bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0179_orf1
Length=157
Score E
Sequences producing significant alignments: (Bits) Value
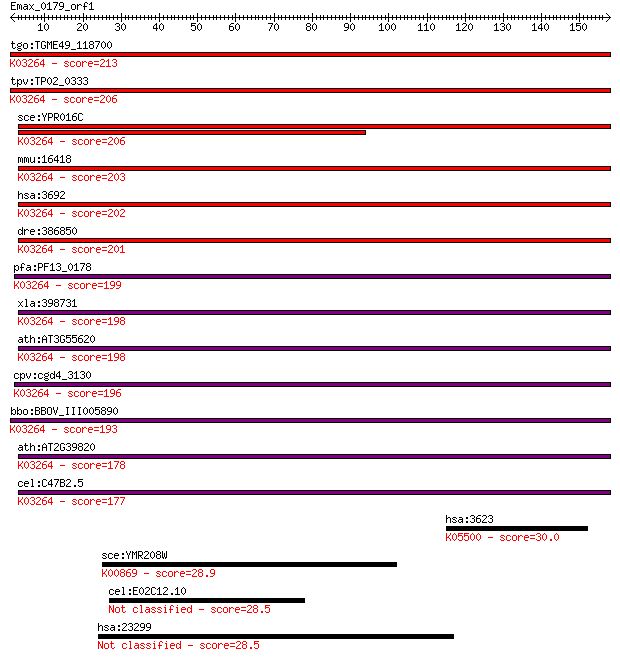
tgo:TGME49_118700 eukaryotic translation initiation factor 6, ... 213 2e-55
tpv:TP02_0333 translation initiation factor 6; K03264 translat... 206 2e-53
sce:YPR016C TIF6, CDC95; Constituent of 66S pre-ribosomal part... 206 2e-53
mmu:16418 Eif6, 1110004P11, AA408895, CAB, Itgb4bp, eIF-6, imc... 203 2e-52
hsa:3692 EIF6, CAB, EIF3A, ITGB4BP, b(2)gcn, eIF-6, p27(BBP), ... 202 4e-52
dre:386850 eif6, Itgb4bp, MGC56562, eIF-6, itgb4bp4, wu:fc28a0... 201 1e-51
pfa:PF13_0178 translation initiation factor 6, putative; K0326... 199 2e-51
xla:398731 eif6, eIF-6, itgb4bp, p27(BBP), p27BBP; eukaryotic ... 198 6e-51
ath:AT3G55620 emb1624 (embryo defective 1624); ribosome bindin... 198 6e-51
cpv:cgd4_3130 eIF6, translation initiation factor 6 ; K03264 t... 196 4e-50
bbo:BBOV_III005890 17.m07522; translation initiation factor eI... 193 2e-49
ath:AT2G39820 eukaryotic translation initiation factor 6, puta... 178 8e-45
cel:C47B2.5 eif-6; Eukaryotic Initiation Factor family member ... 177 9e-45
hsa:3623 INHA; inhibin, alpha; K05500 inhibin, alpha 30.0 3.2
sce:YMR208W ERG12, RAR1; Erg12p (EC:2.7.1.36); K00869 mevalona... 28.9 7.1
cel:E02C12.10 hypothetical protein 28.5 9.9
hsa:23299 BICD2, KIAA0699, bA526D8.1; bicaudal D homolog 2 (Dr... 28.5 10.0
> tgo:TGME49_118700 eukaryotic translation initiation factor 6,
putative ; K03264 translation initiation factor 6
Length=264
Score = 213 bits (543), Expect = 2e-55, Method: Compositional matrix adjust.
