bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0230_orf1
Length=131
Score E
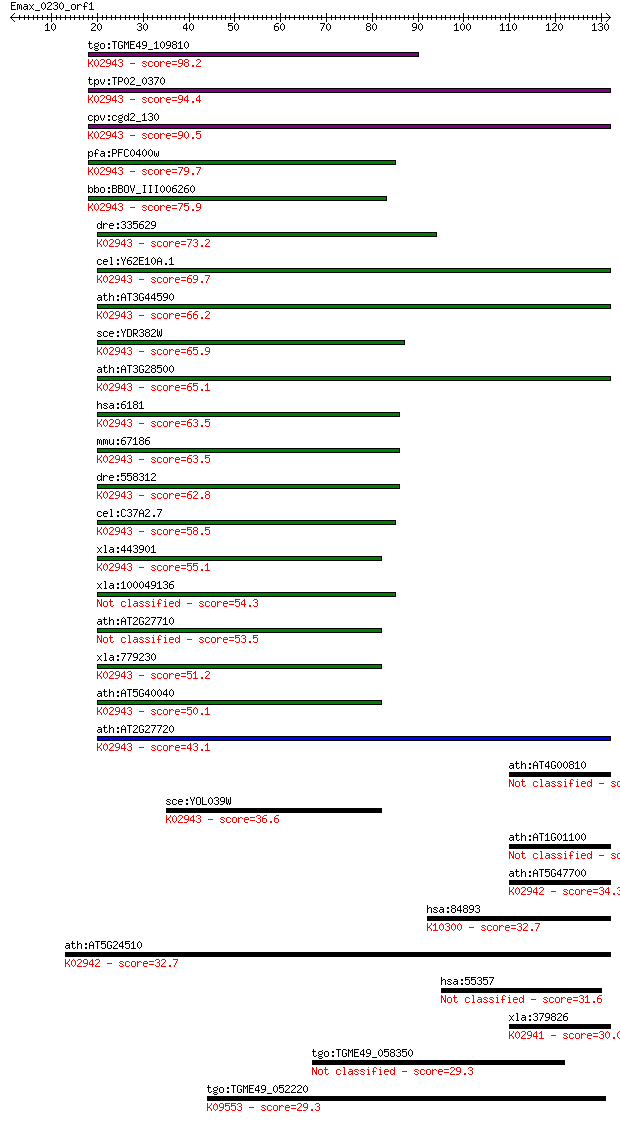
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_109810 60S acidic ribosomal protein P2, putative ; ... 98.2 6e-21
tpv:TP02_0370 60S acidic ribosomal protein P2; K02943 large su... 94.4 8e-20
cpv:cgd2_130 60S acidic ribosomal protein LP2 ; K02943 large s... 90.5 1e-18
pfa:PFC0400w 60S Acidic ribosomal protein P2, putative; K02943... 79.7 2e-15
bbo:BBOV_III006260 17.m07556; 60S acidic ribosomal protein P2 ... 75.9 3e-14
dre:335629 rplp2, RpP1, wu:fb05g05, wu:fj30h02, zgc:73372; rib... 73.2 2e-13
cel:Y62E10A.1 rla-2; Ribosomal protein, Large subunit, Acidic ... 69.7 2e-12
ath:AT3G44590 60S acidic ribosomal protein P2 (RPP2D); K02943 ... 66.2 2e-11
sce:YDR382W RPP2B, RPL45, YPA1; Rpp2bp; K02943 large subunit r... 65.9 3e-11
ath:AT3G28500 60S acidic ribosomal protein P2 (RPP2C); K02943 ... 65.1 5e-11
hsa:6181 RPLP2, D11S2243E, MGC71408, P2, RPP2; ribosomal prote... 63.5 1e-10
mmu:67186 Rplp2, 2700049I22Rik; ribosomal protein, large P2; K... 63.5 2e-10
dre:558312 rplp2l; ribosomal protein, large P2, like; K02943 l... 62.8 3e-10
cel:C37A2.7 hypothetical protein; K02943 large subunit ribosom... 58.5 5e-09
xla:443901 MGC80163 protein; K02943 large subunit ribosomal pr... 55.1 5e-08
xla:100049136 hypothetical protein LOC100049136 54.3 1e-07
ath:AT2G27710 60S acidic ribosomal protein P2 (RPP2B) 53.5 2e-07
xla:779230 rplp2, MGC154377; ribosomal protein, large, P2; K02... 51.2 7e-07
ath:AT5G40040 60S acidic ribosomal protein P2 (RPP2E); K02943 ... 50.1 2e-06
ath:AT2G27720 60S acidic ribosomal protein P2 (RPP2A); K02943 ... 43.1 2e-04
ath:AT4G00810 60S acidic ribosomal protein P1 (RPP1B) 37.4 0.011
sce:YOL039W RPP2A, RPL44, RPLA2; Ribosomal protein P2 alpha, a... 36.6 0.021
ath:AT1G01100 60S acidic ribosomal protein P1 (RPP1A) 34.3 0.092
ath:AT5G47700 60S acidic ribosomal protein P1 (RPP1C); K02942 ... 34.3 0.094
hsa:84893 FBXO18, FBH1, FLJ14590, Fbx18, MGC131916, MGC141935,... 32.7 0.29
ath:AT5G24510 60s acidic ribosomal protein P1, putative; K0294... 32.7 0.29
hsa:55357 TBC1D2, DKFZp761D1823, FLJ10702, FLJ16244, FLJ42782,... 31.6 0.74
xla:379826 rplp0, MGC53408, arbp; ribosomal protein, large, P0... 30.0 1.7
tgo:TGME49_058350 hypothetical protein 29.3 3.3
tgo:TGME49_052220 Hsc70/Hsp90-organizing protein, putative ; K... 29.3 3.7
> tgo:TGME49_109810 60S acidic ribosomal protein P2, putative
