bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0324_orf3
Length=265
Score E
Sequences producing significant alignments: (Bits) Value
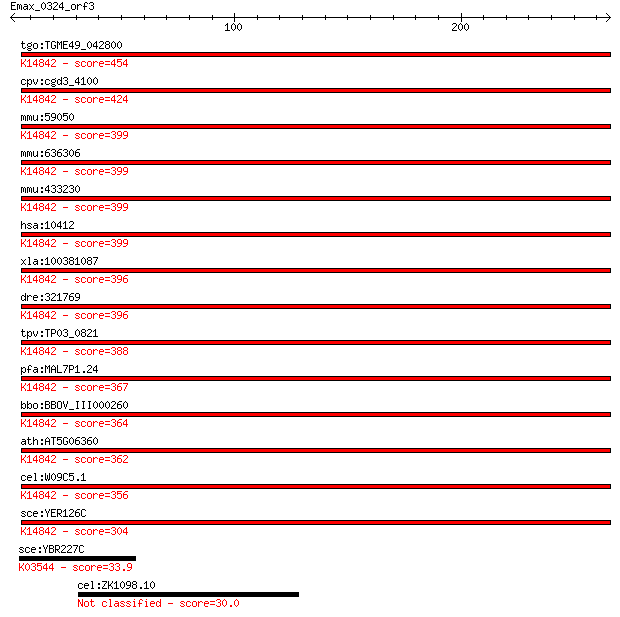
tgo:TGME49_042800 TGF-beta-inducible nuclear protein 1, putati... 454 2e-127
cpv:cgd3_4100 conserved protein, COG SSU ribosomal protein S8E... 424 1e-118
mmu:59050 Nsa2, 5730427N09Rik, LNR42, MGC118054, Nsa2p, Tinp1,... 399 6e-111
mmu:636306 ribosome biogenesis protein NSA2 homolog; K14842 ri... 399 6e-111
mmu:433230 Gm5515, EG433230; predicted gene 5515; K14842 ribos... 399 6e-111
hsa:10412 NSA2, CDK105, HCL-G1, HCLG1, HUSSY29, TINP1, YR-29; ... 399 7e-111
xla:100381087 nsa2, cdk105, hcl-g1, hclg1, hussy-29, hussy29, ... 396 4e-110
dre:321769 nsa2, fb34b10, wu:fb34b10, zgc:56334; NSA2 ribosome... 396 5e-110
tpv:TP03_0821 hypothetical protein; K14842 ribosome biogenesis... 388 1e-107
pfa:MAL7P1.24 cdk105, putative; K14842 ribosome biogenesis pro... 367 2e-101
bbo:BBOV_III000260 17.m07048; ribosomal protein Se8; K14842 ri... 364 2e-100
ath:AT5G06360 ribosomal protein S8e family protein; K14842 rib... 362 1e-99
cel:W09C5.1 hypothetical protein; K14842 ribosome biogenesis p... 356 5e-98
sce:YER126C NSA2; Nsa2p; K14842 ribosome biogenesis protein NSA2 304 3e-82
sce:YBR227C MCX1; Mcx1p; K03544 ATP-dependent Clp protease ATP... 33.9 0.67
cel:ZK1098.10 unc-16; UNCoordinated family member (unc-16) 30.0 9.4
> tgo:TGME49_042800 TGF-beta-inducible nuclear protein 1, putative
; K14842 ribosome biogenesis protein NSA2
Length=260
Score = 454 bits (1167), Expect = 2e-127, Method: Compositional matrix adjust.
Identities = 217/260 (83%), Positives = 237/260 (91%), Gaps = 0/260 (0%)
Query 6 MPQNEYMEQHRKRFGERLDTAERKRKKAARMPHERSKTARKLRGIKAKLFNRRRFVEKAQ 65
MPQNEY+E H+KRFG RLD ER+RKKAAR PH+ SK AR+LRGIK+KLFN++R+VEKA
Sbjct 1 MPQNEYIELHQKRFGRRLDHHERQRKKAAREPHKLSKQARRLRGIKSKLFNKKRYVEKAT 60
Query 66 MKKTIKQHEEKEGKQKEPETVPEGAVPSYLLDREGVSRTKILTNMIKQKRKEKAGKWQVP 125
MKKT+K EEK+ K EPE P+GAVP+YLLDRE V RTK+L+NM+KQKRKEKAGKWQVP
Sbjct 61 MKKTLKHFEEKDAKSAEPEQAPDGAVPAYLLDREAVKRTKVLSNMVKQKRKEKAGKWQVP 120
Query 126 IPKVKAMTEEEMFKVLRSGKRKKKMWKRVVNKVCFVGDDFTRKPPKFERYIRPTSLRFKK 185
+PKVKAMTE+EMFKVLRSGKR+KK WKRVVNKVCFVG+DFTRKPPKFERYIRPT LRFKK
Sbjct 121 LPKVKAMTEDEMFKVLRSGKRRKKAWKRVVNKVCFVGEDFTRKPPKFERYIRPTGLRFKK 180
Query 186 AHVTHPELKATFQLDIVKVKKNPQSQLYTSLGVITKGTIIEVNVSELGLVTQTGKVVWAK 245
AHVTHPELK TF LDIV VKKNPQS LYTSLGVITKGTIIEVNVSELGLVTQTGKVVWAK
Sbjct 181 AHVTHPELKTTFHLDIVGVKKNPQSHLYTSLGVITKGTIIEVNVSELGLVTQTGKVVWAK 240
Query 246 YAQVTNNPELDGCINAVLLV 265
YAQVTNNPELDGCINAVLLV
Sbjct 241 YAQVTNNPELDGCINAVLLV 260
> cpv:cgd3_4100 conserved protein, COG SSU ribosomal protein S8E
; K14842 ribosome biogenesis protein NSA2
Length=260
Score = 424 bits (1090), Expect = 1e-118, Method: Compositional matrix adjust.
