bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0234_orf1
Length=174
Score E
Sequences producing significant alignments: (Bits) Value
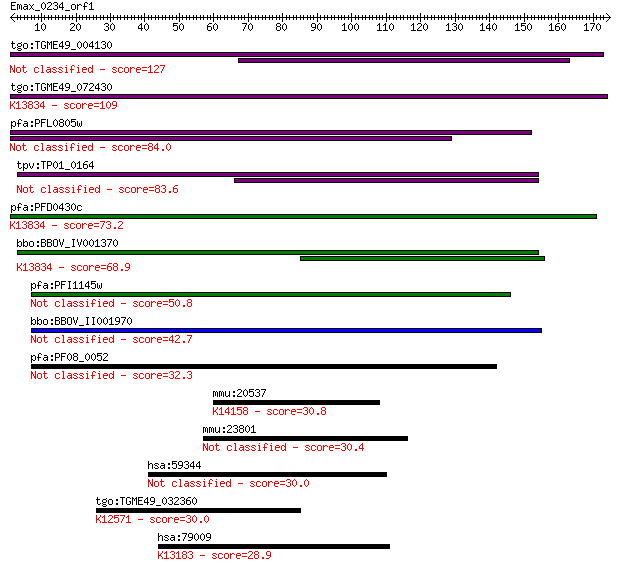
tgo:TGME49_004130 membrane-attack complex / perforin domain-co... 127 3e-29
tgo:TGME49_072430 membrane-attack complex / perforin domain-co... 109 6e-24
pfa:PFL0805w MAC/Perforin, putative 84.0 3e-16
tpv:TP01_0164 hypothetical protein 83.6 3e-16
pfa:PFD0430c MAC/Perforin, putative; K13834 sporozoite microne... 73.2 5e-13
bbo:BBOV_IV001370 21.m02755; MAC/perforin domain containing pr... 68.9 8e-12
pfa:PFI1145w MAC/Perforin, putative 50.8 2e-06
bbo:BBOV_II001970 18.m09950; mac/perforin domain containing me... 42.7 6e-04
pfa:PF08_0052 perforin like protein 5 32.3
mmu:20537 Slc5a1, Sglt1; solute carrier family 5 (sodium/gluco... 30.8 2.7
mmu:23801 Aloxe3, MGC143829, MGC143830, e-LOX-3; arachidonate ... 30.4 3.0
hsa:59344 ALOXE3, E-LOX, MGC119694, MGC119695, MGC119696, eLOX... 30.0 3.9
tgo:TGME49_032360 RNA polymerase Rpb1 C-terminal repeat domain... 30.0 3.9
hsa:79009 DDX50, GU2, GUB, MGC3199, RH-II/GuB; DEAD (Asp-Glu-A... 28.9 9.9
> tgo:TGME49_004130 membrane-attack complex / perforin domain-containing
protein
Length=1054
Score = 127 bits (318), Expect = 3e-29, Method: Composition-based stats.
Identities = 70/179 (39%), Positives = 99/179 (55%), Gaps = 9/179 (5%)
Query 1 PPGFAMCDPHERILFGFAFQVNFLDDGAIAERAEIKSCPAGRDKCDGLERPAKEGDDARI 60
PPGFA C + ++ GFA +NF + G + I SCP GR+KCDG+ + E D+ RI
Sbjct 732 PPGFAKCPEGQVVILGFAMHLNFKEPGT--DNFRIISCPPGREKCDGVGTASSETDEGRI 789
Query 61 FALCGAETITGLEQVVVQSPLKA-----VAVCPQGSLILTGFSLSLTGGREGPLRTGFFP 115
+ LCG E I ++QVV +SP A A CP ++++ GF +S+ GG +G
Sbjct 790 YILCGEEPINEIQQVVAESPAHAGASVLEASCPDETVVVGGFGISVRGGSDGLDSFSIES 849
Query 116 CRAGLPTCTALGVRGTQQNMVWVACVEDTTPGLQRLTNVGA-AVVGVATKRSY-SDGLV 172
C G CT RG+++N +W+ CV+ PGL+ L NV G A KR+ SDG V
Sbjct 850 CTTGQTICTKAPTRGSEKNFLWMMCVDKQYPGLRELVNVAELGSHGNANKRAVNSDGNV 908
Score = 37.0 bits (84), Expect = 0.037, Method: Composition-based stats.