; K02943 large subunit ribosomal protein LP2
Length=113
Score = 98.2 bits (243), Expect = 6e-21, Method: Compositional matrix adjust.
Identities = 46/72 (63%), Positives = 58/72 (80%), Gaps = 0/72 (0%)
Query 18 MAMKYVAAYLMNVLGGKDAPTAKDVAQTIEAVGGDVDEQVLNALITAMEGKTAHEMIAAG 77
MAMKYVAAYLM VL G DAPT K V +T+ +VG DV++ +++A A+EGKT HE+IAAG
Sbjct 1 MAMKYVAAYLMVVLSGTDAPTKKQVEKTLSSVGIDVEDDIMDAFFKAVEGKTPHELIAAG 60
Query 78 MEKLQKIPTGGA 89
MEKLQK+P+GG
Sbjct 61 MEKLQKVPSGGV 72
> tpv:TP02_0370 60S acidic ribosomal protein P2; K02943 large
subunit ribosomal protein LP2
Length=110
Score = 94.4 bits (233), Expect = 8e-20, Method: Compositional matrix adjust.
Identities = 55/114 (48%), Positives = 73/114 (64%), Gaps = 4/114 (3%)
Query 18 MAMKYVAAYLMNVLGGKDAPTAKDVAQTIEAVGGDVDEQVLNALITAMEGKTAHEMIAAG 77
M++KYVA+YL+ V G D+P+ DV + +VG +VDE LNA +A+ GK+ HE I+AG
Sbjct 1 MSLKYVASYLLAVTCGNDSPSKDDVRDVLNSVGSEVDEDALNAFFSAVSGKSVHETISAG 60
Query 78 MEKLQKIPTGGAVAAAPAAGGAAPAAGGSAPAKEEKKEEPEEEEDGDMGLSLFD 131
+ KLQ +P GG VA A A+G S +E KKE EEE+ DMG SLFD
Sbjct 61 LSKLQTLPAGGGVAVAS----TVQASGSSEKQEESKKEPEPEEEEDDMGFSLFD 110
> cpv:cgd2_130 60S acidic ribosomal protein LP2 ; K02943 large
subunit ribosomal protein LP2
Length=112
Score = 90.5 bits (223), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 58/114 (50%), Positives = 82/114 (71%), Gaps = 2/114 (1%)
Query 18 MAMKYVAAYLMNVLGGKDAPTAKDVAQTIEAVGGDVDEQVLNALITAMEGKTAHEMIAAG 77
M MKYVAAYL+ V GK+ PTA D+ + +E+VG + D+ +++ LI+ M GK +HE+IA+G
Sbjct 1 MGMKYVAAYLLCVSSGKEQPTASDIKKVLESVGIEYDQSIIDVLISNMSGKLSHEVIASG 60
Query 78 MEKLQKIPTGGAVAAAPAAGGAAPAAGGSAPAKEEKKEEPEEEEDGDMGLSLFD 131
+ KLQ +PTGG + AA + AA SAPA+++K+EE EEE GD+G SLFD
Sbjct 61 LSKLQSVPTGGVAVSGGAAAASGGAAQDSAPAEKKKEEEEEEE--GDLGFSLFD 112
> pfa:PFC0400w 60S Acidic ribosomal protein P2, putative; K02943
large subunit ribosomal protein LP2
Length=112
Score = 79.7 bits (195), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 36/67 (53%), Positives = 50/67 (74%), Gaps = 0/67 (0%)
Query 18 MAMKYVAAYLMNVLGGKDAPTAKDVAQTIEAVGGDVDEQVLNALITAMEGKTAHEMIAAG 77
MAMKYVAAYLM VLGG + P+ K+V + AV DV+++VLN I +++GK+ HE+I G
Sbjct 1 MAMKYVAAYLMCVLGGNENPSTKEVKNVLGAVNADVEDEVLNNFIDSLKGKSCHELITDG 60
Query 78 MEKLQKI 84
++KLQ I
Sbjct 61 LKKLQNI 67
> bbo:BBOV_III006260 17.m07556; 60S acidic ribosomal protein P2
(L12EI); K02943 large subunit ribosomal protein LP2
Length=112
Score = 75.9 bits (185), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 34/65 (52%), Positives = 47/65 (72%), Gaps = 0/65 (0%)
