bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0207_orf1
Length=107
Score E
Sequences producing significant alignments: (Bits) Value
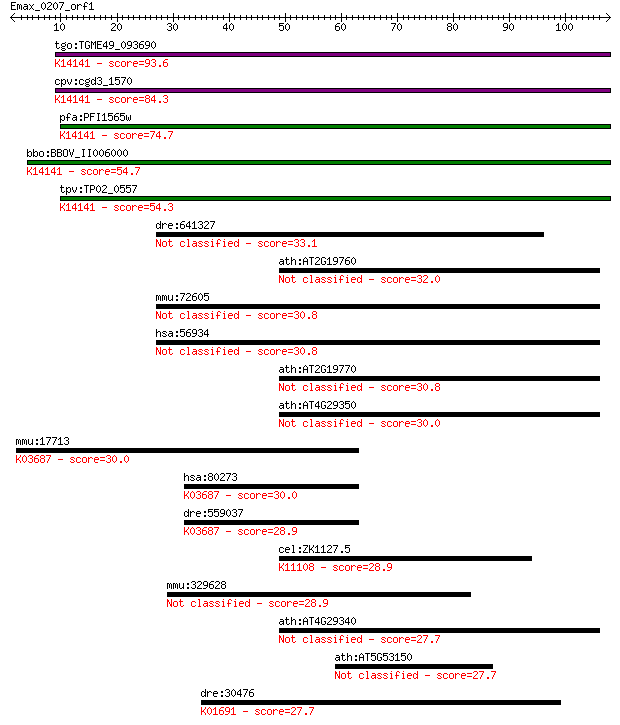
tgo:TGME49_093690 profilin family protein ; K14141 profilin-li... 93.6 1e-19
cpv:cgd3_1570 sporozoite antigen ; K14141 profilin-like protein 84.3 9e-17
pfa:PFI1565w PfPfn; profilin, putative; K14141 profilin-like p... 74.7 7e-14
bbo:BBOV_II006000 18.m06498; hypothetical protein; K14141 prof... 54.7 7e-08
tpv:TP02_0557 hypothetical protein; K14141 profilin-like protein 54.3 8e-08
dre:641327 ca10a, MGC123167, zgc:123167; carbonic anhydrase Xa 33.1 0.20
ath:AT2G19760 PRF1; PRF1 (PROFILIN 1); actin binding 32.0 0.49
mmu:72605 Car10, 2700029L05Rik, BB085816, Ca10; carbonic anhyd... 30.8 0.95
hsa:56934 CA10, CA-RPX, CARPX, HUCEP-15; carbonic anhydrase X 30.8 0.95
ath:AT2G19770 PRF5; PRF5 (PROFILIN5); actin binding / actin mo... 30.8 0.97
ath:AT4G29350 PFN2; PFN2 (PROFILIN 2); actin binding / protein... 30.0 1.9
mmu:17713 Grpel1, AA408748, MGC8152, mt-GrpE#1, mt-Grpel1; Grp... 30.0 2.0
hsa:80273 GRPEL1, FLJ25609, HMGE; GrpE-like 1, mitochondrial (... 30.0 2.1
dre:559037 grpel1, MGC123352, zgc:123352; GrpE-like 1, mitocho... 28.9 3.8
cel:ZK1127.5 hypothetical protein; K11108 RNA 3'-terminal phos... 28.9 4.5
mmu:329628 Fat4, 6030410K14Rik, 9430004M15; FAT tumor suppress... 28.9 4.5
ath:AT4G29340 PRF4; PRF4 (PROFILIN 4); actin binding 27.7 8.3
ath:AT5G53150 heat shock protein binding / unfolded protein bi... 27.7 8.6
dre:30476 wif1, MGC110507, zgc:110507; wnt inhibitory factor 1... 27.7 9.6
> tgo:TGME49_093690 profilin family protein ; K14141 profilin-like
protein
Length=163
Score = 93.6 bits (231), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 45/100 (45%), Positives = 66/100 (66%), Gaps = 1/100 (1%)
Query 9 AWEKLIKNNYKIEMMKEDG-EIELIDCYEHELLRHAIVDGKAPNGVYIGGIKYKLAEVKR 67
W KL K++++ + + EDG + E ++ A+ DG APNGV+IGG KYK+ ++
Sbjct 41 GWSKLYKDDHEEDTIGEDGNACGKVSINEASTIKAAVDDGSAPNGVWIGGQKYKVVRPEK 100
Query 68 DFTYNDQNYDIAILGKNKGGGFLIKTPNDNVVVALYDEEK 107
F YND +DI + ++KGG LIKTPN ++V+ALYDEEK
Sbjct 101 GFEYNDCTFDITMCARSKGGAHLIKTPNGSIVIALYDEEK 140
> cpv:cgd3_1570 sporozoite antigen ; K14141 profilin-like protein
Length=162
Score = 84.3 bits (207), Expect = 9e-17, Method: Compositional matrix adjust.
