bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0319_orf1
Length=118
Score E
Sequences producing significant alignments: (Bits) Value
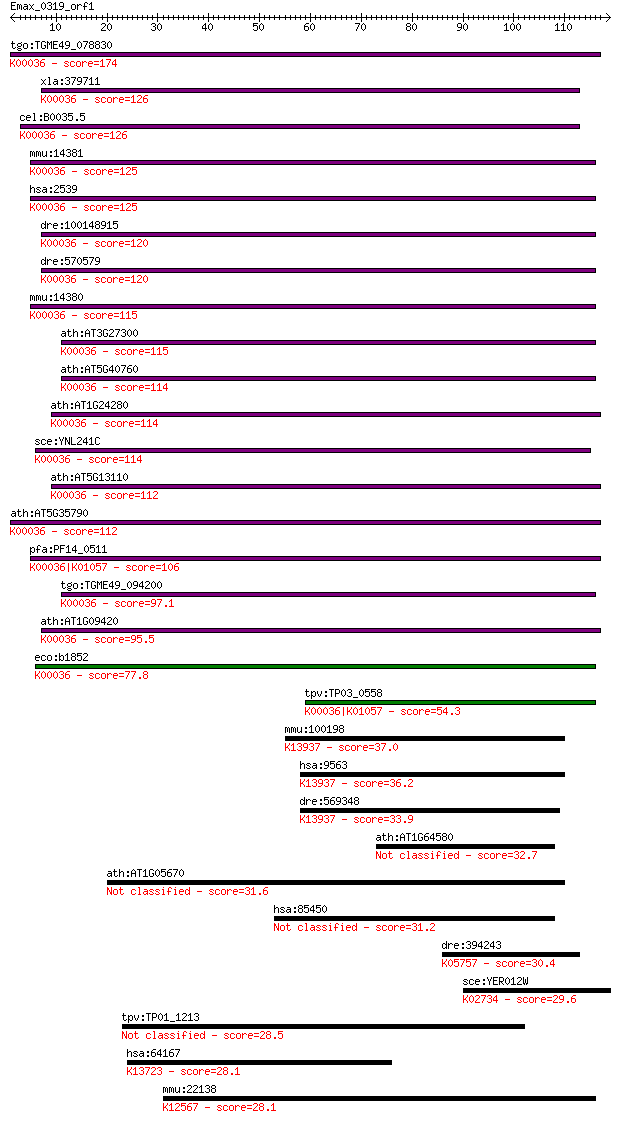
tgo:TGME49_078830 glucose-6-phosphate dehydrogenase, putative ... 174 9e-44
xla:379711 g6pd, MGC69058, g6pdh; glucose-6-phosphate dehydrog... 126 2e-29
cel:B0035.5 glucose-6-phosphate-1-dehydrogenase; K00036 glucos... 126 2e-29
mmu:14381 G6pdx, G28A, G6pd, Gpdx; glucose-6-phosphate dehydro... 125 4e-29
hsa:2539 G6PD, G6PD1; glucose-6-phosphate dehydrogenase (EC:1.... 125 5e-29
dre:100148915 glucose-6-phosphate dehydrogenase-like; K00036 g... 120 1e-27
dre:570579 g6pd, fj78b06, si:dkey-90a13.8, wu:fj78b06; glucose... 120 1e-27
mmu:14380 G6pd2, G6pdx-ps1, Gpd-2, Gpd2; glucose-6-phosphate d... 115 4e-26
ath:AT3G27300 G6PD5; G6PD5 (glucose-6-phosphate dehydrogenase ... 115 5e-26
ath:AT5G40760 G6PD6; G6PD6 (GLUCOSE-6-PHOSPHATE DEHYDROGENASE ... 114 6e-26
ath:AT1G24280 G6PD3; G6PD3 (GLUCOSE-6-PHOSPHATE DEHYDROGENASE ... 114 7e-26
sce:YNL241C ZWF1, MET19, POS10; Glucose-6-phosphate dehydrogen... 114 1e-25
ath:AT5G13110 G6PD2; G6PD2 (GLUCOSE-6-PHOSPHATE DEHYDROGENASE ... 112 2e-25
ath:AT5G35790 G6PD1; G6PD1 (GLUCOSE-6-PHOSPHATE DEHYDROGENASE ... 112 3e-25
pfa:PF14_0511 glucose-6-phosphate dehydrogenase-6-phosphogluco... 106 2e-23
tgo:TGME49_094200 glucose-6-phosphate dehydrogenase (EC:1.1.1.... 97.1 1e-20
ath:AT1G09420 G6PD4; G6PD4 (GLUCOSE-6-PHOSPHATE DEHYDROGENASE ... 95.5 3e-20
eco:b1852 zwf, ECK1853, JW1841; glucose-6-phosphate 1-dehydrog... 77.8 8e-15
tpv:TP03_0558 glucose-6-phosphate dehydrogenase-6-phosphogluco... 54.3 1e-07
mmu:100198 H6pd, AI785303, G6pd1, Gpd-1, Gpd1; hexose-6-phosph... 37.0 0.014
hsa:9563 H6PD, DKFZp686A01246, G6PDH, GDH, MGC87643; hexose-6-... 36.2 0.023
dre:569348 glucose-6-phosphate dehydrogenase X-linked-like; K1... 33.9 0.13
ath:AT1G64580 pentatricopeptide (PPR) repeat-containing protein 32.7 0.26
ath:AT1G05670 UDP-glucoronosyl/UDP-glucosyl transferase family... 31.6 0.68
hsa:85450 ITPRIP, DANGER, KIAA1754, bA127L20, bA127L20.2; inos... 31.2 0.86
dre:394243 arpc1a, wu:fk93e03; actin related protein 2/3 compl... 30.4 1.4
sce:YER012W PRE1; Beta 4 subunit of the 20S proteasome; locali... 29.6 2.3
tpv:TP01_1213 hypothetical protein 28.5 5.8
hsa:64167 ERAP2, FLJ23633, FLJ23701, FLJ23807, L-RAP, LRAP; en... 28.1 6.2