Query 18 MAMKYVAAYLMNVLGGKDAPTAKDVAQTIEAVGGDVDEQVLNALITAMEGKTAHEMIAAG 77
MA+KYV++YL+ V G + P+ D+ + ++AVG DVDE+ L L+ +M GKT HE IAAG
Sbjct 1 MALKYVSSYLLAVAAGNENPSVDDLKKILDAVGSDVDEECLQGLVDSMSGKTVHETIAAG 60
Query 78 MEKLQ 82
M KLQ
Sbjct 61 MTKLQ 65
> dre:335629 rplp2, RpP1, wu:fb05g05, wu:fj30h02, zgc:73372; ribosomal
protein, large P2; K02943 large subunit ribosomal protein
LP2
Length=115
Score = 73.2 bits (178), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 33/74 (44%), Positives = 52/74 (70%), Gaps = 0/74 (0%)
Query 20 MKYVAAYLMNVLGGKDAPTAKDVAQTIEAVGGDVDEQVLNALITAMEGKTAHEMIAAGME 79
M+YVAAYL+ VLGG P+AKD+ + +VG + D++ LN +++ + GK +E++ AG+
Sbjct 1 MRYVAAYLLAVLGGNTNPSAKDIKNILGSVGIEADDERLNKVVSELNGKDINEVMNAGLS 60
Query 80 KLQKIPTGGAVAAA 93
KL +P GGAVA +
Sbjct 61 KLASVPAGGAVAVS 74
> cel:Y62E10A.1 rla-2; Ribosomal protein, Large subunit, Acidic
(P1) family member (rla-2); K02943 large subunit ribosomal
protein LP2
Length=110
Score = 69.7 bits (169), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 49/112 (43%), Positives = 64/112 (57%), Gaps = 2/112 (1%)
Query 20 MKYVAAYLMNVLGGKDAPTAKDVAQTIEAVGGDVDEQVLNALITAMEGKTAHEMIAAGME 79
M+YV+AYL+ VLGG P D+ + AVG D D + +++ + GKT E+IA G
Sbjct 1 MRYVSAYLLAVLGGNANPKVDDLKNILSAVGVDADAETAKLVVSRLAGKTVEELIAEGSA 60
Query 80 KLQKIPTGGAVAAAPAAGGAAPAAGGSAPAKEEKKEEPEEEEDGDMGLSLFD 131
L + G A A AA AA A +A +K KKEEP+EE D DMG LFD
Sbjct 61 GL--VSVSGGAAPAAAAAPAAGGAAPAADSKPAKKEEPKEESDDDMGFGLFD 110
> ath:AT3G44590 60S acidic ribosomal protein P2 (RPP2D); K02943
large subunit ribosomal protein LP2
Length=111
Score = 66.2 bits (160), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 51/112 (45%), Positives = 67/112 (59%), Gaps = 1/112 (0%)
Query 20 MKYVAAYLMNVLGGKDAPTAKDVAQTIEAVGGDVDEQVLNALITAMEGKTAHEMIAAGME 79
MK AA+L+ VLGG P+A ++ I AVG DVD + + L+ + GK E+IA+G E
Sbjct 1 MKVAAAFLLAVLGGNANPSADNIKDIIGAVGADVDGESIELLLKEVSGKDIAELIASGRE 60
Query 80 KLQKIPTGGAVAAAPAAGGAAPAAGGSAPAKEEKKEEPEEEEDGDMGLSLFD 131
KL +P+GG VA + A A A KE KKEE EE +D DMG SLF+
Sbjct 61 KLASVPSGGGVAVSAAPSSGGGGAAAPAEKKEAKKEEKEESDD-DMGFSLFE 111
> sce:YDR382W RPP2B, RPL45, YPA1; Rpp2bp; K02943 large subunit
ribosomal protein LP2
Length=110
Score = 65.9 bits (159), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 32/68 (47%), Positives = 48/68 (70%), Gaps = 1/68 (1%)
Query 20 MKYVAAYLMNVLGGKDAPTAKDVAQTIEAVGGDVDEQVLNALITAMEGK-TAHEMIAAGM 78
MKY+AAYL+ V GG AP+A D+ +E+VG +VDE +N L++++EGK + E+IA G