Identities = 42/102 (41%), Positives = 64/102 (62%), Gaps = 5/102 (4%)
Query 9 AWEKLIKNNYKIEMMKEDG---EIELIDCYEHELLRHAIVDGKAPNGVYIGGIKYKLAEV 65
AW+ L++ +++ +++ DG ELI+ + L AI +GKAPNGV++GG KYK+ V
Sbjct 40 AWKTLVREDHEENVIQSDGVSEAAELIN--DQTTLCQAISEGKAPNGVWVGGNKYKIIRV 97
Query 66 KRDFTYNDQNYDIAILGKNKGGGFLIKTPNDNVVVALYDEEK 107
++DF ND + + +GG FL+ T N VVVA+YDE K
Sbjct 98 EKDFQQNDATVHVTFCNRPQGGCFLVDTQNGTVVVAVYDESK 139
> pfa:PFI1565w PfPfn; profilin, putative; K14141 profilin-like
protein
Length=171
Score = 74.7 bits (182), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 37/98 (37%), Positives = 55/98 (56%), Gaps = 0/98 (0%)
Query 10 WEKLIKNNYKIEMMKEDGEIELIDCYEHELLRHAIVDGKAPNGVYIGGIKYKLAEVKRDF 69
W K +Y IE+ E+G E + + +G AP+GV++GG KY+ ++RD
Sbjct 51 WSLFYKEDYDIEVEDENGTKTTKTINEGQTILVVFNEGYAPDGVWLGGTKYQFINIERDL 110
Query 70 TYNDQNYDIAILGKNKGGGFLIKTPNDNVVVALYDEEK 107
+ N+D+A K KGG L+K P N++V LYDEEK
Sbjct 111 EFEGYNFDVATCAKLKGGLHLVKVPGGNILVVLYDEEK 148
> bbo:BBOV_II006000 18.m06498; hypothetical protein; K14141 profilin-like
protein
Length=164
Score = 54.7 bits (130), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 27/104 (25%), Positives = 49/104 (47%), Gaps = 0/104 (0%)
Query 4 NENESAWEKLIKNNYKIEMMKEDGEIELIDCYEHELLRHAIVDGKAPNGVYIGGIKYKLA 63
+ ++ W+ + ++ Y+ E E+G+ E + ++ G++IGG KY A
Sbjct 38 DHDDLCWDSVYRDPYEFEATDENGQPIKHQITEKATIMEVFEKRRSSIGIFIGGNKYTFA 97
Query 64 EVKRDFTYNDQNYDIAILGKNKGGGFLIKTPNDNVVVALYDEEK 107
D D + KNKGG L+KTP +V+ ++DE +
Sbjct 98 NYDDDCPVGDYTFKCVSAAKNKGGAHLVKTPGGYIVICVFDENR 141
> tpv:TP02_0557 hypothetical protein; K14141 profilin-like protein
Length=164
Score = 54.3 bits (129), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 30/98 (30%), Positives = 46/98 (46%), Gaps = 0/98 (0%)
Query 10 WEKLIKNNYKIEMMKEDGEIELIDCYEHELLRHAIVDGKAPNGVYIGGIKYKLAEVKRDF 69
W+ + K+ Y E + G I+ E +R + G+++GG KY A D
Sbjct 44 WDSVYKDPYVYETFDDAGNPLKINVDEKFTIREVFEKKMSSEGIFLGGEKYTFASYDPDM 103
Query 70 TYNDQNYDIAILGKNKGGGFLIKTPNDNVVVALYDEEK 107
++ KNKGG LIKTP + +VV +YDE +
Sbjct 104 ESGSFKFECVCGAKNKGGCHLIKTPGNYIVVVVYDETR 141
> dre:641327 ca10a, MGC123167, zgc:123167; carbonic anhydrase
Xa
Length=328
Score = 33.1 bits (74), Expect = 0.20, Method: Composition-based stats.
Identities = 22/69 (31%), Positives = 31/69 (44%), Gaps = 1/69 (1%)
Query 27 GEIELIDCYEHELLRHAIVDGKAPNGVYIGGIKYKLAEVKRDFTYNDQNYDIAILGKNKG 86
GE++LI Y HEL + K+PNG+ I I K++E F N D K
Sbjct 154 GEVQLIH-YNHELYTNYTEAAKSPNGLVIVSIFMKISETSNSFLNRMLNRDTITRITYKN 212
Query 87 GGFLIKTPN 95
+L+ N
Sbjct 213 DAYLLSGLN 221
> ath:AT2G19760 PRF1; PRF1 (PROFILIN 1); actin binding
Length=131
Score = 32.0 bits (71), Expect = 0.49, Method: Compositional matrix adjust.