mmu:22138 Ttn, 1100001C23Rik, 2310036G12Rik, 2310057K23Rik, 23... 28.1 7.0
> tgo:TGME49_078830 glucose-6-phosphate dehydrogenase, putative
(EC:3.1.1.31 1.1.1.49); K00036 glucose-6-phosphate 1-dehydrogenase
[EC:1.1.1.49]
Length=878
Score = 174 bits (440), Expect = 9e-44, Method: Compositional matrix adjust.
Identities = 85/117 (72%), Positives = 98/117 (83%), Gaps = 1/117 (0%)
Query 1 SFYPEEELSENELVIIVQPREAIYLKFYTKKPGLAS-GLQLTELDLSVMDRLQVDRLPDA 59
SF+ E L+ NELVI+VQP EA+YLK +TKKPGL S GLQ TELDLSVMDR V+RLPDA
Sbjct 733 SFFHEPNLTPNELVILVQPHEAVYLKIHTKKPGLLSQGLQPTELDLSVMDRFDVERLPDA 792
Query 60 YERLLLDVIKGDRQNFVRTDELREAWRIFTPLLKQIDEPGVKPEPYPYGSHGPESAY 116
YERLLLDVI+GD+QNFVRTDELREAWRIFTPLL +I+E + P PYP GS GP ++Y
Sbjct 793 YERLLLDVIRGDKQNFVRTDELREAWRIFTPLLHEIEEKNIDPLPYPAGSSGPSASY 849
> xla:379711 g6pd, MGC69058, g6pdh; glucose-6-phosphate dehydrogenase
(EC:1.1.1.49); K00036 glucose-6-phosphate 1-dehydrogenase
[EC:1.1.1.49]
Length=518
Score = 126 bits (316), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 60/106 (56%), Positives = 77/106 (72%), Gaps = 0/106 (0%)
Query 7 ELSENELVIIVQPREAIYLKFYTKKPGLASGLQLTELDLSVMDRLQVDRLPDAYERLLLD 66
+ NELVI VQP EA+Y K TKKPG+ + +ELDL+ R + +LPDAYERL+LD
Sbjct 387 QCKRNELVIRVQPNEAVYTKMMTKKPGMFFNPEESELDLTYGSRYKDVKLPDAYERLILD 446
Query 67 VIKGDRQNFVRTDELREAWRIFTPLLKQIDEPGVKPEPYPYGSHGP 112
V G++ +FVR+DELREAWRIFTP+L Q++ +KP PY YGS GP
Sbjct 447 VFCGNQMHFVRSDELREAWRIFTPVLHQLEREKIKPHPYKYGSRGP 492
> cel:B0035.5 glucose-6-phosphate-1-dehydrogenase; K00036 glucose-6-phosphate
1-dehydrogenase [EC:1.1.1.49]
Length=522
Score = 126 bits (316), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 60/110 (54%), Positives = 79/110 (71%), Gaps = 0/110 (0%)
Query 3 YPEEELSENELVIIVQPREAIYLKFYTKKPGLASGLQLTELDLSVMDRLQVDRLPDAYER 62
YP EL +ELV+ VQP EA+Y+K TKKPG+ G++ TELDL+ +R + RLPDAYER
Sbjct 388 YPSGELKRSELVMRVQPNEAVYMKLMTKKPGMGFGVEETELDLTYNNRFKEVRLPDAYER 447
Query 63 LLLDVIKGDRQNFVRTDELREAWRIFTPLLKQIDEPGVKPEPYPYGSHGP 112
L L+V G + NFVRTDEL AWRI TP+L+++ + V+P Y +GS GP
Sbjct 448 LFLEVFMGSQINFVRTDELEYAWRILTPVLEELKKKKVQPVQYKFGSRGP 497
> mmu:14381 G6pdx, G28A, G6pd, Gpdx; glucose-6-phosphate dehydrogenase
X-linked (EC:1.1.1.49); K00036 glucose-6-phosphate
1-dehydrogenase [EC:1.1.1.49]
Length=515
Score = 125 bits (314), Expect = 4e-29, Method: Compositional matrix adjust.