Sbjct 1 MKYLAAYLLLVQGGNAAPSAADIKAVVESVGAEVDEARINELLSSLEGKGSLEEIIAEGQ 60
Query 79 EKLQKIPT 86
+K +PT
Sbjct 61 KKFATVPT 68
> ath:AT3G28500 60S acidic ribosomal protein P2 (RPP2C); K02943
large subunit ribosomal protein LP2
Length=115
Score = 65.1 bits (157), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 44/115 (38%), Positives = 68/115 (59%), Gaps = 3/115 (2%)
Query 20 MKYVAAYLMNVLGGKDAPTAKDVAQTIEAVGGDVDEQVLNALITAMEGKTAHEMIAAGME 79
MK +AA+L+ LGG + PT+ D+ + +E+VG ++DE ++ L + ++ E+IAAG E
Sbjct 1 MKVIAAFLLAKLGGNENPTSNDLKKILESVGAEIDETKIDLLFSLIKDHDVTELIAAGRE 60
Query 80 KLQKIPTGGAVAAAPAAGGAAPAAGGSAPAKEEKK--EEPEEEEDGDMG-LSLFD 131
K+ + +GG A A GG AA + P E KK EE ++E D G + LFD
Sbjct 61 KMSALSSGGPAVAMVAGGGGGGAASAAEPVAESKKKVEEVKDESSDDAGMMGLFD 115
> hsa:6181 RPLP2, D11S2243E, MGC71408, P2, RPP2; ribosomal protein,
large, P2; K02943 large subunit ribosomal protein LP2
Length=115
Score = 63.5 bits (153), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 27/66 (40%), Positives = 46/66 (69%), Gaps = 0/66 (0%)
Query 20 MKYVAAYLMNVLGGKDAPTAKDVAQTIEAVGGDVDEQVLNALITAMEGKTAHEMIAAGME 79
M+YVA+YL+ LGG +P+AKD+ + +++VG + D+ LN +I+ + GK ++IA G+
Sbjct 1 MRYVASYLLAALGGNSSPSAKDIKKILDSVGIEADDDRLNKVISELNGKNIEDVIAQGIG 60
Query 80 KLQKIP 85
KL +P
Sbjct 61 KLASVP 66
> mmu:67186 Rplp2, 2700049I22Rik; ribosomal protein, large P2;
K02943 large subunit ribosomal protein LP2
Length=115
Score = 63.5 bits (153), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 27/66 (40%), Positives = 46/66 (69%), Gaps = 0/66 (0%)
Query 20 MKYVAAYLMNVLGGKDAPTAKDVAQTIEAVGGDVDEQVLNALITAMEGKTAHEMIAAGME 79
M+YVA+YL+ LGG +P+AKD+ + +++VG + D+ LN +I+ + GK ++IA G+
Sbjct 1 MRYVASYLLAALGGNSSPSAKDIKKILDSVGIEADDDRLNKVISELNGKNIEDVIAQGVG 60
Query 80 KLQKIP 85
KL +P
Sbjct 61 KLASVP 66
> dre:558312 rplp2l; ribosomal protein, large P2, like; K02943
large subunit ribosomal protein LP2
Length=114
Score = 62.8 bits (151), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 27/66 (40%), Positives = 44/66 (66%), Gaps = 0/66 (0%)
Query 20 MKYVAAYLMNVLGGKDAPTAKDVAQTIEAVGGDVDEQVLNALITAMEGKTAHEMIAAGME 79
M+YVAAYL+ LGGKD+P+ D+ + +E+VG + D+ ++ ++ + GK E+IA G
Sbjct 1 MRYVAAYLLAALGGKDSPSTGDIKKILESVGIEADDVRMSKVVAELNGKNVEEVIAQGFS 60
Query 80 KLQKIP 85
KL +P
Sbjct 61 KLASVP 66
> cel:C37A2.7 hypothetical protein; K02943 large subunit ribosomal
protein LP2
Length=107
Score = 58.5 bits (140), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 29/65 (44%), Positives = 44/65 (67%), Gaps = 0/65 (0%)