Identities = 201/260 (77%), Positives = 232/260 (89%), Gaps = 0/260 (0%)
Query 6 MPQNEYMEQHRKRFGERLDTAERKRKKAARMPHERSKTARKLRGIKAKLFNRRRFVEKAQ 65
MPQNEY+E HRKR G R D E+KRK +R H+ S+ A+KLRGIKAKL+N++R+ EKA
Sbjct 1 MPQNEYIELHRKRHGYRFDHFEKKRKFESRKVHKDSQHAQKLRGIKAKLYNKKRYAEKAT 60
Query 66 MKKTIKQHEEKEGKQKEPETVPEGAVPSYLLDREGVSRTKILTNMIKQKRKEKAGKWQVP 125
++K +K +EEK+ +K+PE V +GAVPSYLLDREGV+RTKIL+NMIKQKRKEK+ KWQVP
Sbjct 61 LRKLVKANEEKDAVEKDPENVQKGAVPSYLLDREGVNRTKILSNMIKQKRKEKSAKWQVP 120
Query 126 IPKVKAMTEEEMFKVLRSGKRKKKMWKRVVNKVCFVGDDFTRKPPKFERYIRPTSLRFKK 185
IPK+KA+ E+EMFK+LR+GKR+KK WKRVVNKVCFVG+DFTRKPPK ERYIRPT LRFKK
Sbjct 121 IPKIKALNEDEMFKILRTGKRQKKSWKRVVNKVCFVGNDFTRKPPKLERYIRPTGLRFKK 180
Query 186 AHVTHPELKATFQLDIVKVKKNPQSQLYTSLGVITKGTIIEVNVSELGLVTQTGKVVWAK 245
AHVTHPELK TF LDIV +KKNPQSQLYT+LGVITKGTIIEVNVSELGLVTQTGKVVWAK
Sbjct 181 AHVTHPELKTTFYLDIVSIKKNPQSQLYTTLGVITKGTIIEVNVSELGLVTQTGKVVWAK 240
Query 246 YAQVTNNPELDGCINAVLLV 265
YAQVTNNPE+DGCINAVLLV
Sbjct 241 YAQVTNNPEIDGCINAVLLV 260
> mmu:59050 Nsa2, 5730427N09Rik, LNR42, MGC118054, Nsa2p, Tinp1,
Yr29; NSA2 ribosome biogenesis homolog (S. cerevisiae); K14842
ribosome biogenesis protein NSA2
Length=260
Score = 399 bits (1025), Expect = 6e-111, Method: Compositional matrix adjust.