Identities = 62/111 (55%), Positives = 78/111 (70%), Gaps = 0/111 (0%)
Query 5 EEELSENELVIIVQPREAIYLKFYTKKPGLASGLQLTELDLSVMDRLQVDRLPDAYERLL 64
++ NELVI VQP EA+Y K TKKPG+ + +ELDL+ +R + +LPDAYERL+
Sbjct 382 HQQCKRNELVIRVQPNEAVYTKMMTKKPGMFFNPEESELDLTYGNRYKNVKLPDAYERLI 441
Query 65 LDVIKGDRQNFVRTDELREAWRIFTPLLKQIDEPGVKPEPYPYGSHGPESA 115
LDV G + +FVR+DELREAWRIFTPLL +ID +P PY YGS GP A
Sbjct 442 LDVFCGSQMHFVRSDELREAWRIFTPLLHKIDREKPQPIPYVYGSRGPTEA 492
> hsa:2539 G6PD, G6PD1; glucose-6-phosphate dehydrogenase (EC:1.1.1.49);
K00036 glucose-6-phosphate 1-dehydrogenase [EC:1.1.1.49]
Length=545
Score = 125 bits (313), Expect = 5e-29, Method: Compositional matrix adjust.
Identities = 63/111 (56%), Positives = 78/111 (70%), Gaps = 0/111 (0%)
Query 5 EEELSENELVIIVQPREAIYLKFYTKKPGLASGLQLTELDLSVMDRLQVDRLPDAYERLL 64
++ NELVI VQP EA+Y K TKKPG+ + +ELDL+ +R + +LPDAYERL+
Sbjct 412 HQQCKRNELVIRVQPNEAVYTKMMTKKPGMFFNPEESELDLTYGNRYKNVKLPDAYERLI 471
Query 65 LDVIKGDRQNFVRTDELREAWRIFTPLLKQIDEPGVKPEPYPYGSHGPESA 115
LDV G + +FVR+DELREAWRIFTPLL QI+ KP PY YGS GP A
Sbjct 472 LDVFCGSQMHFVRSDELREAWRIFTPLLHQIELEKPKPIPYIYGSRGPTEA 522
> dre:100148915 glucose-6-phosphate dehydrogenase-like; K00036
glucose-6-phosphate 1-dehydrogenase [EC:1.1.1.49]
Length=523
Score = 120 bits (300), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 61/109 (55%), Positives = 74/109 (67%), Gaps = 0/109 (0%)
Query 7 ELSENELVIIVQPREAIYLKFYTKKPGLASGLQLTELDLSVMDRLQVDRLPDAYERLLLD 66
+ NELV+ VQP EAIY K +KKPG+ + TELDL+ R + +LPDAYERL+LD
Sbjct 392 QCRRNELVVRVQPNEAIYAKMMSKKPGVYFSPEETELDLTYHSRYRDVKLPDAYERLILD 451
Query 67 VIKGDRQNFVRTDELREAWRIFTPLLKQIDEPGVKPEPYPYGSHGPESA 115
V G + +FVR+DELREAWRIFTPLL QI+ P Y YGS GP A
Sbjct 452 VFCGSQMHFVRSDELREAWRIFTPLLHQIESEKTPPIKYKYGSRGPAEA 500
> dre:570579 g6pd, fj78b06, si:dkey-90a13.8, wu:fj78b06; glucose-6-phosphate
dehydrogenase; K00036 glucose-6-phosphate 1-dehydrogenase
[EC:1.1.1.49]
Length=523
Score = 120 bits (300), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 61/109 (55%), Positives = 74/109 (67%), Gaps = 0/109 (0%)
Query 7 ELSENELVIIVQPREAIYLKFYTKKPGLASGLQLTELDLSVMDRLQVDRLPDAYERLLLD 66
+ NELV+ VQP EAIY K +KKPG+ + TELDL+ R + +LPDAYERL+LD
Sbjct 392 QCRRNELVVRVQPNEAIYAKMMSKKPGVYFSPEETELDLTYHSRYRDVKLPDAYERLILD 451
Query 67 VIKGDRQNFVRTDELREAWRIFTPLLKQIDEPGVKPEPYPYGSHGPESA 115
V G + +FVR+DELREAWRIFTPLL QI+ P Y YGS GP A
Sbjct 452 VFCGSQMHFVRSDELREAWRIFTPLLHQIESEKTPPIKYKYGSRGPAEA 500
> mmu:14380 G6pd2, G6pdx-ps1, Gpd-2, Gpd2; glucose-6-phosphate
dehydrogenase 2 (EC:1.1.1.49); K00036 glucose-6-phosphate 1-dehydrogenase
[EC:1.1.1.49]
Length=513
Score = 115 bits (288), Expect = 4e-26, Method: Compositional matrix adjust.
