 34.7 bits (78), Expect = 0.074, Method: Composition-based stats.
Identities = 21/67 (31%), Positives = 35/67 (52%), Gaps = 1/67 (1%)
Query 44 DLMWQSVFAALDLVISTISPRKLLYLAADGVAPRAKMNQQRARRYRAAKSAKEAAEAQAK 103
D + + + + + + TI PRK + A DGV P AK+ A+R R +EAA+ ++
Sbjct 156 DAVLRRLVSLISATLRTIHPRKSVVFALDGVPPLAKL-LTTAKRRRKRDDEEEAADRPSE 214
Query 104 QQRAYTE 110
+ A E
Sbjct 215 SEEAEGE 221
> cel:Y54E2A.8 hypothetical protein
Length=485
Score = 31.2 bits (69), Expect = 0.96, Method: Composition-based stats.
Identities = 16/48 (33%), Positives = 25/48 (52%), Gaps = 3/48 (6%)
Query 61 ISPRKLLYLAADGVAPRAKMNQQRARRYRAAKSAKE---AAEAQAKQQ 105
I PRK++ + P+AKM ++R R + K +E A E K+Q
Sbjct 227 IPPRKIIEKPVEKAPPKAKMTRKRMMRAKHEKRLREEQIALEESKKEQ 274
> mmu:15185 Hdac6, Hd6, Hdac5, Sfc6, mHDA2; histone deacetylase
6 (EC:3.5.1.98); K11407 histone deacetylase 6/10 [EC:3.5.1.98]
Length=1149
Score = 29.3 bits (64), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 16/52 (30%), Positives = 25/52 (48%), Gaps = 0/52 (0%)
Query 56 LVISTISPRKLLYLAADGVAPRAKMNQQRARRYRAAKSAKEAAEAQAKQQRA 107
LVI + P A + P+ K+ ++ R+ AA KE+ QAK + A
Sbjct 849 LVIKKLPPTASPVSAKEMTTPKGKVPEESVRKTIAALPGKESTLGQAKSKMA 900
> dre:560447 deleted in malignant brain tumors 1-like
Length=901
Score = 29.3 bits (64), Expect = 3.2, Method: Composition-based stats.
Identities = 16/56 (28%), Positives = 26/56 (46%), Gaps = 3/56 (5%)
Query 28 HNCSHGNSGGMLHANEDLMWQSVFAALDLVISTISPRKLLYLAADGVAPRAKMNQQ 83
HNC+HG G++ ++E Q AL S +SP +++ PR N +
Sbjct 558 HNCNHGEDAGVVCSSE---LQMASLALISTHSAVSPGEIVQFRCSTPKPRCSANAE 610
> mmu:27355 MGC169319, MMPAL, Pald, mKIAA1274; cDNA sequence X99384
Length=859
Score = 28.9 bits (63), Expect = 4.4, Method: Composition-based stats.
Identities = 20/60 (33%), Positives = 28/60 (46%), Gaps = 12/60 (20%)
Query 48 QSVFAALDLVISTISPRKLLYLAADGVAPRAKMNQQRARRYRAAKSAKEAAEAQAKQQRA 107
+ V AALD+V T++P + + YR AK+ KEA EAQ Q R+
Sbjct 727 KEVDAALDIVSETMTPMHY------------HLREIIISTYRQAKATKEAQEAQRLQLRS 774
> tgo:TGME49_111620 WD repeat domain-containing protein (EC:2.7.11.7)
Length=3704
Score = 28.5 bits (62), Expect = 5.3, Method: Composition-based stats.
Identities = 15/48 (31%), Positives = 25/48 (52%), Gaps = 0/48 (0%)
Query 58 ISTISPRKLLYLAADGVAPRAKMNQQRARRYRAAKSAKEAAEAQAKQQ 105
+ T+S R L G+ PR +++++ R R ++AEA A QQ
Sbjct 2965 VHTLSYRLEEELVRKGLLPRGPLHEKKGRPARGKGLGVDSAEADATQQ 3012
Lambda K H
0.319 0.129 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2057481480
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40