bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0191_orf1
Length=121
Score E
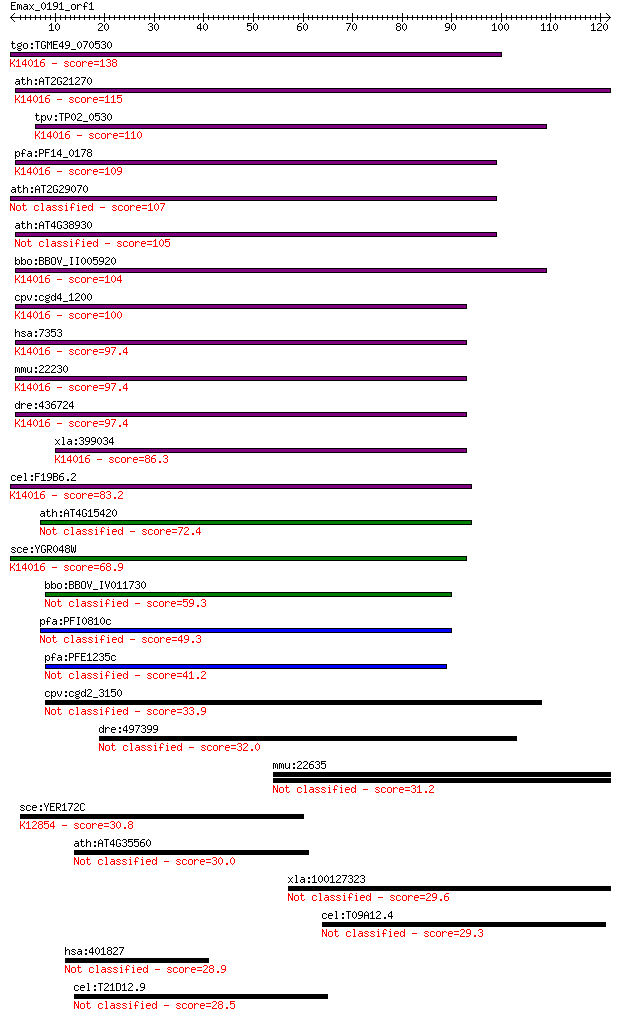
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_070530 ubiquitin fusion degradation domain-containi... 138 4e-33
ath:AT2G21270 ubiquitin fusion degradation UFD1 family protein... 115 3e-26
tpv:TP02_0530 hypothetical protein; K14016 ubiquitin fusion de... 110 8e-25
pfa:PF14_0178 UFD1; Ubiquitin fusion degradation protein UFD1,... 109 3e-24
ath:AT2G29070 ubiquitin fusion degradation UFD1 family protein 107 1e-23
ath:AT4G38930 ubiquitin fusion degradation UFD1 family protein 105 3e-23
bbo:BBOV_II005920 18.m06490; ubiquitin fusion degradation prot... 104 5e-23
cpv:cgd4_1200 ubiquitin fusion degradation protein (UFD1); dou... 100 9e-22
hsa:7353 UFD1L, UFD1; ubiquitin fusion degradation 1 like (yea... 97.4 1e-20
mmu:22230 Ufd1l, Ufd1; ubiquitin fusion degradation 1 like; K1... 97.4 1e-20
dre:436724 ufd1l, wu:fc55f04, zgc:92341; ubiquitin fusion degr... 97.4 1e-20
xla:399034 ufd1l, MGC68571, ufd1; ubiquitin fusion degradation... 86.3 2e-17
cel:F19B6.2 ufd-1; Ubiquitin Fusion Degradation (yeast UFD hom... 83.2 2e-16
ath:AT4G15420 PRLI-interacting factor K 72.4 4e-13
sce:YGR048W UFD1; Protein that interacts with Cdc48p and Npl4p... 68.9 4e-12
bbo:BBOV_IV011730 23.m05946; ubiquitin fusion degradation prot... 59.3 3e-09
pfa:PFI0810c apicoplast Ufd1 precursor 49.3 3e-06
pfa:PFE1235c Ubiquitin fusion degradation protein UFD1, putative 41.2 9e-04
cpv:cgd2_3150 ubiquitin fusion degradation (UFD1) family prote... 33.9 0.14
dre:497399 im:7140212 32.0 0.50
mmu:22635 Zan; zonadhesin 31.2 0.73
sce:YER172C BRR2, PRP44, RSS1, SLT22, SNU246; Brr2p (EC:3.6.1.... 30.8 1.1
ath:AT4G35560 hypothetical protein 30.0 2.1
xla:100127323 hypothetical protein LOC100127323 29.6 2.5
cel:T09A12.4 nhr-66; Nuclear Hormone Receptor family member (n... 29.3 3.0
hsa:401827 MSLNL, C16orf37, MPFL; mesothelin-like 28.9 4.0
cel:T21D12.9 hypothetical protein 28.5 5.8
> tgo:TGME49_070530 ubiquitin fusion degradation domain-containing
protein (EC:3.1.3.48); K14016 ubiquitin fusion degradation
protein 1
Length=335
Score = 138 bits (347), Expect = 4e-33, Method: Compositional matrix adjust.
