bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0295_orf2
Length=171
Score E
Sequences producing significant alignments: (Bits) Value
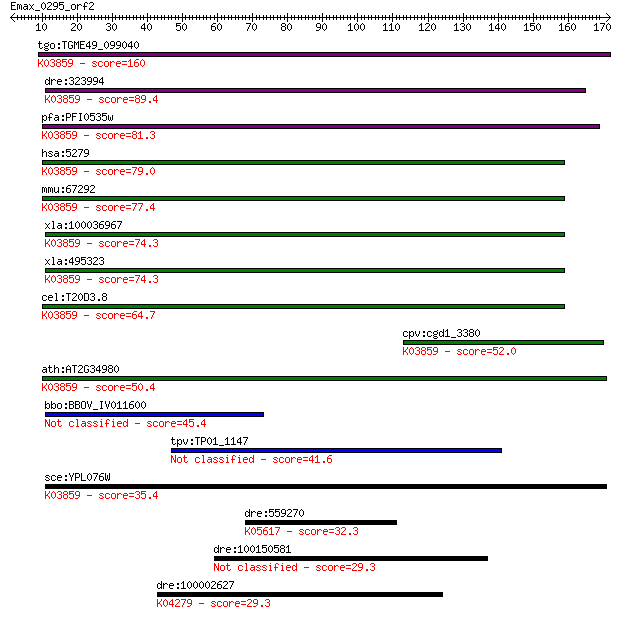
tgo:TGME49_099040 phosphatidylinositol N-acetylglucosaminyltra... 160 1e-39
dre:323994 pigc, wu:fc16d03, zgc:85894; phosphatidylinositol g... 89.4 5e-18
pfa:PFI0535w Phosphatidylinositol N-acetylglucosaminyltransfer... 81.3 1e-15
hsa:5279 PIGC, GPI2, MGC2049; phosphatidylinositol glycan anch... 79.0 7e-15
mmu:67292 Pigc, 3110030E07Rik, AW212108; phosphatidylinositol ... 77.4 2e-14
xla:100036967 hypothetical protein LOC100036967; K03859 phosph... 74.3 2e-13
xla:495323 pigc; phosphatidylinositol glycan anchor biosynthes... 74.3 2e-13
cel:T20D3.8 hypothetical protein; K03859 phosphatidylinositol ... 64.7 1e-10
cpv:cgd1_3380 phosphatidylinositol-glycan-class c, pigC, 8x tr... 52.0 1e-06
ath:AT2G34980 SETH1; SETH1; phosphatidylinositol N-acetylgluco... 50.4 3e-06
bbo:BBOV_IV011600 23.m05886; hypothetical protein 45.4 8e-05
tpv:TP01_1147 hypothetical protein 41.6 0.001
sce:YPL076W GPI2, GCR4; Gpi2p (EC:2.4.1.198); K03859 phosphati... 35.4 0.089
dre:559270 slc1a6, zgc:171923; solute carrier family 1 (high a... 32.3 0.87
dre:100150581 novel protein similar to human and mouse major f... 29.3 6.0
dre:100002627 platelet-activating factor receptor-like; K04279... 29.3 6.4
> tgo:TGME49_099040 phosphatidylinositol N-acetylglucosaminyltransferase
subunit, putative (EC:2.4.1.198); K03859 phosphatidylinositol
glycan, class C
Length=348
Score = 160 bits (406), Expect = 1e-39, Method: Compositional matrix adjust.