Identities = 19/57 (33%), Positives = 27/57 (47%), Gaps = 9/57 (15%)
Query 49 APNGVYIGGIKYKLAEVKRDFTYNDQNYDIAILGKNKGGGFLIKTPNDNVVVALYDE 105
AP G+++GG KY + + ++ I GK GG IK N +V YDE
Sbjct 61 APTGLFLGGEKYMVIQGEQG---------AVIRGKKGPGGVTIKKTNQALVFGFYDE 108
> mmu:72605 Car10, 2700029L05Rik, BB085816, Ca10; carbonic anhydrase
10
Length=328
Score = 30.8 bits (68), Expect = 0.95, Method: Composition-based stats.
Identities = 23/79 (29%), Positives = 36/79 (45%), Gaps = 4/79 (5%)
Query 27 GEIELIDCYEHELLRHAIVDGKAPNGVYIGGIKYKLAEVKRDFTYNDQNYDIAILGKNKG 86
GE++LI Y HEL + K+PNG+ + I K+++ F N D K
Sbjct 154 GEVQLIH-YNHELYTNVTEAAKSPNGLVVVSIFIKVSDSSNPFLNRMLNRDTITRITYKN 212
Query 87 GGFLIKTPNDNVVVALYDE 105
+L++ N + LY E
Sbjct 213 DAYLLQGLN---IEELYPE 228
> hsa:56934 CA10, CA-RPX, CARPX, HUCEP-15; carbonic anhydrase
X
Length=328
Score = 30.8 bits (68), Expect = 0.95, Method: Composition-based stats.
Identities = 23/79 (29%), Positives = 36/79 (45%), Gaps = 4/79 (5%)
Query 27 GEIELIDCYEHELLRHAIVDGKAPNGVYIGGIKYKLAEVKRDFTYNDQNYDIAILGKNKG 86
GE++LI Y HEL + K+PNG+ + I K+++ F N D K
Sbjct 154 GEVQLIH-YNHELYTNVTEAAKSPNGLVVVSIFIKVSDSSNPFLNRMLNRDTITRITYKN 212
Query 87 GGFLIKTPNDNVVVALYDE 105
+L++ N + LY E
Sbjct 213 DAYLLQGLN---IEELYPE 228
> ath:AT2G19770 PRF5; PRF5 (PROFILIN5); actin binding / actin
monomer binding
Length=134
Score = 30.8 bits (68), Expect = 0.97, Method: Compositional matrix adjust.
Identities = 17/57 (29%), Positives = 28/57 (49%), Gaps = 9/57 (15%)
Query 49 APNGVYIGGIKYKLAEVKRDFTYNDQNYDIAILGKNKGGGFLIKTPNDNVVVALYDE 105
AP G+++ G+KY + + + + I GK GG IK ++V LY+E
Sbjct 64 APTGMFLAGLKYMVIQGEPN---------AVIRGKKGAGGITIKKTGQSMVFGLYEE 111
> ath:AT4G29350 PFN2; PFN2 (PROFILIN 2); actin binding / protein
binding
Length=131
Score = 30.0 bits (66), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 18/57 (31%), Positives = 26/57 (45%), Gaps = 9/57 (15%)
Query 49 APNGVYIGGIKYKLAEVKRDFTYNDQNYDIAILGKNKGGGFLIKTPNDNVVVALYDE 105
AP G+++GG KY + + + I GK GG IK +V +YDE
Sbjct 61 APTGLFLGGEKYMVVQGEAG---------AVIRGKKGPGGVTIKKTTQALVFGIYDE 108
> mmu:17713 Grpel1, AA408748, MGC8152, mt-GrpE#1, mt-Grpel1; GrpE-like
1, mitochondrial; K03687 molecular chaperone GrpE
Length=217
Score = 30.0 bits (66), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 24/72 (33%), Positives = 35/72 (48%), Gaps = 11/72 (15%)
Query 2 IDNEN---ESAWEKLIKNNYKIE-MMKEDGEIEL------IDCYEHELLRHAIVDGKAPN 51
I N N +S +E L+ +I+ + + G + L D YEHE L H V+GK P
Sbjct 130 ISNNNPHLKSLYEGLVMTEVQIQKVFTKHGLLRLDPIGAKFDPYEHEALFHTPVEGKEPG 189
Query 52 GV-YIGGIKYKL 62
V + + YKL
Sbjct 190 TVALVSKVGYKL 201
> hsa:80273 GRPEL1, FLJ25609, HMGE; GrpE-like 1, mitochondrial
(E. coli); K03687 molecular chaperone GrpE
Length=217
Score = 30.0 bits (66), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 15/32 (46%), Positives = 18/32 (56%), Gaps = 1/32 (3%)
Query 32 IDCYEHELLRHAIVDGKAPNGV-YIGGIKYKL 62
D YEHE L H V+GK P V + + YKL
Sbjct 170 FDPYEHEALFHTPVEGKEPGTVALVSKVGYKL 201
> dre:559037 grpel1, MGC123352, zgc:123352; GrpE-like 1, mitochondrial;
K03687 molecular chaperone GrpE
Length=217
Score = 28.9 bits (63), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 14/32 (43%), Positives = 19/32 (59%), Gaps = 1/32 (3%)
Query 32 IDCYEHELLRHAIVDGKAPNGV-YIGGIKYKL 62
D YEHE + HA V+GK P + + + YKL
Sbjct 170 FDPYEHEAVFHAPVEGKEPGTIALVTKVGYKL 201
> cel:ZK1127.5 hypothetical protein; K11108 RNA 3'-terminal phosphate
cyclase-like protein
Length=379
Score = 28.9 bits (63), Expect = 4.5, Method: Composition-based stats.