Identities = 67/99 (67%), Positives = 82/99 (82%), Gaps = 0/99 (0%)
Query 1 ALEEGGIVTVTNVSLPKGTFVQLQPLETDFLDISNPKALLEMALRGYAALTKGEVVSLPF 60
LEEG IV V N+SLPKGTFV+LQP+ T+FL +SNP+A+LE+ALRGYAALT G+++ LPF
Sbjct 103 GLEEGDIVRVRNISLPKGTFVELQPVTTEFLQVSNPRAVLEVALRGYAALTVGDLIYLPF 162
Query 61 LDCVYHLRVADLRPAPAVSIVETDVEVEFKAPEEQQNPS 99
LD + L V DLRPAPAVSI+ETD+EVEFKAPE P+
Sbjct 163 LDKGFQLLVTDLRPAPAVSIIETDMEVEFKAPEGYVEPT 201
> ath:AT2G21270 ubiquitin fusion degradation UFD1 family protein;
K14016 ubiquitin fusion degradation protein 1
Length=319
Score = 115 bits (288), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 58/120 (48%), Positives = 82/120 (68%), Gaps = 0/120 (0%)
Query 2 LEEGGIVTVTNVSLPKGTFVQLQPLETDFLDISNPKALLEMALRGYAALTKGEVVSLPFL 61
L+EG IV V NV+LPKGT+V+LQP TDFLDISNPKA+LE ALR Y+ LT G+ + +P+
Sbjct 94 LQEGDIVRVRNVTLPKGTYVKLQPHTTDFLDISNPKAILETALRNYSCLTSGDSIMVPYN 153
Query 62 DCVYHLRVADLRPAPAVSIVETDVEVEFKAPEEQQNPSPVPDPAADGGYISSDEASEPPE 121
+ Y + + + +PA A+SI+ETD EV+F P + + P P+A G ++E + PE
Sbjct 154 NKKYFIDIVETKPANAISIIETDCEVDFAPPLDYKEPERPTAPSAAKGPAKAEEVVDEPE 213
> tpv:TP02_0530 hypothetical protein; K14016 ubiquitin fusion
degradation protein 1
Length=260
Score = 110 bits (276), Expect = 8e-25, Method: Compositional matrix adjust.