Identities = 73/163 (44%), Positives = 115/163 (70%), Gaps = 0/163 (0%)
Query 9 GKWQKILWKKQPHPDCYVDETFLNTLMRNANLTRYMYSDLCKSTVVLTQHLSCILIFLLV 68
+W+K+LW++Q +PD YVD++FL++L+ NAN+ Y+Y+DLC++T +TQH+S ++ F +V
Sbjct 65 ARWEKVLWRRQAYPDNYVDDSFLDSLICNANMRAYVYADLCRATAAVTQHISLLVDFTVV 124
Query 69 YRMIVKRSLAASTLVIFDVVSLPLGYALRWSLRPAPRAARQVLQSAIIVFGVLRILAPVL 128
Y M+ K+ ++ +L D+ L G+ LR A + L S+ + G LRILAP+L
Sbjct 125 YIMLEKKRISVGSLFAVDLALLFFGFVLRLLADEALHKTWRGLWSSFVGMGCLRILAPIL 184
Query 129 QTLTQSFSDDTVISLTSICLLIHIPLNDYSYVYRNPETIDEPL 171
+TLTQ+FS+DTV+ L+ + LL+H L DYSY+YRNP+ +DE L
Sbjct 185 RTLTQTFSEDTVVCLSVVSLLVHTALTDYSYIYRNPDKVDESL 227
> dre:323994 pigc, wu:fc16d03, zgc:85894; phosphatidylinositol
glycan, class C (EC:2.4.1.198); K03859 phosphatidylinositol
glycan, class C
Length=293
Score = 89.4 bits (220), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 52/157 (33%), Positives = 84/157 (53%), Gaps = 7/157 (4%)
Query 11 WQKILWKKQPHPDCYVDETFLNTLMRNANLTRYMYSDLCKSTVVLTQHLSCILIFLLVYR 70
W+K+L+++QP PD YVD FL L RN + +Y Y + + T ++ Q +SC+ +FL ++
Sbjct 15 WRKVLYERQPFPDNYVDRRFLEELRRNIRVRQYRYWAVVRETGLIAQQVSCVALFLTLWS 74
Query 71 MIVKRSLAASTLVIFDVVSLPLGYALRWSLRPAPRAARQV---LQSAIIVFGVLRILAPV 127
+ + L S ++ + GY L L A R LQSA + +PV
Sbjct 75 YMEQGELEPSAVLCVCLGCALFGYGLYEVLGGASDGKRTRLADLQSAAVFLAFTFGFSPV 134
Query 128 LQTLTQSFSDDTVISLTSICLLIHIPLNDYSYVYRNP 164
L+TLT+S S DTV +++++ LL H+ S+ Y P
Sbjct 135 LKTLTESVSTDTVYAMSAVMLLAHL----VSFPYAQP 167
> pfa:PFI0535w Phosphatidylinositol N-acetylglucosaminyltransferase,
putative (EC:2.4.1.198); K03859 phosphatidylinositol
glycan, class C
Length=292
Score = 81.3 bits (199), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 50/159 (31%), Positives = 92/159 (57%), Gaps = 1/159 (0%)
Query 10 KWQKILWKKQPHPDCYVDETFLNTLMRNANLTRYMYSDLCKSTVVLTQHLSCILIFLLVY 69
KW+KIL++ Q + D YV +FL++L+ N + +Y YS +C S + + + +L L+ Y
Sbjct 13 KWKKILYEDQDYADNYVHNSFLSSLLTNFGI-KYKYSHVCHSMLCINHQIMIVLFLLISY 71
Query 70 RMIVKRSLAASTLVIFDVVSLPLGYALRWSLRPAPRAARQVLQSAIIVFGVLRILAPVLQ 129
I K + + + ++V + L L + + + + + AII+ G++ +L+PVL
Sbjct 72 YCIDKNVITQNFIYAVNIVIIILKEILIYQNHNSLNNSLKNVLDAIIIIGIIWLLSPVLI 131
Query 130 TLTQSFSDDTVISLTSICLLIHIPLNDYSYVYRNPETID 168
+LTQ+ SD+TV ++ + LLIH+ + Y ++Y E ID
Sbjct 132 SLTQTHSDNTVYLVSIMLLLIHLMFHKYGFIYEKNENID 170
> hsa:5279 PIGC, GPI2, MGC2049; phosphatidylinositol glycan anchor
biosynthesis, class C (EC:2.4.1.198); K03859 phosphatidylinositol
glycan, class C
Length=297
Score = 79.0 bits (193), Expect = 7e-15, Method: Compositional matrix adjust.
