bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0193_orf1
Length=135
Score E
Sequences producing significant alignments: (Bits) Value
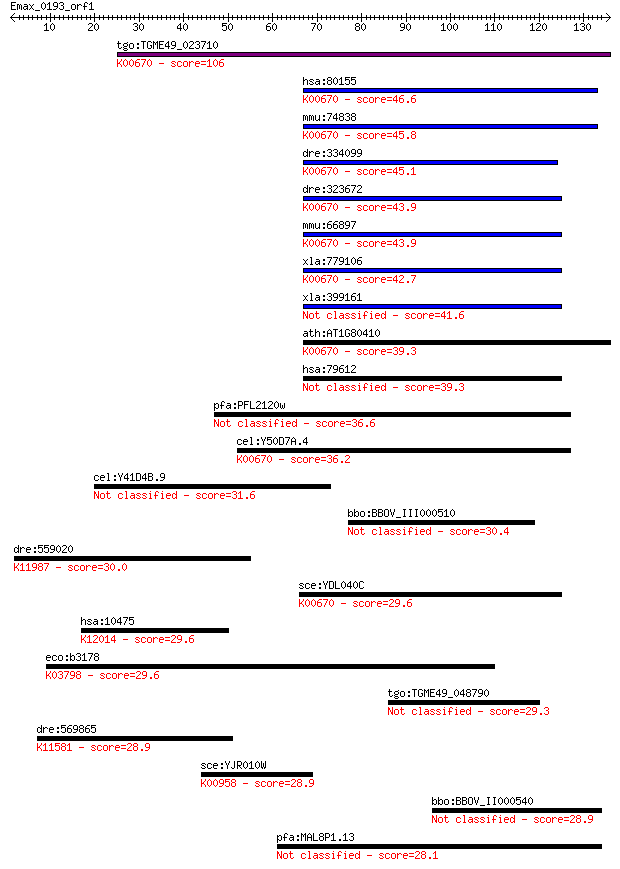
tgo:TGME49_023710 N-terminal acetyltransferase complex subunit... 106 3e-23
hsa:80155 NAA15, Ga19, NARG1, NATH, TBDN100; N(alpha)-acetyltr... 46.6 2e-05
mmu:74838 Naa15, 5730450D16Rik, 6330400I15, MGC29428, Narg1, T... 45.8 3e-05
dre:334099 naa15b, fi31d06, narg1b, wu:fi31d06; N(alpha)-acety... 45.1 7e-05
dre:323672 naa15a, MGC66136, fc06f11, narg1a, wu:fc06f11, wu:f... 43.9 1e-04
mmu:66897 Naa16, 1300019C06Rik, Narg1l; N(alpha)-acetyltransfe... 43.9 2e-04
xla:779106 naa15, ga19, narg1, narg1l, nath, tbdn100; N(alpha)... 42.7 3e-04
xla:399161 naa16, narg1, narg1l; N(alpha)-acetyltransferase 16... 41.6 7e-04
ath:AT1G80410 EMB2753 (EMBRYO DEFECTIVE 2753); binding; K00670... 39.3 0.003
hsa:79612 NAA16, DKFZp686O08147, MGC40612, NARG1L; N(alpha)-ac... 39.3 0.004
pfa:PFL2120w conserved Plasmodium protein 36.6 0.022
cel:Y50D7A.4 hypothetical protein; K00670 peptide alpha-N-acet... 36.2 0.029
cel:Y41D4B.9 nhr-122; Nuclear Hormone Receptor family member (... 31.6 0.74
bbo:BBOV_III000510 17.m07070; patatin-like phospholipase famil... 30.4 1.8
dre:559020 ptgs2b, si:dkey-97o5.6; prostaglandin-endoperoxide ... 30.0 1.9
sce:YDL040C NAT1, AAA1, NAA15; Nat1p; K00670 peptide alpha-N-a... 29.6 2.6
hsa:10475 TRIM38, MGC8946, RNF15, RORET; tripartite motif cont... 29.6 2.8
eco:b3178 ftsH, ECK3167, hflB, JW3145, mrsC, std, tolZ; protea... 29.6 2.9
tgo:TGME49_048790 hypothetical protein 29.3 3.4
dre:569865 si:dkey-57a22.11; K11581 shugoshin-like 2 28.9
sce:YJR010W MET3; ATP sulfurylase, catalyzes the primary step ... 28.9 4.2
bbo:BBOV_II000540 18.m06028; hypothetical protein 28.9 4.3
pfa:MAL8P1.13 folate/biopterin transporter, putative 28.1 8.0
> tgo:TGME49_023710 N-terminal acetyltransferase complex subunit
NARG1, putative ; K00670 peptide alpha-N-acetyltransferase
[EC:2.3.1.88]
Length=964
Score = 106 bits (264), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 59/113 (52%), Positives = 79/113 (69%), Gaps = 2/113 (1%)
Query 25 KRTLTPHSI-RVTREPTEREQELIASYFEHLQRELAPF-PLLDYLPLSFFTGDRFVAALD 82
KR TP + VTR PT+ EQ+ + S+F+ L+ E L +L LSF TGDRF + LD
Sbjct 350 KRNSTPSPLFVVTRIPTDAEQDRLISFFDCLKSEYGKVCSLPSFLVLSFLTGDRFQSRLD 409
Query 83 HFLRPMLRKGVVSIFAAIRRLYTRNRVHLITSVIESYVTNLEKKPSTFGKLLG 135