Identities = 54/112 (48%), Positives = 76/112 (67%), Gaps = 9/112 (8%)
Query 6 GIVTVTNVSLPKGTFVQLQPLETDFLDISNPKALLEMALRGYAALTKGEVVSLPFLDCVY 65
+VT+TNVSLPK T+V+L+PL D+ DISNP+A+LE ALR YA LT G+V+ + ++ +Y
Sbjct 102 NVVTITNVSLPKATWVKLKPLNEDYWDISNPRAVLENALRNYATLTVGDVIPIHYIQTIY 161
Query 66 HLRVADLRPAPAVSIVETDVEVEFKA----PEEQQN-----PSPVPDPAADG 108
+ DL+PA A SI+ETD+EVEF P+E++N P P+ DG
Sbjct 162 LFHIMDLKPAKACSIIETDMEVEFDMPVPEPKEEENDMETDPEPIIGKRLDG 213
> pfa:PF14_0178 UFD1; Ubiquitin fusion degradation protein UFD1,
putative; K14016 ubiquitin fusion degradation protein 1
Length=282
Score = 109 bits (272), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 51/97 (52%), Positives = 72/97 (74%), Gaps = 0/97 (0%)
Query 2 LEEGGIVTVTNVSLPKGTFVQLQPLETDFLDISNPKALLEMALRGYAALTKGEVVSLPFL 61
L+EG IV VT+VSLPKGTFV+L+P DF+++SN +A+LE ALR YA LT G+ + + +L
Sbjct 103 LKEGDIVRVTSVSLPKGTFVKLKPCSKDFMELSNHRAVLETALRNYATLTIGDNIVIHYL 162
Query 62 DCVYHLRVADLRPAPAVSIVETDVEVEFKAPEEQQNP 98
Y +++ DL+PA A +I+ETDVEVEF+ P + P
Sbjct 163 GKTYEIKIVDLKPAFACTIIETDVEVEFEEPIDHSEP 199
> ath:AT2G29070 ubiquitin fusion degradation UFD1 family protein
Length=280
Score = 107 bits (267), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 48/98 (48%), Positives = 71/98 (72%), Gaps = 0/98 (0%)
Query 1 ALEEGGIVTVTNVSLPKGTFVQLQPLETDFLDISNPKALLEMALRGYAALTKGEVVSLPF 60
+LEEG ++ V N+SL KGT+++LQP DFLDISNPKA+LE LR Y+ LT G+ + +P+
Sbjct 56 SLEEGDVMQVKNISLVKGTYIKLQPHTQDFLDISNPKAILETTLRSYSCLTTGDTIMVPY 115
Query 61 LDCVYHLRVADLRPAPAVSIVETDVEVEFKAPEEQQNP 98
+ Y++ V + +P+ AVSI+ETD EV+F P + + P
Sbjct 116 NNKQYYINVVEAKPSSAVSIIETDCEVDFAPPLDYKEP 153
> ath:AT4G38930 ubiquitin fusion degradation UFD1 family protein
Length=311
Score = 105 bits (263), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 48/97 (49%), Positives = 71/97 (73%), Gaps = 0/97 (0%)
Query 2 LEEGGIVTVTNVSLPKGTFVQLQPLETDFLDISNPKALLEMALRGYAALTKGEVVSLPFL 61
L+EG +V V NV+LPKGT+V+LQP TDFLDI+NPKA+LE ALR Y+ LT G+ + +P+
Sbjct 94 LQEGDMVRVRNVTLPKGTYVKLQPHTTDFLDIANPKAILETALRNYSCLTVGDSIMVPYN 153
Query 62 DCVYHLRVADLRPAPAVSIVETDVEVEFKAPEEQQNP 98
+ Y + + + +P+ +SI+ETD EV+F P + + P
Sbjct 154 NKKYFIDIVEAKPSNGISIIETDCEVDFAPPLDYKEP 190
> bbo:BBOV_II005920 18.m06490; ubiquitin fusion degradation protein;
K14016 ubiquitin fusion degradation protein 1
Length=258
Score = 104 bits (260), Expect = 5e-23, Method: Compositional matrix adjust.
Identities = 53/109 (48%), Positives = 72/109 (66%), Gaps = 2/109 (1%)
Query 2 LEEGGIVTVTNVSLPKGTFVQLQPLETDFLDISNPKALLEMALRGYAALTKGEVVSLPFL 61
L+EG +T+ NV LPK +V+ +PL ++ DISNPKA+LE ALR +A LT G+ + + +L
Sbjct 104 LQEGDYLTIRNVRLPKANWVKFRPLNDNYWDISNPKAVLETALRNFATLTIGDRIPIHYL 163
Query 62 DCVYHLRVADLRPAPAVSIVETDVEVEFKAPEEQ--QNPSPVPDPAADG 108
VY L V DLRPA A I+ETD+EVEF +++ Q PVP DG
Sbjct 164 SNVYELDVMDLRPADACCIIETDMEVEFAETKKKKPQLVQPVPGKRLDG 212
> cpv:cgd4_1200 ubiquitin fusion degradation protein (UFD1); double
Psi beta barrel fold ; K14016 ubiquitin fusion degradation
protein 1
Length=322
Score = 100 bits (250), Expect = 9e-22, Method: Compositional matrix adjust.