Query 20 MKYVAAYLMNVLGGKDAPTAKDVAQTIEAVGGDVDEQVLNALITAMEGKTAHEMIAAGME 79
MKY+ AYL+ LGG +P+A+DV + +EA G D D + N+++ A++GKT E+IA G
Sbjct 1 MKYLGAYLLATLGGNASPSAQDVLKVLEAGGLDCDMENANSVVDALKGKTISEVIAQGKV 60
Query 80 KLQKI 84
KL +
Sbjct 61 KLSSV 65
> xla:443901 MGC80163 protein; K02943 large subunit ribosomal
protein LP2
Length=115
Score = 55.1 bits (131), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 26/62 (41%), Positives = 41/62 (66%), Gaps = 0/62 (0%)
Query 20 MKYVAAYLMNVLGGKDAPTAKDVAQTIEAVGGDVDEQVLNALITAMEGKTAHEMIAAGME 79
M+YVAAYL+ VLGG + PT D+ + + +VG + D+Q ++ ++GK+ E+IA G
Sbjct 1 MRYVAAYLLAVLGGSECPTIADLTKILNSVGIETDQQRAEKVVGELKGKSIDEVIAQGNS 60
Query 80 KL 81
KL
Sbjct 61 KL 62
> xla:100049136 hypothetical protein LOC100049136
Length=111
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 24/65 (36%), Positives = 43/65 (66%), Gaps = 0/65 (0%)
Query 20 MKYVAAYLMNVLGGKDAPTAKDVAQTIEAVGGDVDEQVLNALITAMEGKTAHEMIAAGME 79
M+YVAAYL+ LGGK +P+A D+ +++VG D D++ + +I + GK +++ +G+
Sbjct 1 MRYVAAYLLAALGGKTSPSAGDIKSILKSVGIDADDERVKKVIGELGGKDLDDVVNSGLA 60
Query 80 KLQKI 84
KL +
Sbjct 61 KLSSV 65
> ath:AT2G27710 60S acidic ribosomal protein P2 (RPP2B)
Length=115
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 27/62 (43%), Positives = 41/62 (66%), Gaps = 0/62 (0%)
Query 20 MKYVAAYLMNVLGGKDAPTAKDVAQTIEAVGGDVDEQVLNALITAMEGKTAHEMIAAGME 79
MK VAAYL+ VL GK +PT+ D+ + +VG + ++ + L+ ++GK E+IAAG E
Sbjct 1 MKVVAAYLLAVLSGKASPTSADIKTILGSVGAETEDSQIELLLKEVKGKDLAELIAAGRE 60
Query 80 KL 81
KL
Sbjct 61 KL 62
> xla:779230 rplp2, MGC154377; ribosomal protein, large, P2; K02943
large subunit ribosomal protein LP2
Length=115
Score = 51.2 bits (121), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 24/62 (38%), Positives = 40/62 (64%), Gaps = 0/62 (0%)
Query 20 MKYVAAYLMNVLGGKDAPTAKDVAQTIEAVGGDVDEQVLNALITAMEGKTAHEMIAAGME 79
M+YVAAYL+ LGG ++PT D+ + + +VG + D+ ++ ++GK+ E+IA G
Sbjct 1 MRYVAAYLLAALGGTESPTIADLTKILNSVGIETDQHRAEKVVGELKGKSIDEIIAQGNT 60
Query 80 KL 81
KL
Sbjct 61 KL 62
> ath:AT5G40040 60S acidic ribosomal protein P2 (RPP2E); K02943
large subunit ribosomal protein LP2
Length=114
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 21/62 (33%), Positives = 40/62 (64%), Gaps = 0/62 (0%)
Query 20 MKYVAAYLMNVLGGKDAPTAKDVAQTIEAVGGDVDEQVLNALITAMEGKTAHEMIAAGME 79
MK VAAYL+ L G + P+ D+ + +E+VG ++D++ ++ + ++ + E+IA G E
Sbjct 1 MKVVAAYLLAKLSGNENPSVADLKKIVESVGAEIDQEKIDLFFSLIKDRDVTELIAVGRE 60
Query 80 KL 81
K+
Sbjct 61 KM 62
> ath:AT2G27720 60S acidic ribosomal protein P2 (RPP2A); K02943