Identities = 57/111 (51%), Positives = 75/111 (67%), Gaps = 0/111 (0%)
Query 5 EEELSENELVIIVQPREAIYLKFYTKKPGLASGLQLTELDLSVMDRLQVDRLPDAYERLL 64
++ NELVI +QP EA+Y TKKPG+ + +ELDL+ ++ + +LP AYERL+
Sbjct 382 HQKCKRNELVIRMQPNEAVYTTMMTKKPGMFFNPEESELDLTYGNKYKNVKLPGAYERLI 441
Query 65 LDVIKGDRQNFVRTDELREAWRIFTPLLKQIDEPGVKPEPYPYGSHGPESA 115
LDV G + +FVRTDELRE WRIFTPLL +I+ +P PY YGS GP A
Sbjct 442 LDVFCGCQMHFVRTDELREGWRIFTPLLHKIEREKPQPFPYVYGSRGPTEA 492
> ath:AT3G27300 G6PD5; G6PD5 (glucose-6-phosphate dehydrogenase
5); glucose-6-phosphate dehydrogenase (EC:1.1.1.49); K00036
glucose-6-phosphate 1-dehydrogenase [EC:1.1.1.49]
Length=516
Score = 115 bits (287), Expect = 5e-26, Method: Compositional matrix adjust.
Identities = 58/105 (55%), Positives = 72/105 (68%), Gaps = 0/105 (0%)
Query 11 NELVIIVQPREAIYLKFYTKKPGLASGLQLTELDLSVMDRLQVDRLPDAYERLLLDVIKG 70
NE VI +QP EA+Y+K K+PGL +ELDLS R Q +P+AYERL+LD I+G
Sbjct 390 NEFVIRLQPSEAMYMKLTVKQPGLEMQTVQSELDLSYKQRYQDVSIPEAYERLILDTIRG 449
Query 71 DRQNFVRTDELREAWRIFTPLLKQIDEPGVKPEPYPYGSHGPESA 115
D+Q+FVR DEL+ AW IFTPLL +ID+ VK PY GS GP A
Sbjct 450 DQQHFVRRDELKAAWEIFTPLLHRIDKGEVKSVPYKQGSRGPAEA 494
> ath:AT5G40760 G6PD6; G6PD6 (GLUCOSE-6-PHOSPHATE DEHYDROGENASE
6); glucose-6-phosphate dehydrogenase; K00036 glucose-6-phosphate
1-dehydrogenase [EC:1.1.1.49]
Length=515
Score = 114 bits (286), Expect = 6e-26, Method: Compositional matrix adjust.
Identities = 59/105 (56%), Positives = 73/105 (69%), Gaps = 0/105 (0%)
Query 11 NELVIIVQPREAIYLKFYTKKPGLASGLQLTELDLSVMDRLQVDRLPDAYERLLLDVIKG 70
NE VI +QP EA+Y+K K+PGL +ELDLS R Q +P+AYERL+LD IKG
Sbjct 389 NEFVIRLQPSEAMYMKLTVKQPGLDMNTVQSELDLSYGQRYQGVAIPEAYERLILDTIKG 448
Query 71 DRQNFVRTDELREAWRIFTPLLKQIDEPGVKPEPYPYGSHGPESA 115
D+Q+FVR DEL+ AW IFTPLL +ID+ VK PY GS GP+ A
Sbjct 449 DQQHFVRRDELKVAWEIFTPLLHRIDKGEVKSIPYKPGSRGPKEA 493
> ath:AT1G24280 G6PD3; G6PD3 (GLUCOSE-6-PHOSPHATE DEHYDROGENASE
3); glucose-6-phosphate dehydrogenase (EC:1.1.1.49); K00036
glucose-6-phosphate 1-dehydrogenase [EC:1.1.1.49]
Length=599
Score = 114 bits (285), Expect = 7e-26, Method: Compositional matrix adjust.