Identities = 99/157 (63%), Positives = 123/157 (78%), Gaps = 0/157 (0%)
Query 1 PHIPIIHTTISNTHVIGSLSVGNKHGLLLPIGTSDIEMNHIRNSLPEEIRIRRINDKLSA 60
PHIP++H T+ T VIG + VGN+ GL++P T+D E+ H+RNSLP+ + IRR+ ++LSA
Sbjct 59 PHIPVVHATVGGTRVIGRVCVGNRRGLIVPSITTDQELQHLRNSLPDSVEIRRVEERLSA 118
Query 61 LGNCIVTNDYTALIHPDIDAETAEIIQDVLHVEVFRSIIGGQLLVGSYCVLTNQGGLVHS 120
LGN + NDY AL+H D+D ET EI+QDVL VE FR+ IG Q LVGSYC TNQGGLVH
Sbjct 119 LGNNVACNDYVALLHTDMDKETEEIVQDVLGVEAFRATIGKQTLVGSYCHFTNQGGLVHV 178
Query 121 GTSKEEMEDLSQLVQVPFTAGTVNRGSDLVGAGLMVN 157
T E+ME+LSQL+QVP TAGTVNRGSDLVGAGL+ N
Sbjct 179 MTPVEDMEELSQLLQVPLTAGTVNRGSDLVGAGLIAN 215
> tpv:TP02_0333 translation initiation factor 6; K03264 translation
initiation factor 6
Length=249
Score = 206 bits (525), Expect = 2e-53, Method: Compositional matrix adjust.
Identities = 94/157 (59%), Positives = 124/157 (78%), Gaps = 0/157 (0%)
Query 1 PHIPIIHTTISNTHVIGSLSVGNKHGLLLPIGTSDIEMNHIRNSLPEEIRIRRINDKLSA 60
PHIP++HTTI T VIG +SVGNK GLL+ +D E+ H+RNSLP+ + IRRI+++LSA
Sbjct 44 PHIPVVHTTIGGTRVIGRVSVGNKKGLLVSSICTDKELRHLRNSLPDSVEIRRIDERLSA 103
Query 61 LGNCIVTNDYTALIHPDIDAETAEIIQDVLHVEVFRSIIGGQLLVGSYCVLTNQGGLVHS 120
LGNCI NDY LIH D+D ET EI++DVL +EVFR+ I G +L+GSY N+GGLVH
Sbjct 104 LGNCISANDYVGLIHVDMDKETEEIVEDVLGIEVFRASIAGDVLIGSYTRFQNKGGLVHV 163
Query 121 GTSKEEMEDLSQLVQVPFTAGTVNRGSDLVGAGLMVN 157
T+ EME+LSQL+Q+P T+GT+NRGSD++GAG++VN
Sbjct 164 KTTTSEMEELSQLLQIPLTSGTINRGSDVIGAGIVVN 200
> sce:YPR016C TIF6, CDC95; Constituent of 66S pre-ribosomal particles,
has similarity to human translation initiation factor
6 (eIF6); may be involved in the biogenesis and or stability
of 60S ribosomal subunits; K03264 translation initiation
factor 6
Length=245
Score = 206 bits (524), Expect = 2e-53, Method: Compositional matrix adjust.
Identities = 95/155 (61%), Positives = 123/155 (79%), Gaps = 0/155 (0%)
Query 3 IPIIHTTISNTHVIGSLSVGNKHGLLLPIGTSDIEMNHIRNSLPEEIRIRRINDKLSALG 62
IPI+HTTI+ T +IG ++ GN+ GLL+P T+D E+ H+RNSLP+ ++I+R+ ++LSALG
Sbjct 46 IPIVHTTIAGTRIIGRMTAGNRRGLLVPTQTTDQELQHLRNSLPDSVKIQRVEERLSALG 105
Query 63 NCIVTNDYTALIHPDIDAETAEIIQDVLHVEVFRSIIGGQLLVGSYCVLTNQGGLVHSGT 122
N I NDY AL+HPDID ET E+I DVL VEVFR I G +LVGSYC L+NQGGLVH T
Sbjct 106 NVICCNDYVALVHPDIDRETEELISDVLGVEVFRQTISGNILVGSYCSLSNQGGLVHPQT 165
Query 123 SKEEMEDLSQLVQVPFTAGTVNRGSDLVGAGLMVN 157
S ++ E+LS L+QVP AGTVNRGS +VGAG++VN
Sbjct 166 SVQDQEELSSLLQVPLVAGTVNRGSSVVGAGMVVN 200
Score = 29.3 bits (64), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 24/92 (26%), Positives = 44/92 (47%), Gaps = 2/92 (2%)
Query 3 IPIIHTTISNTHVIGSL-SVGNKHGLLLPIGTSDIEMNHIRNSLPEEIRIRRINDKLSAL 61
+ + TIS ++GS S+ N+ GL+ P TS + + + L + +N S +
Sbjct 135 VEVFRQTISGNILVGSYCSLSNQGGLVHP-QTSVQDQEELSSLLQVPLVAGTVNRGSSVV 193
Query 62 GNCIVTNDYTALIHPDIDAETAEIIQDVLHVE 93
G +V NDY A+ D A +I+ + ++
Sbjct 194 GAGMVVNDYLAVTGLDTTAPELSVIESIFRLQ 225
> mmu:16418 Eif6, 1110004P11, AA408895, CAB, Itgb4bp, eIF-6, imc-415,
p27(BBP), p27BBP; eukaryotic translation initiation
factor 6 (EC:3.6.5.3); K03264 translation initiation factor
6
Length=245
Score = 203 bits (516), Expect = 2e-52, Method: Compositional matrix adjust.