FLRP LRKGVVS+F+ +RRLYT +R+ LIT+++ESYV +LE+ STFG + G
Sbjct 410 AFLRPALRKGVVSLFSLLRRLYTPDRIPLITALLESYVYHLEQDVSTFGPVGG 462
> hsa:80155 NAA15, Ga19, NARG1, NATH, TBDN100; N(alpha)-acetyltransferase
15, NatA auxiliary subunit; K00670 peptide alpha-N-acetyltransferase
[EC:2.3.1.88]
Length=866
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 25/71 (35%), Positives = 39/71 (54%), Gaps = 5/71 (7%)
Query 67 LPLSFFTGDRFVAALDHFLRPMLRKGVVSIFAAIRRLY-TRNRVHLITSVIESYVTNLEK 125
LPL+F +G++F LD FLR KG +F +R LY + +V +I ++ Y T+L+
Sbjct 296 LPLNFLSGEKFKECLDKFLRMNFSKGCPPVFNTLRSLYKDKEKVAIIEELVVGYETSLKS 355
Query 126 ----KPSTFGK 132
P+ GK
Sbjct 356 CRLFNPNDDGK 366
> mmu:74838 Naa15, 5730450D16Rik, 6330400I15, MGC29428, Narg1,
Tbdn-1, mNAT1; N(alpha)-acetyltransferase 15, NatA auxiliary
subunit; K00670 peptide alpha-N-acetyltransferase [EC:2.3.1.88]
Length=865
Score = 45.8 bits (107), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 24/71 (33%), Positives = 39/71 (54%), Gaps = 5/71 (7%)
Query 67 LPLSFFTGDRFVAALDHFLRPMLRKGVVSIFAAIRRLY-TRNRVHLITSVIESYVTNLEK 125
LPL+F +G++F LD FLR KG +F +R LY + +V ++ ++ Y T+L+
Sbjct 296 LPLNFLSGEKFKECLDRFLRMNFSKGCPPVFNTLRSLYRDKEKVAIVEELVVGYETSLKS 355
Query 126 ----KPSTFGK 132
P+ GK
Sbjct 356 CRLFNPNDDGK 366
> dre:334099 naa15b, fi31d06, narg1b, wu:fi31d06; N(alpha)-acetyltransferase
15, NatA auxiliary subunit b; K00670 peptide
alpha-N-acetyltransferase [EC:2.3.1.88]
Length=863
Score = 45.1 bits (105), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 22/58 (37%), Positives = 33/58 (56%), Gaps = 1/58 (1%)
Query 67 LPLSFFTGDRFVAALDHFLRPMLRKGVVSIFAAIRRLY-TRNRVHLITSVIESYVTNL 123
LPLSF +GD F LD +LR KG +F +R LY + +V +I ++ + T+L
Sbjct 296 LPLSFLSGDTFRECLDRYLRMNFSKGCPPVFTTLRSLYQDKEKVSIIEELVVGFETSL 353
> dre:323672 naa15a, MGC66136, fc06f11, narg1a, wu:fc06f11, wu:fc23c05,
zgc:66136; N(alpha)-acetyltransferase 15, NatA auxiliary
subunit a; K00670 peptide alpha-N-acetyltransferase [EC:2.3.1.88]
Length=867
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 21/59 (35%), Positives = 35/59 (59%), Gaps = 1/59 (1%)
Query 67 LPLSFFTGDRFVAALDHFLRPMLRKGVVSIFAAIRRLY-TRNRVHLITSVIESYVTNLE 124
LPLSF TG++F LD +LR KG +F ++ LY +++V +I ++ Y +L+
Sbjct 296 LPLSFLTGEKFRECLDRYLRMNFSKGCPPVFTTLKSLYRHKDKVAIIEELVVGYDKSLK 354
> mmu:66897 Naa16, 1300019C06Rik, Narg1l; N(alpha)-acetyltransferase