large subunit ribosomal protein LP2
Length=130
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 48/130 (36%), Positives = 71/130 (54%), Gaps = 18/130 (13%)
Query 20 MKYVAAYLMNVLGGKDAPTAKDVAQTI---------------EAVGGDVDEQVLNALITA 64
MK VAA+L+ VL GK +PT D+ + + G + ++ + L+
Sbjct 1 MKVVAAFLLAVLSGKASPTTGDIKDILGSGIMVLVLDCVVIGDCFGAETEDSQIELLLKE 60
Query 65 MEGKTAHEMIAAGMEKLQKIPTGGAVAAAPAAGGAAPAAGGSAPA---KEEKKEEPEEEE 121
++GK E+IAAG EKL +P+GG A A+ + GG APA K+E+K+E +EE
Sbjct 61 VKGKDLAELIAAGREKLASVPSGGGGGVAVASATSGGGGGGGAPAAESKKEEKKEEKEES 120
Query 122 DGDMGLSLFD 131
D DMG SLF+
Sbjct 121 DDDMGFSLFE 130
> ath:AT4G00810 60S acidic ribosomal protein P1 (RPP1B)
Length=113
Score = 37.4 bits (85), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 16/22 (72%), Positives = 18/22 (81%), Gaps = 0/22 (0%)
Query 110 KEEKKEEPEEEEDGDMGLSLFD 131
KEEKK+EP EE DGD+G LFD
Sbjct 92 KEEKKDEPAEESDGDLGFGLFD 113
> sce:YOL039W RPP2A, RPL44, RPLA2; Ribosomal protein P2 alpha,
a component of the ribosomal stalk, which is involved in the
interaction between translational elongation factors and the
ribosome; regulates the accumulation of P1 (Rpp1Ap and Rpp1Bp)
in the cytoplasm; K02943 large subunit ribosomal protein
LP2
Length=106
Score = 36.6 bits (83), Expect = 0.021, Method: Compositional matrix adjust.
Identities = 15/47 (31%), Positives = 33/47 (70%), Gaps = 0/47 (0%)
Query 35 DAPTAKDVAQTIEAVGGDVDEQVLNALITAMEGKTAHEMIAAGMEKL 81
+ P A + +E+VG +++++ ++++++A+EGK+ E+I G EKL
Sbjct 15 NTPDATKIKAILESVGIEIEDEKVSSVLSALEGKSVDELITEGNEKL 61
> ath:AT1G01100 60S acidic ribosomal protein P1 (RPP1A)
Length=96
Score = 34.3 bits (77), Expect = 0.092, Method: Compositional matrix adjust.
Identities = 14/22 (63%), Positives = 18/22 (81%), Gaps = 0/22 (0%)
Query 110 KEEKKEEPEEEEDGDMGLSLFD 131
+E+KK+EP EE DGD+G LFD
Sbjct 75 EEKKKDEPAEESDGDLGFGLFD 96
> ath:AT5G47700 60S acidic ribosomal protein P1 (RPP1C); K02942
large subunit ribosomal protein LP1
Length=113
Score = 34.3 bits (77), Expect = 0.094, Method: Compositional matrix adjust.
Identities = 14/22 (63%), Positives = 18/22 (81%), Gaps = 0/22 (0%)
Query 110 KEEKKEEPEEEEDGDMGLSLFD 131
+E+KK+EP EE DGD+G LFD
Sbjct 92 EEKKKDEPAEESDGDLGFGLFD 113
> hsa:84893 FBXO18, FBH1, FLJ14590, Fbx18, MGC131916, MGC141935,
MGC141937; F-box protein, helicase, 18 (EC:3.6.4.12); K10300
F-box protein, helicase, 18 [EC:3.6.4.12]
Length=1094
Score = 32.7 bits (73), Expect = 0.29, Method: Composition-based stats.