Identities = 90/155 (58%), Positives = 121/155 (78%), Gaps = 0/155 (0%)
Query 3 IPIIHTTISNTHVIGSLSVGNKHGLLLPIGTSDIEMNHIRNSLPEEIRIRRINDKLSALG 62
IP++H +I+ +IG + VGN+HGLL+P T+D E+ HIRNSLP+ ++IRR+ ++LSALG
Sbjct 46 IPVVHASIAGCRIIGRMCVGNRHGLLVPNNTTDQELQHIRNSLPDSVQIRRVEERLSALG 105
Query 63 NCIVTNDYTALIHPDIDAETAEIIQDVLHVEVFRSIIGGQLLVGSYCVLTNQGGLVHSGT 122
N NDY AL+HPD+D ET EI+ DVL VEVFR + Q+LVGSYCV +NQGGLVH T
Sbjct 106 NVTTCNDYVALVHPDLDRETEEILADVLKVEVFRQTVADQVLVGSYCVFSNQGGLVHPKT 165
Query 123 SKEEMEDLSQLVQVPFTAGTVNRGSDLVGAGLMVN 157
S E+ ++LS L+QVP AGTVNRGS+++ AG++VN
Sbjct 166 SIEDQDELSSLLQVPLVAGTVNRGSEVIAAGMVVN 200
> hsa:3692 EIF6, CAB, EIF3A, ITGB4BP, b(2)gcn, eIF-6, p27(BBP),
p27BBP; eukaryotic translation initiation factor 6 (EC:3.6.5.3);
K03264 translation initiation factor 6
Length=245
Score = 202 bits (514), Expect = 4e-52, Method: Compositional matrix adjust.
Identities = 90/155 (58%), Positives = 121/155 (78%), Gaps = 0/155 (0%)
Query 3 IPIIHTTISNTHVIGSLSVGNKHGLLLPIGTSDIEMNHIRNSLPEEIRIRRINDKLSALG 62
IP++H +I+ +IG + VGN+HGLL+P T+D E+ HIRNSLP+ ++IRR+ ++LSALG
Sbjct 46 IPVVHASIAGCRIIGRMCVGNRHGLLVPNNTTDQELQHIRNSLPDTVQIRRVEERLSALG 105
Query 63 NCIVTNDYTALIHPDIDAETAEIIQDVLHVEVFRSIIGGQLLVGSYCVLTNQGGLVHSGT 122
N NDY AL+HPD+D ET EI+ DVL VEVFR + Q+LVGSYCV +NQGGLVH T
Sbjct 106 NVTTCNDYVALVHPDLDRETEEILADVLKVEVFRQTVADQVLVGSYCVFSNQGGLVHPKT 165
Query 123 SKEEMEDLSQLVQVPFTAGTVNRGSDLVGAGLMVN 157
S E+ ++LS L+QVP AGTVNRGS+++ AG++VN
Sbjct 166 SIEDQDELSSLLQVPLVAGTVNRGSEVIAAGMVVN 200
> dre:386850 eif6, Itgb4bp, MGC56562, eIF-6, itgb4bp4, wu:fc28a04,
wu:ft88f05, zgc:56562; eukaryotic translation initiation
factor 6 (EC:3.6.5.3); K03264 translation initiation factor
6
Length=245
Score = 201 bits (510), Expect = 1e-51, Method: Compositional matrix adjust.