Identities = 192/260 (73%), Positives = 222/260 (85%), Gaps = 0/260 (0%)
Query 6 MPQNEYMEQHRKRFGERLDTAERKRKKAARMPHERSKTARKLRGIKAKLFNRRRFVEKAQ 65
MPQNEY+E HRKR+G RLD E+KRKK R HERSK A+K+ G+KAKL++++R EK Q
Sbjct 1 MPQNEYIELHRKRYGYRLDYHEKKRKKEGREAHERSKKAKKMIGLKAKLYHKQRHAEKIQ 60
Query 66 MKKTIKQHEEKEGKQKEPETVPEGAVPSYLLDREGVSRTKILTNMIKQKRKEKAGKWQVP 125
MKKTIK HE++ KQK+ E P+GAVP+YLLDREG SR K+L+NMIKQKRKEKAGKW+VP
Sbjct 61 MKKTIKMHEKRNTKQKDDEKTPQGAVPAYLLDREGQSRAKVLSNMIKQKRKEKAGKWEVP 120
Query 126 IPKVKAMTEEEMFKVLRSGKRKKKMWKRVVNKVCFVGDDFTRKPPKFERYIRPTSLRFKK 185
+PKV+A E E+ KV+R+GKRKKK WKR+V KVCFVGD FTRKPPK+ER+IRP LRFKK
Sbjct 121 LPKVRAQGETEVLKVIRTGKRKKKAWKRMVTKVCFVGDGFTRKPPKYERFIRPMGLRFKK 180
Query 186 AHVTHPELKATFQLDIVKVKKNPQSQLYTSLGVITKGTIIEVNVSELGLVTQTGKVVWAK 245
AHVTHPELKATF L I+ VKKNP S LYT+LGVITKGT+IEVNVSELGLVTQ GKV+W K
Sbjct 181 AHVTHPELKATFCLPILGVKKNPSSPLYTTLGVITKGTVIEVNVSELGLVTQGGKVIWGK 240
Query 246 YAQVTNNPELDGCINAVLLV 265
YAQVTNNPE DGCINAVLLV
Sbjct 241 YAQVTNNPENDGCINAVLLV 260
> mmu:636306 ribosome biogenesis protein NSA2 homolog; K14842
ribosome biogenesis protein NSA2
Length=260
Score = 399 bits (1025), Expect = 6e-111, Method: Compositional matrix adjust.
Identities = 192/260 (73%), Positives = 222/260 (85%), Gaps = 0/260 (0%)
Query 6 MPQNEYMEQHRKRFGERLDTAERKRKKAARMPHERSKTARKLRGIKAKLFNRRRFVEKAQ 65
MPQNEY+E HRKR+G RLD E+KRKK R HERSK A+K+ G+KAKL++++R EK Q
Sbjct 1 MPQNEYIELHRKRYGYRLDYHEKKRKKEGREAHERSKKAKKMIGLKAKLYHKQRHAEKIQ 60
Query 66 MKKTIKQHEEKEGKQKEPETVPEGAVPSYLLDREGVSRTKILTNMIKQKRKEKAGKWQVP 125
MKKTIK HE++ KQK+ E P+GAVP+YLLDREG SR K+L+NMIKQKRKEKAGKW+VP
Sbjct 61 MKKTIKMHEKRNTKQKDDEKTPQGAVPAYLLDREGQSRAKVLSNMIKQKRKEKAGKWEVP 120
Query 126 IPKVKAMTEEEMFKVLRSGKRKKKMWKRVVNKVCFVGDDFTRKPPKFERYIRPTSLRFKK 185
+PKV+A E E+ KV+R+GKRKKK WKR+V KVCFVGD FTRKPPK+ER+IRP LRFKK
Sbjct 121 LPKVRAQGETEVLKVIRTGKRKKKAWKRMVTKVCFVGDGFTRKPPKYERFIRPMGLRFKK 180
Query 186 AHVTHPELKATFQLDIVKVKKNPQSQLYTSLGVITKGTIIEVNVSELGLVTQTGKVVWAK 245
AHVTHPELKATF L I+ VKKNP S LYT+LGVITKGT+IEVNVSELGLVTQ GKV+W K
Sbjct 181 AHVTHPELKATFCLPILGVKKNPSSPLYTTLGVITKGTVIEVNVSELGLVTQGGKVIWGK 240
Query 246 YAQVTNNPELDGCINAVLLV 265
YAQVTNNPE DGCINAVLLV
Sbjct 241 YAQVTNNPENDGCINAVLLV 260
> mmu:433230 Gm5515, EG433230; predicted gene 5515; K14842 ribosome
biogenesis protein NSA2
Length=260
Score = 399 bits (1025), Expect = 6e-111, Method: Compositional matrix adjust.
Identities = 192/260 (73%), Positives = 222/260 (85%), Gaps = 0/260 (0%)
Query 6 MPQNEYMEQHRKRFGERLDTAERKRKKAARMPHERSKTARKLRGIKAKLFNRRRFVEKAQ 65
MPQNEY+E HRKR+G RLD E+KRKK R HERSK A+K+ G+KAKL++++R EK Q
Sbjct 1 MPQNEYIELHRKRYGYRLDYHEKKRKKEGREAHERSKKAKKMIGLKAKLYHKQRHAEKIQ 60
Query 66 MKKTIKQHEEKEGKQKEPETVPEGAVPSYLLDREGVSRTKILTNMIKQKRKEKAGKWQVP 125
MKKTIK HE++ KQK+ E P+GAVP+YLLDREG SR K+L+NMIKQKRKEKAGKW+VP
Sbjct 61 MKKTIKMHEKRNTKQKDDEKTPQGAVPAYLLDREGQSRAKVLSNMIKQKRKEKAGKWEVP 120
Query 126 IPKVKAMTEEEMFKVLRSGKRKKKMWKRVVNKVCFVGDDFTRKPPKFERYIRPTSLRFKK 185
+PKV+A E E+ KV+R+GKRKKK WKR+V KVCFVGD FTRKPPK+ER+IRP LRFKK
Sbjct 121 LPKVRAQGETEVLKVIRTGKRKKKAWKRMVTKVCFVGDGFTRKPPKYERFIRPMGLRFKK 180
Query 186 AHVTHPELKATFQLDIVKVKKNPQSQLYTSLGVITKGTIIEVNVSELGLVTQTGKVVWAK 245
AHVTHPELKATF L I+ VKKNP S LYT+LGVITKGT+IEVNVSELGLVTQ GKV+W K
Sbjct 181 AHVTHPELKATFCLPILGVKKNPSSPLYTTLGVITKGTVIEVNVSELGLVTQGGKVIWGK 240
Query 246 YAQVTNNPELDGCINAVLLV 265
YAQVTNNPE DGCINAVLLV
Sbjct 241 YAQVTNNPENDGCINAVLLV 260
> hsa:10412 NSA2, CDK105, HCL-G1, HCLG1, HUSSY29, TINP1, YR-29;
NSA2 ribosome biogenesis homolog (S. cerevisiae); K14842 ribosome
biogenesis protein NSA2
Length=260
Score = 399 bits (1024), Expect = 7e-111, Method: Compositional matrix adjust.