Identities = 16/48 (33%), Positives = 24/48 (50%), Gaps = 3/48 (6%)
Query 49 APNGVYIGGIKYKLAEVKRDFTYNDQNYDIAILG---KNKGGGFLIKT 93
AP + + G+K +V F ND+ DI I K +GGG ++ T
Sbjct 121 APGEISVDGMKASWLKVYNKFVLNDEKLDIKIQARGLKPEGGGVVVFT 168
> mmu:329628 Fat4, 6030410K14Rik, 9430004M15; FAT tumor suppressor
homolog 4 (Drosophila)
Length=4981
Score = 28.9 bits (63), Expect = 4.5, Method: Composition-based stats.
Identities = 19/57 (33%), Positives = 32/57 (56%), Gaps = 3/57 (5%)
Query 29 IELIDCYEHELLRHAIVDGKA--PNGVYIGGIKYKLAEVKRD-FTYNDQNYDIAILG 82
+E I+ E+ H+I KA P+ G + Y L + ++ FT N+QN +I++LG
Sbjct 897 VESINAVENWQAGHSIFQAKAVDPDEGVNGRVLYSLKQNPKNLFTINEQNGNISLLG 953
> ath:AT4G29340 PRF4; PRF4 (PROFILIN 4); actin binding
Length=134
Score = 27.7 bits (60), Expect = 8.3, Method: Compositional matrix adjust.
Identities = 16/57 (28%), Positives = 25/57 (43%), Gaps = 9/57 (15%)
Query 49 APNGVYIGGIKYKLAEVKRDFTYNDQNYDIAILGKNKGGGFLIKTPNDNVVVALYDE 105
AP G+++ G KY + + + I GK GG IK + V +Y+E
Sbjct 64 APTGLFMAGAKYMVIQGEPG---------AVIRGKKGAGGITIKKTGQSCVFGIYEE 111
> ath:AT5G53150 heat shock protein binding / unfolded protein
binding
Length=726
Score = 27.7 bits (60), Expect = 8.6, Method: Composition-based stats.
Identities = 12/28 (42%), Positives = 20/28 (71%), Gaps = 0/28 (0%)
Query 59 KYKLAEVKRDFTYNDQNYDIAILGKNKG 86
KY++ EV D+T +DQ+ +A+L K +G
Sbjct 589 KYEMVEVLDDYTEDDQSLTVALLLKAEG 616
> dre:30476 wif1, MGC110507, zgc:110507; wnt inhibitory factor
1; K01691 WNT inhibitory factor 1
Length=378
Score = 27.7 bits (60), Expect = 9.6, Method: Composition-based stats.
Identities = 19/72 (26%), Positives = 37/72 (51%), Gaps = 8/72 (11%)
Query 35 YEHELLRHA---IVDGKAPNGVYIGGIKYKLAEVKRDF-TYNDQN----YDIAILGKNKG 86
YE + LR I+D N +G + +K + V+ F DQ+ +++ IL + G
Sbjct 99 YEFQTLRSLDKDIMDDPTVNVPLLGSVPHKASVVQVGFPCRGDQDGVAAFEVTILVMDAG 158
Query 87 GGFLIKTPNDNV 98
G +++TP++ +
Sbjct 159 GNIILRTPHNAI 170
Lambda K H
0.314 0.138 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2007980300
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40