Identities = 43/91 (47%), Positives = 65/91 (71%), Gaps = 0/91 (0%)
Query 2 LEEGGIVTVTNVSLPKGTFVQLQPLETDFLDISNPKALLEMALRGYAALTKGEVVSLPFL 61
L+EG I ++ N SL KGT+V+ PL DFLDISNPKA+LE LR +A LT G+++++ +
Sbjct 125 LQEGDITSIMNTSLSKGTYVKFMPLSMDFLDISNPKAVLETTLRNFATLTVGDIITIHYN 184
Query 62 DCVYHLRVADLRPAPAVSIVETDVEVEFKAP 92
+ Y + V + +P A+SI+ETD++V+F P
Sbjct 185 NNSYRINVLETKPNNAISIIETDIQVDFAPP 215
> hsa:7353 UFD1L, UFD1; ubiquitin fusion degradation 1 like (yeast);
K14016 ubiquitin fusion degradation protein 1
Length=266
Score = 97.4 bits (241), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 46/91 (50%), Positives = 66/91 (72%), Gaps = 0/91 (0%)
Query 2 LEEGGIVTVTNVSLPKGTFVQLQPLETDFLDISNPKALLEMALRGYAALTKGEVVSLPFL 61
LEEGG+V V +V+L T+ + QP DFLDI+NPKA+LE ALR +A LT G+V+++ +
Sbjct 104 LEEGGLVQVESVNLQVATYSKFQPQSPDFLDITNPKAVLENALRNFACLTTGDVIAINYN 163
Query 62 DCVYHLRVADLRPAPAVSIVETDVEVEFKAP 92
+ +Y LRV + +P AVSI+E D+ V+F AP
Sbjct 164 EKIYELRVMETKPDKAVSIIECDMNVDFDAP 194
> mmu:22230 Ufd1l, Ufd1; ubiquitin fusion degradation 1 like;
K14016 ubiquitin fusion degradation protein 1
Length=307
Score = 97.4 bits (241), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 46/91 (50%), Positives = 66/91 (72%), Gaps = 0/91 (0%)
Query 2 LEEGGIVTVTNVSLPKGTFVQLQPLETDFLDISNPKALLEMALRGYAALTKGEVVSLPFL 61
LEEGG+V V +V+L T+ + QP DFLDI+NPKA+LE ALR +A LT G+V+++ +
Sbjct 104 LEEGGLVQVESVNLQVATYSKFQPQSPDFLDITNPKAVLENALRNFACLTTGDVIAINYN 163
Query 62 DCVYHLRVADLRPAPAVSIVETDVEVEFKAP 92
+ +Y LRV + +P AVSI+E D+ V+F AP
Sbjct 164 EKIYELRVMETKPDKAVSIIECDMNVDFDAP 194
> dre:436724 ufd1l, wu:fc55f04, zgc:92341; ubiquitin fusion degradation
1-like; K14016 ubiquitin fusion degradation protein
1
Length=308
Score = 97.4 bits (241), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 46/91 (50%), Positives = 66/91 (72%), Gaps = 0/91 (0%)
Query 2 LEEGGIVTVTNVSLPKGTFVQLQPLETDFLDISNPKALLEMALRGYAALTKGEVVSLPFL 61
LEEGG+V V +V+L T+ + QP DFLDI+NPKA+LE ALR +A LT G+V+++ +
Sbjct 104 LEEGGLVQVESVNLMVATYSKFQPQSPDFLDITNPKAVLENALRNFACLTTGDVIAINYN 163
Query 62 DCVYHLRVADLRPAPAVSIVETDVEVEFKAP 92
+ +Y LRV + +P AVSI+E D+ V+F AP
Sbjct 164 EKIYELRVMETKPDKAVSIIECDMNVDFDAP 194
> xla:399034 ufd1l, MGC68571, ufd1; ubiquitin fusion degradation
1 like; K14016 ubiquitin fusion degradation protein 1
Length=307
Score = 86.3 bits (212), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 41/83 (49%), Positives = 59/83 (71%), Gaps = 0/83 (0%)
Query 10 VTNVSLPKGTFVQLQPLETDFLDISNPKALLEMALRGYAALTKGEVVSLPFLDCVYHLRV 69
V +V+L T+ + QP DFLDI+NPKA+LE ALR +A LT G+VV++ + + +Y LRV
Sbjct 112 VESVNLQVATYSKFQPQSPDFLDITNPKAVLENALRNFACLTTGDVVAINYNEKIYELRV 171
Query 70 ADLRPAPAVSIVETDVEVEFKAP 92
+ +P AVSI+E D+ V+F AP
Sbjct 172 METKPDKAVSIIECDMNVDFDAP 194
> cel:F19B6.2 ufd-1; Ubiquitin Fusion Degradation (yeast UFD homolog)
family member (ufd-1); K14016 ubiquitin fusion degradation
protein 1