Identities = 192/260 (73%), Positives = 222/260 (85%), Gaps = 0/260 (0%)
Query 6 MPQNEYMEQHRKRFGERLDTAERKRKKAARMPHERSKTARKLRGIKAKLFNRRRFVEKAQ 65
MPQNEY+E HRKR+G RLD E+KRKK +R HERSK A+K+ G+KAKL++++R EK Q
Sbjct 1 MPQNEYIELHRKRYGYRLDYHEKKRKKESREAHERSKKAKKMIGLKAKLYHKQRHAEKIQ 60
Query 66 MKKTIKQHEEKEGKQKEPETVPEGAVPSYLLDREGVSRTKILTNMIKQKRKEKAGKWQVP 125
MKKTIK HE++ KQK E P+GAVP+YLLDREG SR K+L+NMIKQKRKEKAGKW+VP
Sbjct 61 MKKTIKMHEKRNTKQKNDEKTPQGAVPAYLLDREGQSRAKVLSNMIKQKRKEKAGKWEVP 120
Query 126 IPKVKAMTEEEMFKVLRSGKRKKKMWKRVVNKVCFVGDDFTRKPPKFERYIRPTSLRFKK 185
+PKV+A E E+ KV+R+GKRKKK WKR+V KVCFVGD FTRKPPK+ER+IRP LRFKK
Sbjct 121 LPKVRAQGETEVLKVIRTGKRKKKAWKRMVTKVCFVGDGFTRKPPKYERFIRPMGLRFKK 180
Query 186 AHVTHPELKATFQLDIVKVKKNPQSQLYTSLGVITKGTIIEVNVSELGLVTQTGKVVWAK 245
AHVTHPELKATF L I+ VKKNP S LYT+LGVITKGT+IEVNVSELGLVTQ GKV+W K
Sbjct 181 AHVTHPELKATFCLPILGVKKNPSSPLYTTLGVITKGTVIEVNVSELGLVTQGGKVIWGK 240
Query 246 YAQVTNNPELDGCINAVLLV 265
YAQVTNNPE DGCINAVLLV
Sbjct 241 YAQVTNNPENDGCINAVLLV 260
> xla:100381087 nsa2, cdk105, hcl-g1, hclg1, hussy-29, hussy29,
tinp1, yr-29; NSA2 ribosome biogenesis homolog; K14842 ribosome
biogenesis protein NSA2
Length=260
Score = 396 bits (1018), Expect = 4e-110, Method: Compositional matrix adjust.