Identities = 22/98 (22%), Positives = 40/98 (40%), Gaps = 2/98 (2%)
Query 67 ETITGLEQVVVQSPLKAVAVCPQGSLILTGFSLSLTGGREGPLRTGFFPCRAGLPTCTAL 126
+ +T QV P A CP+G +++ GF++ L G C G C +
Sbjct 718 KQLTQATQVAWSGPPPGFAKCPEGQVVILGFAMHLNFKEPGTDNFRIISCPPGREKCDGV 777
Query 127 GVRG--TQQNMVWVACVEDTTPGLQRLTNVGAAVVGVA 162
G T + +++ C E+ +Q++ A G +
Sbjct 778 GTASSETDEGRIYILCGEEPINEIQQVVAESPAHAGAS 815
> tgo:TGME49_072430 membrane-attack complex / perforin domain-containing
protein ; K13834 sporozoite microneme protein 2
Length=854
Score = 109 bits (272), Expect = 6e-24, Method: Compositional matrix adjust.
Identities = 64/179 (35%), Positives = 89/179 (49%), Gaps = 6/179 (3%)
Query 1 PPGFAMCDPHERILFGFAFQVNF-LDDGAIAERAEIKSCPAGRDKCDGLERPAKEGDDAR 59
PPG+A C + +LFGFA + NF + I C AGR+KCDG+ GDD R
Sbjct 595 PPGYARCPREQVVLFGFAMRFNFKVTISNNLANYHIAPCTAGREKCDGIGAEEAAGDDER 654
Query 60 IFALCGAETITGLEQVVVQSPLK---AVAVCPQGSLILTGFSLSLTGGREGPLRTGFFPC 116
I+ CG E + QVV ++ AVA CP+ ++I GF +S+ G T PC
Sbjct 655 IYMACGPEVVNEFYQVVAETEAGENVAVATCPEDTVIAFGFGISIGTGFYSSENTQVEPC 714
Query 117 RAGLPTCTALGVRGTQQNMVWVACVEDTTPGLQRLTNVG-AAVVGVATKR-SYSDGLVR 173
AG CT T ++ VW+ C E + PG+ +L N+ G A R +DG+V
Sbjct 715 TAGQTRCTKARTSNTVKSYVWMVCAEKSFPGIAQLNNIAEVGTRGKANSRMKNTDGIVN 773
> pfa:PFL0805w MAC/Perforin, putative
Length=1073
Score = 84.0 bits (206), Expect = 3e-16, Method: Composition-based stats.
Identities = 54/155 (34%), Positives = 76/155 (49%), Gaps = 7/155 (4%)
Query 1 PPGFAMCDPHERILFGFAFQVNFLDDGAIAERAEIKSCPAGRDKC--DGLERPAKEGDDA 58
PPG C IL GF+ +NF + ++ I C ++ C +G E K D
Sbjct 823 PPGLLTCPIGTTILMGFSINLNFYKNKYLSSTNGITLCEPMKESCSGNGFE---KNYSDI 879
Query 59 RIFALCGAETITGLEQVVVQSPL-KAVAVCPQGSLILTGFSLSLTGGREGPLRTGFFPCR 117
RIFALC + + QVV Q K A CP +IL GF+L G + +PCR
Sbjct 880 RIFALCTNKPFDFITQVVQQGEAPKISASCPGELVILFGFALMKGIGSSSANKIDIYPCR 939
Query 118 AGLPTCTA-LGVRGTQQNMVWVACVEDTTPGLQRL 151
G +C A L +Q+M+++ACV+ TT GL+ L
Sbjct 940 TGQNSCEAVLQNHKFKQSMIYLACVDKTTNGLEYL 974
Score = 37.4 bits (85), Expect = 0.028, Method: Composition-based stats.
Identities = 35/143 (24%), Positives = 53/143 (37%), Gaps = 19/143 (13%)
Query 1 PPGFAMCDPHERILFGFAFQVNFLDDGAIAERAEIKSCPAGRDKCDGLERPAKEGDDARI 60
P A C ILFGFA + A + +I C G++ C+ + + K + I
Sbjct 903 PKISASCPGELVILFGFALMKGI--GSSSANKIDIYPCRTGQNSCEAVLQNHK-FKQSMI 959
Query 61 FALCGAETITGLEQVVVQSPLKAVA---------------VCPQGSLILTGFSLSLTGGR 105
+ C +T GLE + S K + CPQ + ++ GFSL
Sbjct 960 YLACVDKTTNGLEYLQTYSKTKNLGDVISDKYKSDGYLNFSCPQNNTLVFGFSLEFHTNF 1019
Query 106 EGPLRTGFFPCRAGLPTCTALGV 128
+ R F C C G+
Sbjct 1020 QAT-RNNFLNCSKYTNICEISGI 1041
> tpv:TP01_0164 hypothetical protein
Length=1182
Score = 83.6 bits (205), Expect = 3e-16, Method: Composition-based stats.