Length=342
Score = 83.2 bits (204), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 38/93 (40%), Positives = 60/93 (64%), Gaps = 0/93 (0%)
Query 1 ALEEGGIVTVTNVSLPKGTFVQLQPLETDFLDISNPKALLEMALRGYAALTKGEVVSLPF 60
L++G + + + +LPK TF +L+P+ +FL+I+NPKA+LE+ LR YA LTK + + +
Sbjct 107 GLDDGDTIRIESATLPKATFAKLKPMSLEFLNITNPKAVLEVELRKYACLTKNDRIPTSY 166
Query 61 LDCVYHLRVADLRPAPAVSIVETDVEVEFKAPE 93
V DL+PA +V I+E DV ++F PE
Sbjct 167 AGQTLEFLVVDLKPANSVCIIECDVNLDFDPPE 199
> ath:AT4G15420 PRLI-interacting factor K
Length=561
Score = 72.4 bits (176), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 34/87 (39%), Positives = 58/87 (66%), Gaps = 0/87 (0%)
Query 7 IVTVTNVSLPKGTFVQLQPLETDFLDISNPKALLEMALRGYAALTKGEVVSLPFLDCVYH 66
+V + + LPKG++ +LQP F D+ N KA+LE LR +A L+ +V+ + + Y
Sbjct 166 LVEIRYIRLPKGSYAKLQPDNLGFSDLPNHKAILETILRQHATLSLDDVLLVNYGQVSYK 225
Query 67 LRVADLRPAPAVSIVETDVEVEFKAPE 93
L+V +LRPA ++S++ETD+EV+ +P+
Sbjct 226 LQVLELRPATSISVLETDIEVDIVSPD 252
> sce:YGR048W UFD1; Protein that interacts with Cdc48p and Npl4p,
involved in recognition of polyubiquitinated proteins and
their presentation to the 26S proteasome for degradation;
involved in transporting proteins from the ER to the cytosol;
K14016 ubiquitin fusion degradation protein 1
Length=361
Score = 68.9 bits (167), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 29/95 (30%), Positives = 61/95 (64%), Gaps = 3/95 (3%)
Query 1 ALEEGGIVTVTNVSLPKGTFVQLQPLETDFLDISNPKALLEMALRGYAALTKGEVVSLPF 60
++ G ++ +++ +P G FV+L+P DFLDIS+PKA+LE LR ++ LT +V+ + +
Sbjct 105 GIQPGSLLQISSTDVPLGQFVKLEPQSVDFLDISDPKAVLENVLRNFSTLTVDDVIEISY 164
Query 61 LDCVYHLRVADLRP---APAVSIVETDVEVEFKAP 92
+ +++ +++P + ++ ++ETD+ +F P
Sbjct 165 NGKTFKIKILEVKPESSSKSICVIETDLVTDFAPP 199
> bbo:BBOV_IV011730 23.m05946; ubiquitin fusion degradation protein
UFD1
Length=297
Score = 59.3 bits (142), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 31/87 (35%), Positives = 48/87 (55%), Gaps = 5/87 (5%)
Query 8 VTVTNVSLPKGTFVQLQPLETDFLDISNPKALLEMALRGYAALTKGEVVSLPFLDCVYHL 67
V VT + L FV + P+E+ F +S PKA+LE L+ Y++LT+G + + YHL
Sbjct 208 VYVTQLKLQDAIFVSISPVESSFFALSAPKAVLEEHLKQYSSLTRGTTIQITHEGITYHL 267
Query 68 RVADL-----RPAPAVSIVETDVEVEF 89
RV + + A SI +TDV ++
Sbjct 268 RVNRIETEHCKDAECASIQDTDVSIDL 294
> pfa:PFI0810c apicoplast Ufd1 precursor
Length=296
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 28/88 (31%), Positives = 44/88 (50%), Gaps = 5/88 (5%)
Query 7 IVTVTNVSLPKGTFVQLQPLETDFLDISNPKALLEMALRGYAALTKGEVVSLPFLDCVYH 66
++ + V L + V LQP E F D+ NPK +LE LR Y+ +T+ +S+ D VY+
Sbjct 202 VIRLRFVKLETASSVVLQPHEKKFFDLENPKKILEEKLRYYSCITRNSTISIKHDDVVYY 261
Query 67 LRVADL-----RPAPAVSIVETDVEVEF 89
V + + SI + DV +F
Sbjct 262 FDVIRIDSEKKKDTEVASIQDADVIFDF 289
> pfa:PFE1235c Ubiquitin fusion degradation protein UFD1, putative
Length=700
Score = 41.2 bits (95), Expect = 9e-04, Method: Composition-based stats.