Identities = 48/156 (30%), Positives = 83/156 (53%), Gaps = 8/156 (5%)
Query 10 KWQKILWKKQPHPDCYVDETFLNTLMRNANLTRYMYSDLCKSTVVLTQHLSCILIFLLVY 69
KWQK+L+++QP PD YVD FL L +N + +Y Y + + V+ Q L + +F++++
Sbjct 13 KWQKVLYERQPFPDNYVDRRFLEELRKNIHARKYQYWAVVFESSVVIQQLCSVCVFVVIW 72
Query 70 RMIVKRSLAASTLVIFDVVSLPLGYALRWSLRPAPRAARQV-------LQSAIIVFGVLR 122
+ + LA L+ + S +GY L + L ++ L+SA++
Sbjct 73 WYMDEGLLAPHWLLGTGLASSLIGYVL-FDLIDGGEGRKKSGQTRWADLKSALVFITFTY 131
Query 123 ILAPVLQTLTQSFSDDTVISLTSICLLIHIPLNDYS 158
+PVL+TLT+S S DT+ +++ LL H+ DY
Sbjct 132 GFSPVLKTLTESVSTDTIYAMSVFMLLGHLIFFDYG 167
> mmu:67292 Pigc, 3110030E07Rik, AW212108; phosphatidylinositol
glycan anchor biosynthesis, class C (EC:2.4.1.198); K03859
phosphatidylinositol glycan, class C
Length=297
Score = 77.4 bits (189), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 47/155 (30%), Positives = 79/155 (50%), Gaps = 6/155 (3%)
Query 10 KWQKILWKKQPHPDCYVDETFLNTLMRNANLTRYMYSDLCKSTVVLTQHLSCILIFLLVY 69
KWQK+L+++QP PD YVD+ FL L +N +Y Y + + V+ Q L + +F++++
Sbjct 13 KWQKVLYERQPFPDNYVDQRFLEELRKNIYARKYQYWAVVFESSVVIQQLCSVCVFVVIW 72
Query 70 RMIVKRSLAASTLVIFDVVSLPLGYAL------RWSLRPAPRAARQVLQSAIIVFGVLRI 123
+ + LA L + S +GY L + + R L+S ++
Sbjct 73 WYMDEGLLAPQWLFGTGLASSLVGYVLFDLIDGGDGRKKSGRTRWADLKSTLVFITFTYG 132
Query 124 LAPVLQTLTQSFSDDTVISLTSICLLIHIPLNDYS 158
+PVL+TLT+S S DT+ ++ LL H+ DY
Sbjct 133 FSPVLKTLTESVSTDTIYAMAVFMLLGHLIFFDYG 167
> xla:100036967 hypothetical protein LOC100036967; K03859 phosphatidylinositol
glycan, class C
Length=317
Score = 74.3 bits (181), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 47/154 (30%), Positives = 78/154 (50%), Gaps = 6/154 (3%)
Query 11 WQKILWKKQPHPDCYVDETFLNTLMRNANLTRYMYSDLCKSTVVLTQHLSCILIFLLVYR 70
W+K+L++ QP PD YVD FL L +N + RY Y + V+ Q L + +F +++
Sbjct 34 WKKVLYEHQPFPDNYVDNRFLEELRKNIYVRRYHYWAVVFEAGVVIQQLCSVCVFSVIWW 93
Query 71 MIVKRSLAASTLVIFDVVSLPLGYAL------RWSLRPAPRAARQVLQSAIIVFGVLRIL 124
+ + L+ L ++ LGY L R + R L+SA++
Sbjct 94 YMDQDLLSPQKLCGVSLLLTLLGYILFDAVDKGEGRRDSGRTHWADLKSALVFVAFTYGF 153
Query 125 APVLQTLTQSFSDDTVISLTSICLLIHIPLNDYS 158
+PVL+TLT+S S DT+ +++ + LL H+ DY
Sbjct 154 SPVLKTLTESISTDTIYAMSVLMLLGHLVFFDYG 187
> xla:495323 pigc; phosphatidylinositol glycan anchor biosynthesis,
class C (EC:2.4.1.198); K03859 phosphatidylinositol glycan,
class C
Length=319
Score = 74.3 bits (181), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 46/154 (29%), Positives = 78/154 (50%), Gaps = 6/154 (3%)
Query 11 WQKILWKKQPHPDCYVDETFLNTLMRNANLTRYMYSDLCKSTVVLTQHLSCILIFLLVYR 70
W+K+L++ QP PD YVD FL L +N + RY Y + V+ Q L + +F ++
Sbjct 36 WKKVLYEHQPFPDNYVDNRFLEELRKNIYVRRYHYWAVVFEAGVVIQQLCSVCVFSFIWW 95
Query 71 MIVKRSLAASTLVIFDVVSLPLGYALRWSL------RPAPRAARQVLQSAIIVFGVLRIL 124
+ + L+ L + LGY L ++ R + R L+SA++
Sbjct 96 YMDQDLLSPQKLCGVGLAFTLLGYILFDAVDQGEGRRDSGRTHWADLKSALVFVAFTYGF 155
Query 125 APVLQTLTQSFSDDTVISLTSICLLIHIPLNDYS 158
+PVL+TLT+S S DT+ +++ + LL H+ D+
Sbjct 156 SPVLKTLTESISTDTIYAMSVLMLLGHLVFFDFG 189
> cel:T20D3.8 hypothetical protein; K03859 phosphatidylinositol
glycan, class C
Length=282