Identities = 191/260 (73%), Positives = 222/260 (85%), Gaps = 0/260 (0%)
Query 6 MPQNEYMEQHRKRFGERLDTAERKRKKAARMPHERSKTARKLRGIKAKLFNRRRFVEKAQ 65
MPQN+Y+E HRKR+G RLD E+KRKK +R HERS+ A+KL G+KAKL++++R EK Q
Sbjct 1 MPQNDYIELHRKRYGYRLDYHEKKRKKESREAHERSQKAKKLIGLKAKLYHKQRHAEKIQ 60
Query 66 MKKTIKQHEEKEGKQKEPETVPEGAVPSYLLDREGVSRTKILTNMIKQKRKEKAGKWQVP 125
MKKT+K HE++ KQK E PEGAVP+YLLDREG +R K+L+NMIKQKRKEKAGKW+VP
Sbjct 61 MKKTLKMHEKRNTKQKNEEKTPEGAVPAYLLDREGQTRAKVLSNMIKQKRKEKAGKWEVP 120
Query 126 IPKVKAMTEEEMFKVLRSGKRKKKMWKRVVNKVCFVGDDFTRKPPKFERYIRPTSLRFKK 185
+PKV+A E E+ KV+R+GKRKKK WKR+V KVCFVGD FTRKPPK+ER+IRP LRFKK
Sbjct 121 LPKVRAQGETEVLKVIRTGKRKKKAWKRMVTKVCFVGDGFTRKPPKYERFIRPMGLRFKK 180
Query 186 AHVTHPELKATFQLDIVKVKKNPQSQLYTSLGVITKGTIIEVNVSELGLVTQTGKVVWAK 245
AHVTHPELKATF L I+ VKKNP S LYT+LGVITKGT+IEVNVSELGLVTQ GKVVW K
Sbjct 181 AHVTHPELKATFCLPILGVKKNPSSPLYTTLGVITKGTVIEVNVSELGLVTQGGKVVWGK 240
Query 246 YAQVTNNPELDGCINAVLLV 265
YAQVTNNPE DGCINAVLLV
Sbjct 241 YAQVTNNPENDGCINAVLLV 260
> dre:321769 nsa2, fb34b10, wu:fb34b10, zgc:56334; NSA2 ribosome
biogenesis homolog (S. cerevisiae); K14842 ribosome biogenesis
protein NSA2
Length=260
Score = 396 bits (1017), Expect = 5e-110, Method: Compositional matrix adjust.
Identities = 189/260 (72%), Positives = 221/260 (85%), Gaps = 0/260 (0%)
Query 6 MPQNEYMEQHRKRFGERLDTAERKRKKAARMPHERSKTARKLRGIKAKLFNRRRFVEKAQ 65
MPQNE++E HRKR G RLD E+KRKK +R HERS ARK+ G+KAKL++++R EK Q
Sbjct 1 MPQNEHIELHRKRHGYRLDHHEKKRKKESREAHERSHKARKMIGLKAKLYHKQRHAEKIQ 60
Query 66 MKKTIKQHEEKEGKQKEPETVPEGAVPSYLLDREGVSRTKILTNMIKQKRKEKAGKWQVP 125
MKKTIK HE+++ KQK+ + PEGAVP+YLLDREG SR K+L+NMIKQKRKEKAGKW+VP
Sbjct 61 MKKTIKMHEQRKSKQKDDDKTPEGAVPAYLLDREGQSRAKVLSNMIKQKRKEKAGKWEVP 120
Query 126 IPKVKAMTEEEMFKVLRSGKRKKKMWKRVVNKVCFVGDDFTRKPPKFERYIRPTSLRFKK 185
+PKV+A E E+ KV+R+GKR+KK WKR+V KVCFVGD FTRKPPK+ER+IRP LRF K
Sbjct 121 LPKVRAQGETEVLKVIRTGKRQKKAWKRMVTKVCFVGDGFTRKPPKYERFIRPMGLRFNK 180
Query 186 AHVTHPELKATFQLDIVKVKKNPQSQLYTSLGVITKGTIIEVNVSELGLVTQTGKVVWAK 245
AHVTHPELKATF L I+ VKKNP S LYT+LGVITKGT+IEVNVSELGLVTQ GKV+W K
Sbjct 181 AHVTHPELKATFCLPILGVKKNPSSSLYTTLGVITKGTVIEVNVSELGLVTQGGKVIWGK 240
Query 246 YAQVTNNPELDGCINAVLLV 265
YAQVTNNPE DGCINAVLLV
Sbjct 241 YAQVTNNPENDGCINAVLLV 260
> tpv:TP03_0821 hypothetical protein; K14842 ribosome biogenesis
protein NSA2
Length=259
Score = 388 bits (997), Expect = 1e-107, Method: Compositional matrix adjust.