Identities = 88/155 (56%), Positives = 120/155 (77%), Gaps = 0/155 (0%)
Query 3 IPIIHTTISNTHVIGSLSVGNKHGLLLPIGTSDIEMNHIRNSLPEEIRIRRINDKLSALG 62
+P+IH +I+ +IG + VGN+HGLL+P T+D E+ HIRN LP+ +RI+R+ ++LSALG
Sbjct 46 MPVIHASIAGCRIIGRMCVGNRHGLLVPNNTTDQELQHIRNCLPDSVRIQRVEERLSALG 105
Query 63 NCIVTNDYTALIHPDIDAETAEIIQDVLHVEVFRSIIGGQLLVGSYCVLTNQGGLVHSGT 122
N I NDY AL+HPD+D ET EI+ D L VEVFR + Q+LVGSYC +NQGGLVH+ T
Sbjct 106 NVIACNDYVALVHPDLDRETEEILADTLKVEVFRQTVAEQVLVGSYCAFSNQGGLVHAKT 165
Query 123 SKEEMEDLSQLVQVPFTAGTVNRGSDLVGAGLMVN 157
S E+ ++LS L+QVP AGTVNRGS+++ AG++VN
Sbjct 166 SIEDQDELSSLLQVPLVAGTVNRGSEVIAAGMVVN 200
> pfa:PF13_0178 translation initiation factor 6, putative; K03264
translation initiation factor 6
Length=247
Score = 199 bits (507), Expect = 2e-51, Method: Compositional matrix adjust.
Identities = 92/156 (58%), Positives = 121/156 (77%), Gaps = 0/156 (0%)
Query 2 HIPIIHTTISNTHVIGSLSVGNKHGLLLPIGTSDIEMNHIRNSLPEEIRIRRINDKLSAL 61
HIP+++TTI T VIG + VGN+ GLL+ +D E+ H+RN LPE ++I+RI ++LSAL
Sbjct 45 HIPLVYTTIGGTRVIGRVCVGNRKGLLVSSICTDQELLHLRNCLPENVKIKRIEERLSAL 104
Query 62 GNCIVTNDYTALIHPDIDAETAEIIQDVLHVEVFRSIIGGQLLVGSYCVLTNQGGLVHSG 121
GNCI TNDY LIH DID ET EIIQDVL +EVFR+ I G LLVG+Y TN GGL+H+
Sbjct 105 GNCITTNDYVGLIHTDIDKETEEIIQDVLDIEVFRTSIAGNLLVGTYSYFTNNGGLLHAM 164
Query 122 TSKEEMEDLSQLVQVPFTAGTVNRGSDLVGAGLMVN 157
TS +E+E+LS+L+Q+P GT+NRGSDL+G+GL+ N
Sbjct 165 TSSQEIEELSELLQIPLITGTINRGSDLIGSGLVAN 200
> xla:398731 eif6, eIF-6, itgb4bp, p27(BBP), p27BBP; eukaryotic
translation initiation factor 6 (EC:3.6.5.3); K03264 translation
initiation factor 6
Length=245
Score = 198 bits (504), Expect = 6e-51, Method: Compositional matrix adjust.
Identities = 87/155 (56%), Positives = 120/155 (77%), Gaps = 0/155 (0%)
Query 3 IPIIHTTISNTHVIGSLSVGNKHGLLLPIGTSDIEMNHIRNSLPEEIRIRRINDKLSALG 62
IP++H +I+ +IG + VGN+HGL++P T+D E+ H+RNSLP+ +RI+R+ ++LSALG
Sbjct 46 IPVVHASIAGCRIIGRMCVGNRHGLMVPNNTTDQELQHMRNSLPDSVRIQRVEERLSALG 105
Query 63 NCIVTNDYTALIHPDIDAETAEIIQDVLHVEVFRSIIGGQLLVGSYCVLTNQGGLVHSGT 122
N I NDY AL+HPD+D ET EI+ DVL VEVFR I Q+LVGSYC +NQGGL+H T
Sbjct 106 NVIACNDYVALVHPDLDRETEEILADVLKVEVFRQTIAEQVLVGSYCAFSNQGGLLHPKT 165
Query 123 SKEEMEDLSQLVQVPFTAGTVNRGSDLVGAGLMVN 157
S E+ ++LS L+QVP GTVNRGS+++ AG++VN
Sbjct 166 SIEDQDELSSLLQVPLVTGTVNRGSEVIAAGMVVN 200
> ath:AT3G55620 emb1624 (embryo defective 1624); ribosome binding
/ translation initiation factor; K03264 translation initiation
factor 6
Length=245
Score = 198 bits (504), Expect = 6e-51, Method: Compositional matrix adjust.