Score = 64.7 bits (156), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 44/153 (28%), Positives = 75/153 (49%), Gaps = 6/153 (3%)
Query 10 KWQKILWKKQPHPDCYV--DETFLNTLMRNANLTRYMYSDLCKSTVVLTQHLSCILIFLL 67
WQKIL++KQP PD Y D FL L +N ++ Y Y + HL I ++ +
Sbjct 6 GWQKILYRKQPFPDNYSGGDAQFLKELRKNVSVVHYDYKSAVFGCMNFLTHLDMITMYFV 65
Query 68 VYRMIVKRSLAASTLVIFDVVSLPLGYALRWS--LRPAPRAARQVLQSAIIVFGVLRILA 125
++ I+ + + + +++ V SL + L + L P P A++ ++ +F
Sbjct 66 LFLNILHSNWSIN--ILYSVFSLTIVLYLFFCKFLIPNPANAKEHARTIFTLFIFAYAFT 123
Query 126 PVLQTLTQSFSDDTVISLTSICLLIHIPLNDYS 158
PV++TLT S S DT+ S + I + +DY
Sbjct 124 PVIRTLTTSISTDTIYSTSIITAIFSCFFHDYG 156
> cpv:cgd1_3380 phosphatidylinositol-glycan-class c, pigC, 8x
transmembrane domains (EC:3.5.1.89); K03859 phosphatidylinositol
glycan, class C
Length=274
Score = 52.0 bits (123), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 22/57 (38%), Positives = 43/57 (75%), Gaps = 0/57 (0%)
Query 113 SAIIVFGVLRILAPVLQTLTQSFSDDTVISLTSICLLIHIPLNDYSYVYRNPETIDE 169
+ +++FG L +++P+L TLT SFSDDT+I+L +I L+++ +DY+ +Y++ + I+
Sbjct 101 NLLVIFGFLWLVSPILITLTASFSDDTIIALCTIAFLLYLLSHDYTIIYKDLKEIEH 157
> ath:AT2G34980 SETH1; SETH1; phosphatidylinositol N-acetylglucosaminyltransferase/
transferase; K03859 phosphatidylinositol
glycan, class C
Length=303
Score = 50.4 bits (119), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 50/168 (29%), Positives = 90/168 (53%), Gaps = 8/168 (4%)
Query 10 KWQKILWKKQ--PHPDCYVDETFLNTLMRNANLTRYMYSDLCKSTVVLTQHLSCILIFLL 67
KW+K+ + + D Y DE+FL ++ NAN+ R + K +V ++Q+L + + +L
Sbjct 14 KWRKVAYGGMQIGYDDNYTDESFLEEMVMNANVVRRDLLKVMKDSVSISQYLCIVALVVL 73
Query 68 VYRMIVKRSLAASTLVIFDVVSLPLGYAL----RWSLRPAPRAARQVLQSAIIVFGVLRI 123
V+ ++ SL ++L++ D+ L G+ + + R ++ + G L I
Sbjct 74 VWVHTLESSLDENSLLLLDLSLLASGFLILLLTEEKMLSLSLLLRYLVNISFFTTG-LYI 132
Query 124 LAPVLQTLTQSFSDDTVISLTSICLLIHIPLNDYS-YVYRNPETIDEP 170
LAP+ QTLT+S S D++ ++T LL+H+ L+DYS R P + P
Sbjct 133 LAPIYQTLTRSISSDSIWAVTVSLLLLHLFLHDYSGSTIRAPGALKTP 180
> bbo:BBOV_IV011600 23.m05886; hypothetical protein
Length=106
Score = 45.4 bits (106), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 22/62 (35%), Positives = 37/62 (59%), Gaps = 1/62 (1%)
Query 11 WQKILWKKQPHPDCYVDETFLNTLMRNANLTRYMYSDLCKSTVVLTQHLSCILIFLLVYR 70
W+++++K+Q +P YV FL L N + Y DLC T++LTQH S +L+ ++
Sbjct 38 WERVMYKRQAYPPNYVSSDFLRGLSENGS-NCYTLWDLCPKTMMLTQHCSTMLLMARMFL 96
Query 71 MI 72
+I
Sbjct 97 LI 98
> tpv:TP01_1147 hypothetical protein
Length=115
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 29/95 (30%), Positives = 52/95 (54%), Gaps = 2/95 (2%)
Query 47 DLCKSTVVLTQHLSCILIFLLVYRMIVKRSLAASTLVIFDVVSLPLGYALRWSLRPAPRA 106
DL ++ +V+T H+S I++ +Y ++ + S + + ++S+ L + + L AP+
Sbjct 12 DLLRNMLVVTDHISTIIMLSKLYFLMKNNKIQLSLIYVLILISI-LVFLILMLLNKAPKT 70
Query 107 ARQV-LQSAIIVFGVLRILAPVLQTLTQSFSDDTV 140
L+ I++ L + VLQTL SFSDDTV
Sbjct 71 EYITHLRMFFIMYFTLELFQGVLQTLISSFSDDTV 105
> sce:YPL076W GPI2, GCR4; Gpi2p (EC:2.4.1.198); K03859 phosphatidylinositol
glycan, class C
Length=280
Score = 35.4 bits (80), Expect = 0.089, Method: Compositional matrix adjust.