Identities = 27/82 (32%), Positives = 43/82 (52%), Gaps = 5/82 (6%)
Query 8 VTVTNVSLPKGTFVQLQPLETDFLDISNPKALLEMALR-GYAALTKGEVVSLPFLDCVYH 66
V +T L K F++L L + DI K LLE L Y+ LT G+ + + L+
Sbjct 158 VLITYCILSKCDFIKLDSLNNNINDIKYMKNLLENELSLNYSTLTLGDYIHINHLN---- 213
Query 67 LRVADLRPAPAVSIVETDVEVE 88
+++L P AVS++ TD+ V+
Sbjct 214 FYISELEPDNAVSLINTDINVD 235
> cpv:cgd2_3150 ubiquitin fusion degradation (UFD1) family protein,
double Psi beta barrel fold
Length=658
Score = 33.9 bits (76), Expect = 0.14, Method: Composition-based stats.
Identities = 26/108 (24%), Positives = 54/108 (50%), Gaps = 9/108 (8%)
Query 8 VTVTNVSLPKGTFVQLQPLET-DFLDISNPKALLEMALRG-YAALTKGE--VVSLPFLD- 62
+ +T L KG+F + L D + + ++LLE LR + LT G+ +++ P
Sbjct 196 IQITYKKLLKGSFASFEILNNQDIFKMHDIESLLESYLRNHFLTLTIGDTLMINQPNYSS 255
Query 63 ---CVYHLRVADLRPAPAVSIVETDVEVEFKAPEEQQNPSPVPDPAAD 107
C+ ++V L P ++S++ TD+ ++ ++ +N + D A +
Sbjct 256 NNYCISLIKVKHLEPDNSISLINTDISLDITY-KDSKNEQTIQDHAEN 302
> dre:497399 im:7140212
Length=825
Score = 32.0 bits (71), Expect = 0.50, Method: Composition-based stats.
Identities = 24/90 (26%), Positives = 39/90 (43%), Gaps = 6/90 (6%)
Query 19 TFVQLQPLETDFLDISNPKALLEMALRGYAALTKGEVVSLPFLDCVYHLRVADLRPAPAV 78
T+ L+P F+D L+ G L+K +V LP L+ +L A RP ++
Sbjct 16 TYRILKPWWDVFMDYIGVIMLMLAIFSGTMQLSKDQVACLPILEKNINLEGAQNRPECSL 75
Query 79 SIVETDVEVEFKAPEEQQN------PSPVP 102
S++ T P+ N P+P+P
Sbjct 76 SVLGTGPSTTKDLPDSVANELLNTQPAPLP 105
> mmu:22635 Zan; zonadhesin
Length=5420
Score = 31.2 bits (69), Expect = 0.73, Method: Composition-based stats.
Identities = 23/70 (32%), Positives = 34/70 (48%), Gaps = 2/70 (2%)
Query 54 EVVSLPFLDCVYHLRVADLRPA-PAVSIVET-DVEVEFKAPEEQQNPSPVPDPAADGGYI 111
EV ++P H V ++ P +V ET EV +PEE P+ VP + + +
Sbjct 708 EVPTVPTEVTGVHTEVTNVSPEETSVPTEETISTEVTTVSPEETTLPTEVPTVSTEVTNV 767
Query 112 SSDEASEPPE 121
S +E S PPE
Sbjct 768 SPEETSVPPE 777
Score = 31.2 bits (69), Expect = 0.73, Method: Composition-based stats.