16, NatA auxiliary subunit; K00670 peptide alpha-N-acetyltransferase
[EC:2.3.1.88]
Length=864
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 23/58 (39%), Positives = 30/58 (51%), Gaps = 3/58 (5%)
Query 67 LPLSFFTGDRFVAALDHFLRPMLRKGVVSIFAAIRRLYTRNRVHLITSVIESYVTNLE 124
LPLSF G +F +D FLRP KG +F ++ LY S+I+ VTN E
Sbjct 296 LPLSFAPGKKFRELMDKFLRPNFSKGCPPLFTTLKSLYCDTEK---VSIIQELVTNYE 350
> xla:779106 naa15, ga19, narg1, narg1l, nath, tbdn100; N(alpha)-acetyltransferase
15, NatA auxiliary subunit; K00670 peptide
alpha-N-acetyltransferase [EC:2.3.1.88]
Length=864
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 21/59 (35%), Positives = 35/59 (59%), Gaps = 1/59 (1%)
Query 67 LPLSFFTGDRFVAALDHFLRPMLRKGVVSIFAAIRRLYT-RNRVHLITSVIESYVTNLE 124
LPL+F +G +F LD +LR KG +F +R LY+ + +V +I ++ Y T+L+
Sbjct 296 LPLNFLSGLKFRECLDKYLRMNFSKGCPPVFNTLRPLYSDKEKVEIIEDLVVGYETSLK 354
> xla:399161 naa16, narg1, narg1l; N(alpha)-acetyltransferase
16, NatA auxiliary subunit
Length=846
Score = 41.6 bits (96), Expect = 7e-04, Method: Composition-based stats.
Identities = 21/59 (35%), Positives = 35/59 (59%), Gaps = 1/59 (1%)
Query 67 LPLSFFTGDRFVAALDHFLRPMLRKGVVSIFAAIRRLYT-RNRVHLITSVIESYVTNLE 124
LPL+F +G +F LD +LR KG +F +R LY+ + +V +I ++ Y T+L+
Sbjct 278 LPLNFLSGLKFRECLDKYLRMNFSKGCPPVFNTLRSLYSDKEKVEIIEDLVVGYETSLK 336
> ath:AT1G80410 EMB2753 (EMBRYO DEFECTIVE 2753); binding; K00670
peptide alpha-N-acetyltransferase [EC:2.3.1.88]
Length=897
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 20/69 (28%), Positives = 34/69 (49%), Gaps = 3/69 (4%)
Query 67 LPLSFFTGDRFVAALDHFLRPMLRKGVVSIFAAIRRLYTRNRVHLITSVIESYVTNLEKK 126
+PL F + F A+ +++P+L KGV S+F+ + LY R ++E V ++
Sbjct 304 IPLDFLQDENFKEAVAKYIKPLLTKGVPSLFSDLSSLYDHPRK---PDILEQLVVEMKHS 360
Query 127 PSTFGKLLG 135
T G G
Sbjct 361 IGTTGSFPG 369
> hsa:79612 NAA16, DKFZp686O08147, MGC40612, NARG1L; N(alpha)-acetyltransferase
16, NatA auxiliary subunit
Length=429
Score = 39.3 bits (90), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 21/58 (36%), Positives = 29/58 (50%), Gaps = 3/58 (5%)
Query 67 LPLSFFTGDRFVAALDHFLRPMLRKGVVSIFAAIRRLYTRNRVHLITSVIESYVTNLE 124
LPL+ G+RF +D FLR KG +F ++ LY S+I+ VTN E
Sbjct 296 LPLTLVPGERFRELMDKFLRVNFSKGCPPLFTTLKSLYYNTEK---VSIIQELVTNYE 350
> pfa:PFL2120w conserved Plasmodium protein
Length=1296
Score = 36.6 bits (83), Expect = 0.022, Method: Composition-based stats.