Identities = 60/108 (55%), Positives = 74/108 (68%), Gaps = 1/108 (0%)
Query 9 SENELVIIVQPREAIYLKFYTKKPGLASGLQLTELDLSVMDRLQVDRLPDAYERLLLDVI 68
+ NELVI VQP EAIYLK K PGL L + L+L R + +PDAYERLLLD I
Sbjct 476 TTNELVIRVQPDEAIYLKINNKVPGLGMRLDQSNLNLLYSARYSKE-IPDAYERLLLDAI 534
Query 69 KGDRQNFVRTDELREAWRIFTPLLKQIDEPGVKPEPYPYGSHGPESAY 116
+G+R+ F+R+DEL AW +FTPLLK+I+E PE YPYGS GP A+
Sbjct 535 EGERRLFIRSDELDAAWALFTPLLKEIEEKKTTPEFYPYGSRGPVGAH 582
> sce:YNL241C ZWF1, MET19, POS10; Glucose-6-phosphate dehydrogenase
(G6PD), catalyzes the first step of the pentose phosphate
pathway; involved in adapting to oxidatve stress; homolog
of the human G6PD which is deficient in patients with hemolytic
anemia (EC:1.1.1.49); K00036 glucose-6-phosphate 1-dehydrogenase
[EC:1.1.1.49]
Length=505
Score = 114 bits (284), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 58/110 (52%), Positives = 75/110 (68%), Gaps = 1/110 (0%)
Query 6 EELSENELVIIVQPREAIYLKFYTKKPGLASGLQLTELDLSVMDRLQVDRLPDAYERLLL 65
+++ NELVI VQP A+YLKF K PGL++ Q+T+L+L+ R Q +P+AYE L+
Sbjct 365 KDIPNNELVIRVQPDAAVYLKFNAKTPGLSNATQVTDLNLTYASRYQDFWIPEAYEVLIR 424
Query 66 DVIKGDRQNFVRTDELREAWRIFTPLLKQIDEP-GVKPEPYPYGSHGPES 114
D + GD NFVR DEL +W IFTPLLK I+ P G PE YPYGS GP+
Sbjct 425 DALLGDHSNFVRDDELDISWGIFTPLLKHIERPDGPTPEIYPYGSRGPKG 474
> ath:AT5G13110 G6PD2; G6PD2 (GLUCOSE-6-PHOSPHATE DEHYDROGENASE
2); glucose-6-phosphate dehydrogenase (EC:1.1.1.49); K00036
glucose-6-phosphate 1-dehydrogenase [EC:1.1.1.49]
Length=596
Score = 112 bits (281), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 60/108 (55%), Positives = 74/108 (68%), Gaps = 1/108 (0%)
Query 9 SENELVIIVQPREAIYLKFYTKKPGLASGLQLTELDLSVMDRLQVDRLPDAYERLLLDVI 68
+ NELVI VQP EAIYLK K PGL L + L+L R + +PDAYERLLLD I
Sbjct 473 ATNELVIRVQPDEAIYLKINNKVPGLGMRLDRSNLNLLYSARYSKE-IPDAYERLLLDAI 531
Query 69 KGDRQNFVRTDELREAWRIFTPLLKQIDEPGVKPEPYPYGSHGPESAY 116
+G+R+ F+R+DEL AW +FTPLLK+I+E PE YPYGS GP A+
Sbjct 532 EGERRLFIRSDELDAAWSLFTPLLKEIEEKKRIPEYYPYGSRGPVGAH 579
> ath:AT5G35790 G6PD1; G6PD1 (GLUCOSE-6-PHOSPHATE DEHYDROGENASE
1); glucose-6-phosphate dehydrogenase (EC:1.1.1.49); K00036
glucose-6-phosphate 1-dehydrogenase [EC:1.1.1.49]
Length=576
Score = 112 bits (280), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 60/117 (51%), Positives = 81/117 (69%), Gaps = 3/117 (2%)
Query 1 SFYPEEELSENELVIIVQPREAIYLKFYTKKPGLASGLQLTELDLSVMDRLQVDR-LPDA 59
SF + + NELVI VQP E IYL+ K PGL G++L DL+++ R + R +PDA
Sbjct 446 SFATNLDNATNELVIRVQPDEGIYLRINNKVPGL--GMRLDRSDLNLLYRSRYPREIPDA 503
Query 60 YERLLLDVIKGDRQNFVRTDELREAWRIFTPLLKQIDEPGVKPEPYPYGSHGPESAY 116
YERLLLD I+G+R+ F+R+DEL AW +FTP LK+++E + PE YPYGS GP A+
Sbjct 504 YERLLLDAIEGERRLFIRSDELDAAWDLFTPALKELEEKKIIPELYPYGSRGPVGAH 560
> pfa:PF14_0511 glucose-6-phosphate dehydrogenase-6-phosphogluconolactonase
(EC:1.1.1.49); K00036 glucose-6-phosphate 1-dehydrogenase
[EC:1.1.1.49]; K01057 6-phosphogluconolactonase
[EC:3.1.1.31]
Length=910
Score = 106 bits (264), Expect = 2e-23, Method: Composition-based stats.