Identities = 56/159 (35%), Positives = 77/159 (48%), Gaps = 16/159 (10%)
Query 3 GFAMCDPHERILFGFAFQVNFLDDGAIAERAE----IKSCPAGRDKCD-GLERPAKEGDD 57
G A+C I+ GF+ + L I + E I CP G +KC + P E
Sbjct 944 GSAICPNKTVIIMGFSLSI--LKKKNIVGKNEFTLHITQCPVGEEKCIVSSDNPMSE--- 998
Query 58 ARIFALCGAETITGLEQVV---VQSPLKAVAVCPQGSLILTGFSLSLTGGREGPLRTGFF 114
+RI+A+CG +TI L Q + P A A CP G I GF+LS+ G L T +
Sbjct 999 SRIWAVCGEDTIPLLNQQTSSEIDEP--ATATCPVGYSIAYGFALSVPKGNVA-LNTDSY 1055
Query 115 PCRAGLPTCTALGVRGTQQNMVWVACVEDTTPGLQRLTN 153
CR+G +CT T N VW+ACVE+ P L ++N
Sbjct 1056 ACRSGTQSCTHESTDKTATNAVWIACVENGAPQLSEISN 1094
Score = 40.8 bits (94), Expect = 0.002, Method: Composition-based stats.
Identities = 27/94 (28%), Positives = 38/94 (40%), Gaps = 8/94 (8%)
Query 66 AETITGLEQVVVQSPLKAVAVCPQGSLILTGFSLS------LTGGREGPLRTGFFPCRAG 119
A I Q+V A+CP ++I+ GFSLS + G E L C G
Sbjct 927 ASVIEKGHQIVYSGNKSGSAICPNKTVIIMGFSLSILKKKNIVGKNEFTLH--ITQCPVG 984
Query 120 LPTCTALGVRGTQQNMVWVACVEDTTPGLQRLTN 153
C ++ +W C EDT P L + T+
Sbjct 985 EEKCIVSSDNPMSESRIWAVCGEDTIPLLNQQTS 1018
> pfa:PFD0430c MAC/Perforin, putative; K13834 sporozoite microneme
protein 2
Length=842
Score = 73.2 bits (178), Expect = 5e-13, Method: Composition-based stats.
Identities = 52/176 (29%), Positives = 85/176 (48%), Gaps = 7/176 (3%)
Query 1 PPGFAMCDPHERI-LFGFAFQVNFLDDGAIAERAEIKSCPAGRDKCDGLERPAKEGDDAR 59
PP A C PH ++ +FGF+ + NF D+ + I+ C AG + C + + + D +
Sbjct 600 PPINAQC-PHGKVVMFGFSLKQNFWDNTNALKGYNIEVCEAGSNSCTSKQGSSNKYDTSY 658
Query 60 IFALCGAETITGLEQVVVQSPLKAVAV-CPQGSLILTGFSLSLTGGREGPLRTGF-FPCR 117
++ CG + + EQV+ +S V CP IL GF +S + GR + PC
Sbjct 659 LYMECGDQPLPFSEQVISESTSTYNTVKCPNDYSILLGFGISSSSGRINSAEYVYSTPCI 718
Query 118 AGLPTCTALGVRGTQQNMVWVACVEDTT-PGLQRLTNVG--AAVVGVATKRSYSDG 170
G+ +C+ Q++ ++V CV+ T G+ L+ V A V + YSDG
Sbjct 719 PGMKSCSLNMNNDNQKSYIYVLCVDTTIWSGVNNLSLVALDGAHGKVNRSKKYSDG 774
> bbo:BBOV_IV001370 21.m02755; MAC/perforin domain containing
protein; K13834 sporozoite microneme protein 2
Length=978
Score = 68.9 bits (167), Expect = 8e-12, Method: Composition-based stats.
Identities = 52/163 (31%), Positives = 78/163 (47%), Gaps = 25/163 (15%)
Query 3 GFAMCDPHERILFGFAFQVNFLDDGAIAERAEIKS----------CPAGRDKCDGLERPA 52
G A+C + I+ GF+ ++ + RA + S CP G++KC P
Sbjct 685 GSAVCPNGKVIMMGFSVVIS-------SSRATVFSKPQYTISMTPCPIGQEKCMVSVPPG 737
Query 53 KEGDDARIFALCGAETITGL-EQVVVQSPLKAVAVCPQGSLILTGFSLSL-TGGREGPLR 110
EG R++ LCG+E+I L ++ V + A A CP I GF LS+ G + P+
Sbjct 738 AEG---RVWILCGSESIPLLIQETNVANNEAATAQCPDEYAIAFGFGLSIPDGAKLTPVD 794
Query 111 TGFFPCRAGLPTCTALGVRGTQQNMVWVACVEDTTPGLQRLTN 153
+ CRAG +CT + + N VW+ACVE P L + N
Sbjct 795 C--YACRAGQQSCTQASPK-SPYNAVWIACVEKNAPELGSIVN 834
Score = 40.0 bits (92), Expect = 0.004, Method: Composition-based stats.