Identities = 15/40 (37%), Positives = 20/40 (50%), Gaps = 0/40 (0%)
Query 92 AAPAAGGAAPAAGGSAPAKEEKKEEPEEEEDGDMGLSLFD 131
A P G A P + GSAP ++ EEE + G S +D
Sbjct 153 ALPQEGSAGPGSPGSAPPSRKRSWSSEEESNQATGTSRWD 192
> ath:AT5G24510 60s acidic ribosomal protein P1, putative; K02942
large subunit ribosomal protein LP1
Length=111
Score = 32.7 bits (73), Expect = 0.29, Method: Compositional matrix adjust.
Identities = 34/122 (27%), Positives = 55/122 (45%), Gaps = 14/122 (11%)
Query 13 LTPSKMAMKYVAAYLMNVLGGKDAPTAKDVAQTIEAVGGDVDEQVLNALITAMEGKTAHE 72
++ S++A Y A L + G + TA+++++ ++ +V+ + E K +
Sbjct 1 MSTSELACTYAALILHD--DGIEI-TAENISKLVKTANVNVESYWPSLFAKLCEKKNIDD 57
Query 73 MIAAGMEKLQKIPTGGAVAAAPAAGGAAPAAGGSAPAKEEKKEEPE---EEEDGDMGLSL 129
+I + GG A P AAP A S EEKK E E EE + DM + L
Sbjct 58 LI-------MNVGAGGCGVARPVTT-AAPTASQSVSIPEEKKNEMEVIKEESEDDMIIGL 109
Query 130 FD 131
FD
Sbjct 110 FD 111
> hsa:55357 TBC1D2, DKFZp761D1823, FLJ10702, FLJ16244, FLJ42782,
PARIS-1, PARIS1, TBC1D2A; TBC1 domain family, member 2
Length=917
Score = 31.6 bits (70), Expect = 0.74, Method: Composition-based stats.
Identities = 17/39 (43%), Positives = 20/39 (51%), Gaps = 4/39 (10%)
Query 95 AAGGAAPAAGGSAPAKEEKKEEPE----EEEDGDMGLSL 129
AG AP + SAP EE +P+ EEE GD SL
Sbjct 3 GAGENAPESSSSAPGSEESARDPQVPPPEEESGDCARSL 41
> xla:379826 rplp0, MGC53408, arbp; ribosomal protein, large,
P0; K02941 large subunit ribosomal protein LP0
Length=315
Score = 30.0 bits (66), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 14/22 (63%), Positives = 17/22 (77%), Gaps = 1/22 (4%)
Query 110 KEEKKEEPEEEEDGDMGLSLFD 131
K+EK+EE EE +D DMG LFD
Sbjct 295 KDEKQEESEESDD-DMGFGLFD 315
> tgo:TGME49_058350 hypothetical protein
Length=839
Score = 29.3 bits (64), Expect = 3.3, Method: Composition-based stats.
Identities = 19/55 (34%), Positives = 26/55 (47%), Gaps = 1/55 (1%)
Query 67 GKTAHEMIAAGMEKLQKIPTGGAVAAAPAAGGAAPAAGGSAPAKEEKKEEPEEEE 121
GKTA + +AA L P GA + + G AP+A +PA+E E E
Sbjct 18 GKTASKRVAASAFDLSS-PKAGAELSPDSPEGTAPSARAGSPAEEPDAVPSRESE 71
> tgo:TGME49_052220 Hsc70/Hsp90-organizing protein, putative ;
K09553 stress-induced-phosphoprotein 1
Length=565
Score = 29.3 bits (64), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 29/93 (31%), Positives = 37/93 (39%), Gaps = 13/93 (13%)
Query 44 QTIEAVGGDVDEQVLNALITAMEGKTA------HEMIAAGMEKLQKIPTGGAVAAAPAAG 97
Q+++ + D +V +I AM G AAG PT G+ A A
Sbjct 170 QSLKLIMAQPDVRVKEGVIAAMGGDLEEEEELPQSRTAAG-------PTRGSAENASATK 222
Query 98 GAAPAAGGSAPAKEEKKEEPEEEEDGDMGLSLF 130
AA PAKE KEE E EE G L+
Sbjct 223 AAAGVKAAEQPAKELTKEEQEAEELKQKGNELY 255
Lambda K H
0.309 0.127 0.352
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2105161088
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40