Identities = 51/112 (45%), Positives = 76/112 (67%), Gaps = 1/112 (0%)
Query 5 EEELSENELVIIVQPREAIYLKFYTKKPGLASGLQLTELDLSVMDRLQVDRLPDAYERLL 64
+E ++ NE VII+QP EAIYLK KK G ++ +L+L+V ++ + +P+AYE LL
Sbjct 766 DENMNNNEFVIILQPVEAIYLKMMIKKTGCEE-MEEVQLNLTVNEKNKKINVPEAYETLL 824
Query 65 LDVIKGDRQNFVRTDELREAWRIFTPLLKQIDEPGVKPEPYPYGSHGPESAY 116
L+ KG ++ F+ +EL E+WRIFTPLLK++ E VKP Y +GS GP+ +
Sbjct 825 LECFKGHKKKFISDEELYESWRIFTPLLKELQEKQVKPLKYSFGSSGPKEVF 876
> tgo:TGME49_094200 glucose-6-phosphate dehydrogenase (EC:1.1.1.49);
K00036 glucose-6-phosphate 1-dehydrogenase [EC:1.1.1.49]
Length=560
Score = 97.1 bits (240), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 48/106 (45%), Positives = 70/106 (66%), Gaps = 4/106 (3%)
Query 11 NELVIIVQPREAIYLKFYTKKPGLASGLQLTELDLSVM-DRLQVDRLPDAYERLLLDVIK 69
N L++ VQP ++ + + PGL + L L + V D + R+PDAYE LLL+V+
Sbjct 421 NSLILEVQPHPSVRFEVNARAPGLGATLGRNVLKMDVNPDGV---RIPDAYENLLLNVLS 477
Query 70 GDRQNFVRTDELREAWRIFTPLLKQIDEPGVKPEPYPYGSHGPESA 115
GD+ +FVRTDELRE+WRIFTP+L +++ V+P YP+G+ GP A
Sbjct 478 GDKSHFVRTDELRESWRIFTPMLHALEDLQVQPRLYPFGTPGPSGA 523
> ath:AT1G09420 G6PD4; G6PD4 (GLUCOSE-6-PHOSPHATE DEHYDROGENASE
4); glucose-6-phosphate dehydrogenase; K00036 glucose-6-phosphate
1-dehydrogenase [EC:1.1.1.49]
Length=625
Score = 95.5 bits (236), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 48/110 (43%), Positives = 70/110 (63%), Gaps = 1/110 (0%)
Query 7 ELSENELVIIVQPREAIYLKFYTKKPGLASGLQLTELDLSVMDRLQVDRLPDAYERLLLD 66
+L NEL++ +P EAI +K K PGL L +EL+L DR + + +PD+YE L+ D
Sbjct 504 DLGTNELILRDEPDEAILVKINNKVPGLGLQLDASELNLLYKDRYKTE-VPDSYEHLIHD 562
Query 67 VIKGDRQNFVRTDELREAWRIFTPLLKQIDEPGVKPEPYPYGSHGPESAY 116
VI GD F+R+DE+ AW I +P+L++ID+ PE Y +G GP +AY
Sbjct 563 VIDGDNHLFMRSDEVAAAWNILSPVLEEIDKHHTAPELYEFGGRGPVAAY 612
> eco:b1852 zwf, ECK1853, JW1841; glucose-6-phosphate 1-dehydrogenase
(EC:1.1.1.49); K00036 glucose-6-phosphate 1-dehydrogenase
[EC:1.1.1.49]
Length=491
Score = 77.8 bits (190), Expect = 8e-15, Method: Compositional matrix adjust.