Identities = 25/76 (32%), Positives = 36/76 (47%), Gaps = 6/76 (7%)
Query 85 AVCPQGSLILTGFSLSLTGGR----EGPLRT-GFFPCRAGLPTCTALGVRGTQQNMVWVA 139
AVCP G +I+ GFS+ ++ R P T PC G C + V + VW+
Sbjct 687 AVCPNGKVIMMGFSVVISSSRATVFSKPQYTISMTPCPIGQEKCM-VSVPPGAEGRVWIL 745
Query 140 CVEDTTPGLQRLTNVG 155
C ++ P L + TNV
Sbjct 746 CGSESIPLLIQETNVA 761
> pfa:PFI1145w MAC/Perforin, putative
Length=821
Score = 50.8 bits (120), Expect = 2e-06, Method: Composition-based stats.
Identities = 36/142 (25%), Positives = 62/142 (43%), Gaps = 11/142 (7%)
Query 7 CDPHERILFGFAFQVNFLDDGAIAERAEIKSCPAGRDKCDGLERPAKEGDDAR--IFALC 64
C+ + L GF+ + +D + + + SC DKC +K D+A IFA+C
Sbjct 605 CEEKQNFLLGFSLSIP--NDLSNLKDFYLNSCDEDSDKC-----YSKMSDNAYSYIFAMC 657
Query 65 GAETITGLEQVVVQS-PLKAVAVCPQGSLILTGFSLSLTGGREGPLRTGFFPCRAGLPTC 123
E I EQ V L + + +IL GF +S+ + P+ +PC+ G +C
Sbjct 658 KEEMIPFFEQKVKSGVGLLTLECSEKNQVILFGFGISVLNTND-PISISLYPCKYGKASC 716
Query 124 TALGVRGTQQNMVWVACVEDTT 145
+ G +W+ C + +
Sbjct 717 SMQGSTDQSAVGLWIVCAHEES 738
> bbo:BBOV_II001970 18.m09950; mac/perforin domain containing
membrane protein
Length=559
Score = 42.7 bits (99), Expect = 6e-04, Method: Composition-based stats.
Identities = 36/152 (23%), Positives = 62/152 (40%), Gaps = 17/152 (11%)
Query 7 CDPHERILFGFAFQVNFLDDGAIAERA-EIKSCPAGRDKCDGLERPAKEGDDARIFALCG 65
C +++++FGF ++ I +R + CP C ER K D I+ LCG
Sbjct 399 CPHNDKVIFGFILEME------IEQRTFSVYQCPTDAYSCSK-ERQKK--CDIVIWMLCG 449
Query 66 AETITGLEQVVVQ---SPLKAVAVCPQGSLILTGFSLSLTGGREGPLRTGFFPCRAGLPT 122
+ + Q S + C G +LTGF + ++ L PC G
Sbjct 450 SSMGLNVMQYAHNFDASNSEKEVKCLSGYKLLTGFIAESSPEKDKSL-ANLIPCHTGADL 508
Query 123 CTALGVRGTQQNMVWVACVEDTTPGLQRLTNV 154
C + + + +W C+++ PGL R + +
Sbjct 509 CRS---NSSLETHIWAVCIDERLPGLGRTSTL 537
> pfa:PF08_0052 perforin like protein 5
Length=676
Score = 32.3 bits (72), Expect = 0.85, Method: Composition-based stats.