Identities = 190/260 (73%), Positives = 218/260 (83%), Gaps = 1/260 (0%)
Query 6 MPQNEYMEQHRKRFGERLDTAERKRKKAARMPHERSKTARKLRGIKAKLFNRRRFVEKAQ 65
MPQNEY+E+H K G R D + RKKAAR P SK A+ LRGIK+K+F + ++ EK Q
Sbjct 1 MPQNEYIERHIKLHGRRFDHLTKTRKKAARAPLRESKRAKTLRGIKSKIFKKHKYAEKIQ 60
Query 66 MKKTIKQHEEKEGKQKEPETVPEGAVPSYLLDREGVSRTKILTNMIKQKRKEKAGKWQVP 125
+K ++Q EEKE K+K E +GA+P+YLLDR ++RTKIL+NMIKQKRKEKAGKW VP
Sbjct 61 FRKLMRQREEKEVKEKIVEKT-DGAIPAYLLDRGDINRTKILSNMIKQKRKEKAGKWTVP 119
Query 126 IPKVKAMTEEEMFKVLRSGKRKKKMWKRVVNKVCFVGDDFTRKPPKFERYIRPTSLRFKK 185
IPKVKA+ E EMFKVL++GKRKKKMWKRV++K CFV +DFTRKPPK+ERYIRPT LRFKK
Sbjct 120 IPKVKALNEMEMFKVLKTGKRKKKMWKRVIDKACFVPEDFTRKPPKYERYIRPTGLRFKK 179
Query 186 AHVTHPELKATFQLDIVKVKKNPQSQLYTSLGVITKGTIIEVNVSELGLVTQTGKVVWAK 245
AHVTHPELK TF LDI+ VKKNPQS LYTSL VITKGTIIEVNVSELGLVTQTGKV+WAK
Sbjct 180 AHVTHPELKTTFYLDIISVKKNPQSTLYTSLNVITKGTIIEVNVSELGLVTQTGKVIWAK 239
Query 246 YAQVTNNPELDGCINAVLLV 265
YAQVTNNPE DGCINAVLLV
Sbjct 240 YAQVTNNPENDGCINAVLLV 259
> pfa:MAL7P1.24 cdk105, putative; K14842 ribosome biogenesis protein
NSA2
Length=259
Score = 367 bits (943), Expect = 2e-101, Method: Compositional matrix adjust.
Identities = 172/260 (66%), Positives = 215/260 (82%), Gaps = 1/260 (0%)
Query 6 MPQNEYMEQHRKRFGERLDTAERKRKKAARMPHERSKTARKLRGIKAKLFNRRRFVEKAQ 65
MPQN+Y+E HRKR+G R D E++RKK AR H+ S A+KLRGIKAK++N++++ EK
Sbjct 1 MPQNDYIELHRKRYGYRFDHFEKERKKEARKVHKDSLKAKKLRGIKAKIYNKKKYTEKVN 60
Query 66 MKKTIKQHEEKEGKQKEPETVPEGAVPSYLLDREGVSRTKILTNMIKQKRKEKAGKWQVP 125
+KKT+K HE K+ K E G +PSYLLDR+ T+ILTN+IKQKRK K GKWQ+P
Sbjct 61 LKKTLKSHELKDIKSTETLKDDNG-LPSYLLDRQQTKHTQILTNLIKQKRKSKCGKWQLP 119
Query 126 IPKVKAMTEEEMFKVLRSGKRKKKMWKRVVNKVCFVGDDFTRKPPKFERYIRPTSLRFKK 185
IPK++A+ E EM +V++SGKR++K WKR+++K+ FVG+DFTRK PKFERYIRP+SLRFKK
Sbjct 120 IPKIQALNEAEMLRVVKSGKRRRKTWKRLIDKISFVGNDFTRKNPKFERYIRPSSLRFKK 179
Query 186 AHVTHPELKATFQLDIVKVKKNPQSQLYTSLGVITKGTIIEVNVSELGLVTQTGKVVWAK 245
A+V H ELK+TF LDI+ VK NPQS LYT+LG+ITKGTIIEVNVSELGLVTQ+GKV+WAK
Sbjct 180 ANVYHSELKSTFSLDIISVKVNPQSNLYTNLGIITKGTIIEVNVSELGLVTQSGKVIWAK 239
Query 246 YAQVTNNPELDGCINAVLLV 265
+AQVTNNPELDGCINA LLV
Sbjct 240 FAQVTNNPELDGCINATLLV 259
> bbo:BBOV_III000260 17.m07048; ribosomal protein Se8; K14842
ribosome biogenesis protein NSA2
Length=259
Score = 364 bits (934), Expect = 2e-100, Method: Compositional matrix adjust.
Identities = 186/261 (71%), Positives = 217/261 (83%), Gaps = 3/261 (1%)
Query 6 MPQNEYMEQHRKRFGERLDTAERKRKKAARMPHERSKTARKLRGIKAKLFNRRRFVEKAQ 65
MPQNEY+E+H G R D R RKK AR H SK +K+RG+KAK+F +RR+ EK +
Sbjct 1 MPQNEYIERHIALHGRRFDHETRMRKKTARAAHTLSKKMQKMRGLKAKIFRKRRYAEKIE 60
Query 66 MKKTIKQHEEKEGKQKEPETVP-EGAVPSYLLDREGVSRTKILTNMIKQKRKEKAGKWQV 124
MKK IK +EEKE K++ P +GA+P+YLL+R V+RTKIL+NMIKQKRKEKAGKW V
Sbjct 61 MKKQIKNNEEKESKEEV--VAPGDGAIPAYLLERGEVNRTKILSNMIKQKRKEKAGKWDV 118
Query 125 PIPKVKAMTEEEMFKVLRSGKRKKKMWKRVVNKVCFVGDDFTRKPPKFERYIRPTSLRFK 184
IPKV+ M E EMF+V++SGKRKKK WKR+V+K+CF+ +D+TRKPPK+E+YIRPT LRFK
Sbjct 119 VIPKVRQMNEAEMFRVMKSGKRKKKQWKRMVDKICFIPEDYTRKPPKYEKYIRPTGLRFK 178
Query 185 KAHVTHPELKATFQLDIVKVKKNPQSQLYTSLGVITKGTIIEVNVSELGLVTQTGKVVWA 244
KAHVTHPELK TF LDI+ VKKNPQS LYTSLGVITKGTIIEVNVSELGLVTQTGKVVWA
Sbjct 179 KAHVTHPELKTTFFLDIISVKKNPQSNLYTSLGVITKGTIIEVNVSELGLVTQTGKVVWA 238
Query 245 KYAQVTNNPELDGCINAVLLV 265
KYAQVTNNPE DGCINAVLLV
Sbjct 239 KYAQVTNNPENDGCINAVLLV 259
> ath:AT5G06360 ribosomal protein S8e family protein; K14842 ribosome
biogenesis protein NSA2
Length=260
Score = 362 bits (928), Expect = 1e-99, Method: Compositional matrix adjust.