Identities = 19/80 (23%), Positives = 41/80 (51%), Gaps = 0/80 (0%)
Query 47 IASYFEHLQRELAPFPLLDYLPLSFFTGDRFVAALDHFLRPMLRKGVVSIFAAIRRLYTR 106
+ YF +LQ+ LL +PL FF ++F ++ +R + + ++IF + L T
Sbjct 493 LEKYFNNLQQLYKNSNLLKIIPLYFFNENKFSLYVEQLIRSLCFQKSLTIFNYFKPLLTF 552
Query 107 NRVHLITSVIESYVTNLEKK 126
+++I ++ Y+ +K+
Sbjct 553 RNINIILFLLHKYIDYYDKQ 572
> cel:Y50D7A.4 hypothetical protein; K00670 peptide alpha-N-acetyltransferase
[EC:2.3.1.88]
Length=852
Score = 36.2 bits (82), Expect = 0.029, Method: Composition-based stats.
Identities = 27/75 (36%), Positives = 37/75 (49%), Gaps = 7/75 (9%)
Query 52 EHLQRELAPFPLLDYLPLSFFTGDRFVAALDHFLRPMLRKGVVSIFAAIRRLYTRNRVHL 111
E +R AP L YL G+ L ++ PMLRKG S+FA++ LY +
Sbjct 291 EKFKRAAAPRRLALYL----VEGEELRRRLHEWMIPMLRKGAPSLFASLVPLYKYPQKQ- 345
Query 112 ITSVIESYVTNLEKK 126
+VIES +T KK
Sbjct 346 --AVIESLITEYVKK 358
> cel:Y41D4B.9 nhr-122; Nuclear Hormone Receptor family member
(nhr-122)
Length=374
Score = 31.6 bits (70), Expect = 0.74, Method: Compositional matrix adjust.
Identities = 19/59 (32%), Positives = 30/59 (50%), Gaps = 6/59 (10%)
Query 20 TKLYSKRTLTPHSIRVTREPTEREQELIASYFEHLQRELAPFP------LLDYLPLSFF 72
T ++SKR PH+ R T + RE +++ + + +LA FP LL + L FF
Sbjct 140 TSMFSKRIPKPHTFRDTDKLMFREYDMVTDWAMNSFPQLADFPNDQRKILLKHFYLQFF 198
> bbo:BBOV_III000510 17.m07070; patatin-like phospholipase family
protein
Length=1263
Score = 30.4 bits (67), Expect = 1.8, Method: Composition-based stats.
Identities = 16/42 (38%), Positives = 23/42 (54%), Gaps = 1/42 (2%)
Query 77 FVAALDHFLRPMLRKGVVSIFAAIRRLYTRNRVHLITSVIES 118
FV + HF R L++GV S+ + + Y R R H I +ES
Sbjct 47 FVTSRQHFRRTSLKEGVTSV-SPLEYNYNRGRRHTIGGNVES 87
> dre:559020 ptgs2b, si:dkey-97o5.6; prostaglandin-endoperoxide
synthase 2b (EC:1.14.99.1); K11987 prostaglandin-endoperoxide
synthase 2 [EC:1.14.99.1]
Length=606
Score = 30.0 bits (66), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 16/58 (27%), Positives = 29/58 (50%), Gaps = 5/58 (8%)
Query 2 EERRVCTEAKQNGKECNITK--LYSKRTLTPH---SIRVTREPTEREQELIASYFEHL 54
+ R VCTE + EC+ T+ Y + TP ++V+ +P+ I ++F+ L
Sbjct 29 QNRGVCTEMGSDAYECDCTRTGYYGQNCTTPEFLTWVKVSLKPSPNTVHYILTHFKSL 86
> sce:YDL040C NAT1, AAA1, NAA15; Nat1p; K00670 peptide alpha-N-acetyltransferase
[EC:2.3.1.88]
Length=854
Score = 29.6 bits (65), Expect = 2.6, Method: Composition-based stats.
Identities = 15/63 (23%), Positives = 33/63 (52%), Gaps = 4/63 (6%)
Query 66 YLPLSFFTG-DRFVAALDHFLRPMLRKGVVSIFAAIRRLYTRNRVH---LITSVIESYVT 121
++PL+F + L ++ P L +GV + F+ ++ LY R + L+ ++ Y++
Sbjct 313 FIPLTFLQDKEELSKKLREYVLPQLERGVPATFSNVKPLYQRRKSKVSPLLEKIVLDYLS 372
Query 122 NLE 124
L+
Sbjct 373 GLD 375
> hsa:10475 TRIM38, MGC8946, RNF15, RORET; tripartite motif containing
38; K12014 tripartite motif-containing protein 38
Length=465
Score = 29.6 bits (65), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 15/35 (42%), Positives = 20/35 (57%), Gaps = 2/35 (5%)
Query 17 CNITKLY--SKRTLTPHSIRVTREPTEREQELIAS 49
CN++KLY K+ L H + VT +P ELI S
Sbjct 274 CNVSKLYFDVKKMLRSHQVSVTLDPDTAHHELILS 308
> eco:b3178 ftsH, ECK3167, hflB, JW3145, mrsC, std, tolZ; protease,
ATP-dependent zinc-metallo (EC:3.4.24.-); K03798 cell
division protease FtsH [EC:3.4.24.-]
Length=644
Score = 29.6 bits (65), Expect = 2.9, Method: Composition-based stats.