Identities = 33/141 (23%), Positives = 53/141 (37%), Gaps = 16/141 (11%)
Query 7 CDPHERILFGFAF--QVNFLDDGAIAERAEIKSCPAGRDKCDGLERPAKEGDDARIFA-- 62
C ++IL GF + +D I I CP+ G+ E D F+
Sbjct 455 CKNGDKILSGFILTNKKKSYEDNHI-----IHMCPSNTVCSSGIN---IESDKNFEFSWI 506
Query 63 LCGAETITGLEQVVVQSPLKA--VAVCPQGSLILTGFSLSLTGGREGPLRTGFFPCRAGL 120
LC E + + Q++ ++ + A CP I GF SLT + PC +
Sbjct 507 LCSKENRSEIHQILTKNTFQGNGKASCPYNMKI--GFGFSLTFQKSINTNIKIEPCESNK 564
Query 121 PTCTALGVRGTQQNMVWVACV 141
C + + Q W+ C+
Sbjct 565 KECKRTNLASSSQTYFWINCL 585
> mmu:20537 Slc5a1, Sglt1; solute carrier family 5 (sodium/glucose
cotransporter), member 1; K14158 solute carrier family
5 (sodium/glucose cotransporter), member 1
Length=665
Score = 30.8 bits (68), Expect = 2.7, Method: Composition-based stats.
Identities = 17/48 (35%), Positives = 24/48 (50%), Gaps = 0/48 (0%)
Query 60 IFALCGAETITGLEQVVVQSPLKAVAVCPQGSLILTGFSLSLTGGREG 107
+ A+ TITG V+ + A+ GS ILTGF+ + GG E
Sbjct 184 LLAITALYTITGGLAAVIYTDTLQTAIMLVGSFILTGFAFNEVGGYEA 231
> mmu:23801 Aloxe3, MGC143829, MGC143830, e-LOX-3; arachidonate
lipoxygenase 3
Length=711
Score = 30.4 bits (67), Expect = 3.0, Method: Composition-based stats.
Identities = 22/59 (37%), Positives = 28/59 (47%), Gaps = 4/59 (6%)
Query 57 DARIFALCGAETITGLEQVVVQSPLKAVAVCPQGSLILTGFSLSLTGGREGPLRTGFFP 115
D I A I GL+Q V +PL + + PQG L+ LS T G E P+ F P
Sbjct 327 DYWILAEAPVHCINGLQQYVT-APLCLLWLNPQGVLLPLAIQLSQTPGPESPI---FLP 381
> hsa:59344 ALOXE3, E-LOX, MGC119694, MGC119695, MGC119696, eLOX3;
arachidonate lipoxygenase 3 (EC:1.13.11.-)
Length=843
Score = 30.0 bits (66), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 23/71 (32%), Positives = 33/71 (46%), Gaps = 3/71 (4%)
Query 41 GRDKC--DGLERPAKEGDDARIFALCGAETITGLEQVVVQSPLKAVAVCPQGSLILTGFS 98
G+D C LER D I A + G +Q V +PL + + PQG+L+
Sbjct 441 GQDTCLQTELERGNIFLADYWILAEAPTHCLNGRQQYVA-APLCLLWLSPQGALVPLAIQ 499
Query 99 LSLTGGREGPL 109
LS T G + P+
Sbjct 500 LSQTPGPDSPI 510
> tgo:TGME49_032360 RNA polymerase Rpb1 C-terminal repeat domain-containing
protein / exonuclease domain-containing protein
(EC:3.1.13.4); K12571 PAB-dependent poly(A)-specific ribonuclease
subunit 2 [EC:3.1.13.4]
Length=2155
Score = 30.0 bits (66), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 21/59 (35%), Positives = 28/59 (47%), Gaps = 1/59 (1%)
Query 26 DGAIAERAEIKSCPAGRDKCDGLERPAKEGDDARIFALCGAETITGLEQVVVQSPLKAV 84
DG + E A S + RD G ER +E +R + G ET G E+ +SPL A
Sbjct 1095 DGGLREEAAAVSA-SSRDTVTGEERSVREEKRSREQDIGGRETRNGEEKEQDESPLSAY 1152
> hsa:79009 DDX50, GU2, GUB, MGC3199, RH-II/GuB; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 50 (EC:3.6.4.13); K13183 ATP-dependent
RNA helicase DDX50 [EC:3.6.4.13]
Length=737
Score = 28.9 bits (63), Expect = 9.9, Method: Composition-based stats.
Identities = 18/73 (24%), Positives = 34/73 (46%), Gaps = 6/73 (8%)
Query 44 KCDGLERPAKEGDDARIF------ALCGAETITGLEQVVVQSPLKAVAVCPQGSLILTGF 97
+C +RPA GD +++ A+ ET + ++ + +K A C G + +
Sbjct 368 QCHWSQRPAVIGDVLQVYSGSEGRAIIFCETKKNVTEMAMNPHIKQNAQCLHGDIAQSQR 427
Query 98 SLSLTGGREGPLR 110
++L G REG +
Sbjct 428 EITLKGFREGSFK 440
Lambda K H
0.322 0.139 0.433
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4471152252
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40