Identities = 43/171 (25%), Positives = 76/171 (44%), Gaps = 22/171 (12%)
Query 11 WQKILWKKQPHPDCYVDETFLNTLMRNANLTRYMYSDLCKSTVVLTQHLSCILIFLLVYR 70
W+++LW KQ +PD Y D +F+ R + SD S Q L F+ Y+
Sbjct 6 WKRLLWLKQEYPDNYTDPSFIELRARQKAESN-QKSDRKLSEAARAQ---IRLDFISFYQ 61
Query 71 MIVKRSLAASTLVI-----FDVVSLPLGYAL------RWSLRPAPRAARQVLQSAIIVFG 119
I+ S T FD + + + R + P + V S II F
Sbjct 62 TILNTSFIYITFTYIYYYGFDPIPPTIFLSFITLIISRTKVDPLLSSFMDVKSSLIITFA 121
Query 120 VLRILAPVLQTLTQSFSDDTVISLTSICLLIHIPLNDYSYVYRNPETIDEP 170
+L L+PVL++L+++ + D++ +L+ L +I +V + ++ D+P
Sbjct 122 MLT-LSPVLKSLSKTTASDSIWTLSFWLTLWYI------FVISSTKSKDKP 165
> dre:559270 slc1a6, zgc:171923; solute carrier family 1 (high
affinity aspartate/glutamate transporter), member 6; K05617
solute carrier family 1 (high affinity aspartate/glutamate
transporter), member 6
Length=560
Score = 32.3 bits (72), Expect = 0.87, Method: Composition-based stats.
Identities = 18/43 (41%), Positives = 25/43 (58%), Gaps = 0/43 (0%)
Query 68 VYRMIVKRSLAASTLVIFDVVSLPLGYALRWSLRPAPRAARQV 110
+ R VK L + VIF V ++ LG AL ++LRP + RQV
Sbjct 48 ITRNSVKTFLRRNAFVIFTVAAVALGVALGFALRPHNLSIRQV 90
> dre:100150581 novel protein similar to human and mouse major
facilitator superfamily domain containing 9 (MFSD9)
Length=490
Score = 29.3 bits (64), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 26/81 (32%), Positives = 38/81 (46%), Gaps = 6/81 (7%)
Query 59 LSCILIFLLVYRMIVKRSLAASTLVIFDVVSLPLGYALRWSL---RPAPRAARQVLQSAI 115
L+C LIFL V + + ST F +S +G L R +A+ ++ +
Sbjct 376 LTCTLIFLYAAAPNVWQVVLTST---FFAISTTIGRTCITDLELQRGGSQASGTLIGAGQ 432
Query 116 IVFGVLRILAPVLQTLTQSFS 136
V V R+LAP+L L Q FS
Sbjct 433 SVTAVGRVLAPLLSGLAQEFS 453
> dre:100002627 platelet-activating factor receptor-like; K04279
platelet-activating factor receptor
Length=365
Score = 29.3 bits (64), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 22/81 (27%), Positives = 38/81 (46%), Gaps = 4/81 (4%)
Query 43 YMYSDLCKSTVVLTQHLSCILIFLLVYRMIVKRSLAASTLVIFDVVSLPLGYALRWSLRP 102
Y Y+D V++ H S I +F LV+ M++ +++ + ++ +S P R RP
Sbjct 185 YHYTDENTKNVLVITHFSIIGLFFLVFLMVIICNISIARALLSQPISQPRASTGR---RP 241
Query 103 APRAARQVLQSAIIVFGVLRI 123
R+ L+ V GV I
Sbjct 242 GG-TKRRALRMLCAVIGVFVI 261
Lambda K H
0.327 0.138 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4276754328
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40