Identities = 174/260 (66%), Positives = 207/260 (79%), Gaps = 0/260 (0%)
Query 6 MPQNEYMEQHRKRFGERLDTAERKRKKAARMPHERSKTARKLRGIKAKLFNRRRFVEKAQ 65
MPQ +Y++ HRKR G RLD ERKRKK AR H+ S A+K GIK K+ ++ + EKA
Sbjct 1 MPQGDYIDLHRKRNGYRLDHFERKRKKEAREVHKHSTMAQKSLGIKGKMIAKKNYAEKAL 60
Query 66 MKKTIKQHEEKEGKQKEPETVPEGAVPSYLLDREGVSRTKILTNMIKQKRKEKAGKWQVP 125
MKKT+K HEE ++K E V EGAVP+YLLDRE +R K+L+N IKQKRKEKAGKW+VP
Sbjct 61 MKKTLKMHEESSSRRKADENVQEGAVPAYLLDREDTTRAKVLSNTIKQKRKEKAGKWEVP 120
Query 126 IPKVKAMTEEEMFKVLRSGKRKKKMWKRVVNKVCFVGDDFTRKPPKFERYIRPTSLRFKK 185
+PKV+ + E+EMF+V+RSGKRK K WKR+V K FVG FTRKPPK+ER+IRP+ LRF K
Sbjct 121 LPKVRPVAEDEMFRVIRSGKRKTKQWKRMVTKATFVGPAFTRKPPKYERFIRPSGLRFTK 180
Query 186 AHVTHPELKATFQLDIVKVKKNPQSQLYTSLGVITKGTIIEVNVSELGLVTQTGKVVWAK 245
AHVTHPELK TF L+I+ +KKNP +YTSLGV+T+GTIIEVNVSELGLVT GKVVW K
Sbjct 181 AHVTHPELKCTFCLEIIGIKKNPNGPMYTSLGVMTRGTIIEVNVSELGLVTPAGKVVWGK 240
Query 246 YAQVTNNPELDGCINAVLLV 265
YAQVTNNPE DGCINAVLLV
Sbjct 241 YAQVTNNPENDGCINAVLLV 260
> cel:W09C5.1 hypothetical protein; K14842 ribosome biogenesis
protein NSA2
Length=259
Score = 356 bits (914), Expect = 5e-98, Method: Compositional matrix adjust.
Identities = 173/261 (66%), Positives = 214/261 (81%), Gaps = 3/261 (1%)
Query 6 MPQNEYMEQHRKRFGERLDTAERKRKKAARMPHERSKTARKLRGIKAKLFNRRRFVEKAQ 65
MPQNE++E HRKR G RLD ER+RKK AR H+RS+ A+ LRG KAKL++++R+ EK +
Sbjct 1 MPQNEHIELHRKRHGRRLDHEERQRKKLARAAHDRSQMAKTLRGHKAKLYHKKRYSEKVE 60
Query 66 MKKTIKQHEEKEGKQKEPETVPE-GAVPSYLLDREGVSRTKILTNMIKQKRKEKAGKWQV 124
M+K +KQHEEK+ QK P+ GAVP+YLLDR+ + +L+NMIKQKRK+KAGK+ V
Sbjct 61 MRKLLKQHEEKD--QKNTVEQPDKGAVPAYLLDRQQQTSGTVLSNMIKQKRKQKAGKFNV 118
Query 125 PIPKVKAMTEEEMFKVLRSGKRKKKMWKRVVNKVCFVGDDFTRKPPKFERYIRPTSLRFK 184
PIP+V+A+++ E FKV+++GK +K WKR+V KV FVG+ FTRKP KFER+IRP LRFK
Sbjct 119 PIPQVRAVSDAEAFKVVKTGKTNRKGWKRMVTKVTFVGESFTRKPAKFERFIRPMGLRFK 178
Query 185 KAHVTHPELKATFQLDIVKVKKNPQSQLYTSLGVITKGTIIEVNVSELGLVTQTGKVVWA 244
KAHVTHPEL+ TF L IV VKKNP SQ+YTSLGVITKGTI+EVNVSELG+VTQ GKVVW
Sbjct 179 KAHVTHPELQTTFHLPIVGVKKNPSSQMYTSLGVITKGTILEVNVSELGMVTQGGKVVWG 238
Query 245 KYAQVTNNPELDGCINAVLLV 265
K+AQVTNNPE DGCINAVLL+
Sbjct 239 KFAQVTNNPENDGCINAVLLI 259
> sce:YER126C NSA2; Nsa2p; K14842 ribosome biogenesis protein
NSA2
Length=261
Score = 304 bits (778), Expect = 3e-82, Method: Compositional matrix adjust.