Identities = 23/70 (32%), Positives = 34/70 (48%), Gaps = 2/70 (2%)
Query 54 EVVSLPFLDCVYHLRVADLRPA-PAVSIVET-DVEVEFKAPEEQQNPSPVPDPAADGGYI 111
EV ++P H V ++ P +V ET EV +PEE P+ VP + + +
Sbjct 787 EVPTVPTEVTGVHTEVTNVSPEETSVPTEETISTEVTTVSPEETTLPTEVPTVSTEVTNV 846
Query 112 SSDEASEPPE 121
S +E S PPE
Sbjct 847 SPEETSVPPE 856
> sce:YER172C BRR2, PRP44, RSS1, SLT22, SNU246; Brr2p (EC:3.6.1.-);
K12854 pre-mRNA-splicing helicase BRR2 [EC:3.6.4.13]
Length=2163
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 20/58 (34%), Positives = 34/58 (58%), Gaps = 10/58 (17%)
Query 3 EEGGIVTVTNVSLPKGTFVQLQPLETDFLDISNP-KALLEMALRGYAALTKGEVVSLP 59
E ++TVT VSLP+G+F +++P + D + I P K +++ L+ E+ SLP
Sbjct 436 ESSKLMTVTKVSLPEGSFKRVKP-QYDEIHIPAPSKPVIDYELK--------EITSLP 484
> ath:AT4G35560 hypothetical protein
Length=1049
Score = 30.0 bits (66), Expect = 2.1, Method: Composition-based stats.
Identities = 19/51 (37%), Positives = 27/51 (52%), Gaps = 4/51 (7%)
Query 14 SLPKGTFVQLQPLETDFLDI----SNPKALLEMALRGYAALTKGEVVSLPF 60
SLPK T V+L ++ + + +NP LL ++ YA L K V LPF
Sbjct 387 SLPKETVVKLPFSDSSSITVGKFLTNPSHLLNLSDEDYAQLAKDAVPFLPF 437
> xla:100127323 hypothetical protein LOC100127323
Length=426
Score = 29.6 bits (65), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 22/67 (32%), Positives = 29/67 (43%), Gaps = 2/67 (2%)
Query 57 SLPFLDCVYHLRVADLRPAPAVSIVETDVEVEFKAP-EEQQNPSPVPDPAADGGY-ISSD 114
S PFLD V H R + P+ A + D V F P + N + D GGY S
Sbjct 149 SYPFLDTVDHTRRPKILPSSATDLPRIDAIVLFGEPIRWETNLQLIIDVLLTGGYPASHH 208
Query 115 EASEPPE 121
+A+ P
Sbjct 209 QAASYPH 215
> cel:T09A12.4 nhr-66; Nuclear Hormone Receptor family member
(nhr-66)
Length=733
Score = 29.3 bits (64), Expect = 3.0, Method: Composition-based stats.
Identities = 24/63 (38%), Positives = 29/63 (46%), Gaps = 8/63 (12%)
Query 64 VYHLRVADLRPAPAVSIVETDVEVEFKAP------EEQQNPSPVPDPAADGGYISSDEAS 117
++ LRV DL PA A DVE +P E +Q PSPV P A G D+
Sbjct 665 MHMLRVFDLIPADACFNQMLDVESVNVSPDGQKDSEAEQGPSPVSVPEAARGSYQDDDM- 723
Query 118 EPP 120
PP
Sbjct 724 -PP 725
> hsa:401827 MSLNL, C16orf37, MPFL; mesothelin-like
Length=1053
Score = 28.9 bits (63), Expect = 4.0, Method: Composition-based stats.
Identities = 14/29 (48%), Positives = 17/29 (58%), Gaps = 0/29 (0%)
Query 12 NVSLPKGTFVQLQPLETDFLDISNPKALL 40
N+S+ TF L PLE LD+ N ALL
Sbjct 881 NISMDIDTFTSLNPLELQSLDVGNVTALL 909
> cel:T21D12.9 hypothetical protein
Length=881
Score = 28.5 bits (62), Expect = 5.8, Method: Composition-based stats.
Identities = 18/51 (35%), Positives = 28/51 (54%), Gaps = 0/51 (0%)
Query 14 SLPKGTFVQLQPLETDFLDISNPKALLEMALRGYAALTKGEVVSLPFLDCV 64
SLP G+F L+ LE L ++ +L + AL G ++L K ++ S CV
Sbjct 329 SLPSGSFRVLRQLEELILSANSIDSLHKFALVGMSSLHKLDLSSNTLAVCV 379
Lambda K H
0.312 0.133 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2013067560
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40