Identities = 42/112 (37%), Positives = 65/112 (58%), Gaps = 2/112 (1%)
Query 6 EELSENELVIIVQPREAIYLKFYTKKPGL--ASGLQLTELDLSVMDRLQVDRLPDAYERL 63
++L +N+L I +QP E + ++ K PGL LQ+T+LDLS + L DAYERL
Sbjct 365 QDLPQNKLTIRLQPDEGVDIQVLNKVPGLDHKHNLQITKLDLSYSETFNQTHLADAYERL 424
Query 64 LLDVIKGDRQNFVRTDELREAWRIFTPLLKQIDEPGVKPEPYPYGSHGPESA 115
LL+ ++G + FVR DE+ EAW+ + + P+PY G+ GP ++
Sbjct 425 LLETMRGIQALFVRRDEVEEAWKWVDSITEAWAMDNDAPKPYQAGTWGPVAS 476
> tpv:TP03_0558 glucose-6-phosphate dehydrogenase-6-phosphogluconolactonase
(EC:1.1.1.49); K00036 glucose-6-phosphate 1-dehydrogenase
[EC:1.1.1.49]; K01057 6-phosphogluconolactonase
[EC:3.1.1.31]
Length=869
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 25/57 (43%), Positives = 33/57 (57%), Gaps = 0/57 (0%)
Query 59 AYERLLLDVIKGDRQNFVRTDELREAWRIFTPLLKQIDEPGVKPEPYPYGSHGPESA 115
AYE L + R F ++ EAWRIFTP+L +I E ++P YP GS GP+ A
Sbjct 806 AYELLFFNAFSNKRDLFPTIQQVNEAWRIFTPVLHEIAEKKIEPFLYPRGSQGPKEA 862
> mmu:100198 H6pd, AI785303, G6pd1, Gpd-1, Gpd1; hexose-6-phosphate
dehydrogenase (glucose 1-dehydrogenase) (EC:1.1.1.47 3.1.1.31);
K13937 hexose-6-phosphate dehydrogenase [EC:1.1.1.47
3.1.1.31]
Length=797
Score = 37.0 bits (84), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 19/55 (34%), Positives = 29/55 (52%), Gaps = 2/55 (3%)
Query 55 RLPDAYERLLLDVIKGDRQNFVRTDELREAWRIFTPLLKQIDEPGVKPEPYPYGS 109
R DAY LL + +++F+ T+ L +W +TPLL + P PYP G+
Sbjct 459 REQDAYSTLLSHIFHCRKESFITTENLLASWVFWTPLLDSLAFE--VPRPYPGGA 511
> hsa:9563 H6PD, DKFZp686A01246, G6PDH, GDH, MGC87643; hexose-6-phosphate
dehydrogenase (glucose 1-dehydrogenase) (EC:1.1.1.47
3.1.1.31); K13937 hexose-6-phosphate dehydrogenase [EC:1.1.1.47
3.1.1.31]
Length=791
Score = 36.2 bits (82), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 17/52 (32%), Positives = 27/52 (51%), Gaps = 2/52 (3%)
Query 58 DAYERLLLDVIKGDRQNFVRTDELREAWRIFTPLLKQIDEPGVKPEPYPYGS 109
DA+ LL + G + F+ T+ L +W +TPLL+ + P YP G+
Sbjct 456 DAHSVLLSHIFHGRKNFFITTENLLASWNFWTPLLESLAHKA--PRLYPGGA 505
> dre:569348 glucose-6-phosphate dehydrogenase X-linked-like;
K13937 hexose-6-phosphate dehydrogenase [EC:1.1.1.47 3.1.1.31]
Length=781
Score = 33.9 bits (76), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 17/51 (33%), Positives = 27/51 (52%), Gaps = 2/51 (3%)
Query 58 DAYERLLLDVIKGDRQNFVRTDELREAWRIFTPLLKQIDEPGVKPEPYPYG 108
+AY L+ V G + +F+ + L +W +TPLL+ + V P YP G
Sbjct 450 EAYGELISHVFFGRKDSFISAEGLLASWAFWTPLLESLSH--VYPRIYPGG 498
> ath:AT1G64580 pentatricopeptide (PPR) repeat-containing protein
Length=1052
Score = 32.7 bits (73), Expect = 0.26, Method: Composition-based stats.
Identities = 15/35 (42%), Positives = 21/35 (60%), Gaps = 4/35 (11%)
Query 73 QNFVRTDELREAWRIFTPLLKQIDEPGVKPEPYPY 107
Q RTD+L+EAW +F L ++ GVKP+ Y
Sbjct 966 QGLCRTDKLKEAWCLFRSLTRK----GVKPDAIAY 996
> ath:AT1G05670 UDP-glucoronosyl/UDP-glucosyl transferase family
protein
Length=1184
Score = 31.6 bits (70), Expect = 0.68, Method: Compositional matrix adjust.
Identities = 22/100 (22%), Positives = 43/100 (43%), Gaps = 14/100 (14%)
Query 20 REAIYLKFYTKKPGLASGLQLTELDLSVMDRLQVDRLPDAYERLLLDVIKGDR------- 72
+ A + + + P + + ++ + Q+ R+ +A+ LLL +KG
Sbjct 669 KTATAIIVFREFPEVGVCWNVASYNIVIHFVCQLGRIKEAHHLLLLMELKGYTPDVISYS 728
Query 73 ---QNFVRTDELREAWRIFTPLLKQIDEPGVKPEPYPYGS 109
+ R EL + W+ L++ + G+KP Y YGS
Sbjct 729 TVVNGYCRFGELDKVWK----LIEVMKRKGLKPNSYIYGS 764
> hsa:85450 ITPRIP, DANGER, KIAA1754, bA127L20, bA127L20.2; inositol
1,4,5-triphosphate receptor interacting protein
Length=547
Score = 31.2 bits (69), Expect = 0.86, Method: Compositional matrix adjust.