Identities = 153/261 (58%), Positives = 197/261 (75%), Gaps = 1/261 (0%)
Query 6 MPQNEYMEQHRKRFGERLDTAERKRKKAARMPHERSKTARKLRGIKAKLFNRRRFVEKAQ 65
MPQN+Y+E+H K+ G+RLD ERKRK+ AR H+ S+ A+KL G K K F ++R+ EK
Sbjct 1 MPQNDYIERHIKQHGKRLDHEERKRKREARESHKISERAQKLTGWKGKQFAKKRYAEKVS 60
Query 66 MKKTIKQHEEKEGK-QKEPETVPEGAVPSYLLDREGVSRTKILTNMIKQKRKEKAGKWQV 124
M+K IK HE+ + K +P A+P+YLLDRE + K +++ IKQKR EKA K+ V
Sbjct 61 MRKKIKAHEQSKVKGSSKPLDTDGDALPTYLLDREQNNTAKAISSSIKQKRLEKADKFSV 120
Query 125 PIPKVKAMTEEEMFKVLRSGKRKKKMWKRVVNKVCFVGDDFTRKPPKFERYIRPTSLRFK 184
P+PKV+ ++EEEMFKV+++GK + K WKR++ K FVG+ FTR+P K ER IRP++LR K
Sbjct 121 PLPKVRGISEEEMFKVIKTGKSRSKSWKRMITKHTFVGEGFTRRPVKMERIIRPSALRQK 180
Query 185 KAHVTHPELKATFQLDIVKVKKNPQSQLYTSLGVITKGTIIEVNVSELGLVTQTGKVVWA 244
KA+VTHPEL T L I+ VKKNPQS +YT LGV+TKGTIIEVNVSELG+VT GKVVW
Sbjct 181 KANVTHPELGVTVFLPILAVKKNPQSPMYTQLGVLTKGTIIEVNVSELGMVTAGGKVVWG 240
Query 245 KYAQVTNNPELDGCINAVLLV 265
KYAQVTN P+ DGC+NAVLLV
Sbjct 241 KYAQVTNEPDRDGCVNAVLLV 261
> sce:YBR227C MCX1; Mcx1p; K03544 ATP-dependent Clp protease ATP-binding
subunit ClpX
Length=520
Score = 33.9 bits (76), Expect = 0.67, Method: Compositional matrix adjust.
Identities = 19/54 (35%), Positives = 29/54 (53%), Gaps = 3/54 (5%)
Query 5 KMPQNEYMEQHR---KRFGERLDTAERKRKKAARMPHERSKTARKLRGIKAKLF 55
K P+N ++Q+ K+FG RL ++ KK A+ + AR LRGI +L
Sbjct 382 KEPKNALLDQYEYIFKQFGVRLCVTQKALKKVAQFALKEGTGARGLRGIMERLL 435
> cel:ZK1098.10 unc-16; UNCoordinated family member (unc-16)
Length=1185
Score = 30.0 bits (66), Expect = 9.4, Method: Compositional matrix adjust.
Identities = 24/109 (22%), Positives = 44/109 (40%), Gaps = 12/109 (11%)
Query 31 KKAARMPHERSKTARKLRGIKAKLFNRRRFVEKAQMKKTIKQHEEKEGK----------- 79
+ R R K R + I + + RR E+ + K +++H +KE
Sbjct 496 SRGNRASSSRGKMTRSVEYIDPDMISERRAAERREQYKLVREHVKKEDGRIEAYGWSLPN 555
Query 80 -QKEPETVPEGAVPSYLLDREGVSRTKILTNMIKQKRKEKAGKWQVPIP 127
+ E +VP LLD E + T ++ + +++ G+W V P
Sbjct 556 VEAEVSSVPIPVCCRPLLDNEPSLKIWCATGVVLRGGRDERGQWIVGDP 604
Lambda K H
0.317 0.132 0.375
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 9827099128
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40