Identities = 32/118 (27%), Positives = 51/118 (43%), Gaps = 35/118 (29%)
Query 9 EAKQNGKECNITKLYSKR--------------TLTPHSIRVTREPTEREQELIASYFEHL 54
EA+ NG+E N+TK S R L +++V EP E E L+AS F
Sbjct 48 EARINGREINVTKKDSNRYTTYIPVQDPKLLDNLLTKNVKVVGEPPE-EPSLLASIF--- 103
Query 55 QRELAPFPLLDYLPLSFFTGDRFVAALDHFLRPML---RKGVVSIFAAIRRLYTRNRV 109
++ FP+L + + F F+R M KG +S + R+ T +++
Sbjct 104 ---ISWFPMLLLIGVWIF-----------FMRQMQGGGGKGAMSFGKSKARMLTEDQI 147
> tgo:TGME49_048790 hypothetical protein
Length=243
Score = 29.3 bits (64), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 11/34 (32%), Positives = 22/34 (64%), Gaps = 0/34 (0%)
Query 86 RPMLRKGVVSIFAAIRRLYTRNRVHLITSVIESY 119
R + R ++FA+++R RN VH +T ++++Y
Sbjct 5 RHVFRAAPAAVFASVQRWGRRNNVHKLTLLVQTY 38
> dre:569865 si:dkey-57a22.11; K11581 shugoshin-like 2
Length=847
Score = 28.9 bits (63), Expect = 4.1, Method: Composition-based stats.
Identities = 15/44 (34%), Positives = 22/44 (50%), Gaps = 0/44 (0%)
Query 7 CTEAKQNGKECNITKLYSKRTLTPHSIRVTREPTEREQELIASY 50
C+ QNG+E + K +T S R T +P E Q+L +Y
Sbjct 280 CSTTVQNGQETELLSARRKTHITSRSKRRTCKPKEPNQDLRKTY 323
> sce:YJR010W MET3; ATP sulfurylase, catalyzes the primary step
of intracellular sulfate activation, essential for assimilatory
reduction of sulfate to sulfide, involved in methionine
metabolism (EC:2.7.7.4); K00958 sulfate adenylyltransferase
[EC:2.7.7.4]
Length=511
Score = 28.9 bits (63), Expect = 4.2, Method: Composition-based stats.
Identities = 12/25 (48%), Positives = 16/25 (64%), Gaps = 0/25 (0%)
Query 44 QELIASYFEHLQRELAPFPLLDYLP 68
QEL+ SY L E+ PF ++ YLP
Sbjct 311 QELVESYKHELDIEVVPFRMVTYLP 335
> bbo:BBOV_II000540 18.m06028; hypothetical protein
Length=300
Score = 28.9 bits (63), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 16/42 (38%), Positives = 24/42 (57%), Gaps = 4/42 (9%)
Query 96 IFAAIRRLYTRNR----VHLITSVIESYVTNLEKKPSTFGKL 133
IFA IR+ T N VH+I ++ E Y T L ++ +FG +
Sbjct 160 IFAVIRKCLTNNTEINIVHIIETIREKYETTLIEEVPSFGSI 201
> pfa:MAL8P1.13 folate/biopterin transporter, putative
Length=505
Score = 28.1 bits (61), Expect = 8.0, Method: Composition-based stats.
Identities = 18/83 (21%), Positives = 40/83 (48%), Gaps = 10/83 (12%)
Query 61 FPLLDYLPLSFFTGDRFVAALDHFLRPMLRKGVVSIFAAIRRLYTRNRVHLITSVIES-- 118
F + +LP+ FT F+ ++ + ++ + I++ I+ Y +N + I ++ +
Sbjct 218 FLIGSFLPICVFTSGFFIIEKRNYTKSSIKDQIKCIYSIIKLSYLKNFIIFIFIMMSTPS 277
Query 119 -------YVTN-LEKKPSTFGKL 133
Y+TN L+ P+ GK+
Sbjct 278 CGNTLFFYITNELKFSPNLLGKM 300
Lambda K H
0.324 0.138 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2296762580
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40