Identities = 17/55 (30%), Positives = 27/55 (49%), Gaps = 0/55 (0%)
Query 53 VDRLPDAYERLLLDVIKGDRQNFVRTDELREAWRIFTPLLKQIDEPGVKPEPYPY 107
VD L +A L + ++F+ D + E W++ PLL + P PEPY +
Sbjct 168 VDDLLEALRSLCNRDTDMEVEDFIGVDSMYENWQVDRPLLCHLFVPFTPPEPYRF 222
> dre:394243 arpc1a, wu:fk93e03; actin related protein 2/3 complex,
subunit 1A; K05757 actin related protein 2/3 complex,
subunit 1A/1B
Length=370
Score = 30.4 bits (67), Expect = 1.4, Method: Composition-based stats.
Identities = 12/27 (44%), Positives = 18/27 (66%), Gaps = 3/27 (11%)
Query 86 RIFTPLLKQIDEPGVKPEPYPYGSHGP 112
R+F+ +K++DE KP P P+GS P
Sbjct 167 RVFSAYIKEVDE---KPAPTPWGSKMP 190
> sce:YER012W PRE1; Beta 4 subunit of the 20S proteasome; localizes
to the nucleus throughout the cell cycle (EC:3.4.25.1);
K02734 20S proteasome subunit beta 4 [EC:3.4.25.1]
Length=198
Score = 29.6 bits (65), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 16/29 (55%), Positives = 17/29 (58%), Gaps = 1/29 (3%)
Query 90 PLLKQIDEPGVKPEPYPYGSHGPESAYTF 118
P L QID G K E PYG+HG YTF
Sbjct 114 PELYQIDYLGTKVE-LPYGAHGYSGFYTF 141
> tpv:TP01_1213 hypothetical protein
Length=605
Score = 28.5 bits (62), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 28/88 (31%), Positives = 40/88 (45%), Gaps = 30/88 (34%)
Query 23 IYLKFYTKKPGLASGLQL------TELD---LSVMDRLQVDRLPDAYERLLLDVIKGDRQ 73
+Y FY+ +AS L L ELD LS ++ L VDRL D+IK
Sbjct 300 LYSPFYSSDIIIASPLGLRPLISGNELDYDFLSSIEVLVVDRL---------DIIKF--- 347
Query 74 NFVRTDELREAWRIFTPLLKQIDEPGVK 101
+ WR+FT L K +++P +K
Sbjct 348 ---------QNWRLFTDLFKMVNKPLMK 366
> hsa:64167 ERAP2, FLJ23633, FLJ23701, FLJ23807, L-RAP, LRAP;
endoplasmic reticulum aminopeptidase 2 (EC:3.4.11.-); K13723
endoplasmic reticulum aminopeptidase 2 [EC:3.4.11.-]
Length=960
Score = 28.1 bits (61), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 18/54 (33%), Positives = 26/54 (48%), Gaps = 2/54 (3%)
Query 24 YLKFYTKKPGLASGLQLTELDLSVMDRLQVDRLPDAYERLLLDVIK--GDRQNF 75
YL+ T P L GL E +MDR + + + +R LL K DRQ++
Sbjct 686 YLQHETSSPALLEGLSYLESFYHMMDRRNISDISENLKRYLLQYFKPVIDRQSW 739
> mmu:22138 Ttn, 1100001C23Rik, 2310036G12Rik, 2310057K23Rik,
2310074I15Rik, AF006999, AV006427, D330041I19Rik, D830007G01Rik,
L56, mdm, shru; titin (EC:2.7.11.1); K12567 titin [EC:2.7.11.1]
Length=33467
Score = 28.1 bits (61), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 21/85 (24%), Positives = 32/85 (37%), Gaps = 0/85 (0%)
Query 31 KPGLASGLQLTELDLSVMDRLQVDRLPDAYERLLLDVIKGDRQNFVRTDELREAWRIFTP 90
+P +L E+ L DR+ + D + D ++GD + T E + FT
Sbjct 28717 RPAPKVTWKLEEMRLKETDRMSIATTKDRTTLTVKDSMRGDSGRYFLTLENTAGVKTFTI 28776
Query 91 LLKQIDEPGVKPEPYPYGSHGPESA 115
+ I PG P S ES
Sbjct 28777 TVVVIGRPGPVTGPIEVSSVSAESC 28801
Lambda K H
0.318 0.140 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2027872200
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40