Identities = 92/155 (59%), Positives = 121/155 (78%), Gaps = 0/155 (0%)
Query 3 IPIIHTTISNTHVIGSLSVGNKHGLLLPIGTSDIEMNHIRNSLPEEIRIRRINDKLSALG 62
IPI+ T+I T +IG L GNK+GLL+P T+D E+ H+RNSLP+++ ++RI+++LSALG
Sbjct 46 IPIVKTSIGGTRIIGRLCAGNKNGLLVPHTTTDQELQHLRNSLPDQVVVQRIDERLSALG 105
Query 63 NCIVTNDYTALIHPDIDAETAEIIQDVLHVEVFRSIIGGQLLVGSYCVLTNQGGLVHSGT 122
NCI NDY AL H D+D ET EII DVL VEVFR I G +LVGSYC L+N+GG+VH T
Sbjct 106 NCIACNDYVALAHTDLDKETEEIIADVLGVEVFRQTIAGNILVGSYCALSNKGGMVHPHT 165
Query 123 SKEEMEDLSQLVQVPFTAGTVNRGSDLVGAGLMVN 157
S E++E+LS L+QVP AGTVNRGS+++ AG+ VN
Sbjct 166 SVEDLEELSTLLQVPLVAGTVNRGSEVIAAGMTVN 200
> cpv:cgd4_3130 eIF6, translation initiation factor 6 ; K03264
translation initiation factor 6
Length=252
Score = 196 bits (497), Expect = 4e-50, Method: Compositional matrix adjust.
Identities = 91/156 (58%), Positives = 122/156 (78%), Gaps = 0/156 (0%)
Query 2 HIPIIHTTISNTHVIGSLSVGNKHGLLLPIGTSDIEMNHIRNSLPEEIRIRRINDKLSAL 61
HIPII+T I T ++G +VGN++GLL+ +D E+ H+RNSLP+ ++++RI ++LSAL
Sbjct 45 HIPIINTLIGGTRLVGRCTVGNRNGLLVSNMATDQELQHLRNSLPDNVKVQRIEERLSAL 104
Query 62 GNCIVTNDYTALIHPDIDAETAEIIQDVLHVEVFRSIIGGQLLVGSYCVLTNQGGLVHSG 121
GNCI NDY ALIH D+D E+ EIIQDVL VEVFR+ I G +LVGSY TNQGG+VH+
Sbjct 105 GNCIACNDYVALIHTDMDKESEEIIQDVLGVEVFRTTIAGHVLVGSYAKFTNQGGIVHTL 164
Query 122 TSKEEMEDLSQLVQVPFTAGTVNRGSDLVGAGLMVN 157
S++EM +LS L+Q+P T+GTVNRGSD+V AG +VN
Sbjct 165 ASEDEMNELSALLQIPITSGTVNRGSDVVSAGCVVN 200
> bbo:BBOV_III005890 17.m07522; translation initiation factor
eIF-6 family protein; K03264 translation initiation factor 6
Length=262
Score = 193 bits (491), Expect = 2e-49, Method: Compositional matrix adjust.
Identities = 92/170 (54%), Positives = 122/170 (71%), Gaps = 13/170 (7%)
Query 1 PHIPIIHTTISNTHVIGSLSVG-------------NKHGLLLPIGTSDIEMNHIRNSLPE 47
P IP++ TTI T V+GS++VG N+ GLL+ +D E+ H+RNSLP+
Sbjct 44 PQIPVVQTTIGGTRVVGSVTVGMLYSNYSVILFTGNRKGLLVSSICTDTELRHLRNSLPD 103
Query 48 EIRIRRINDKLSALGNCIVTNDYTALIHPDIDAETAEIIQDVLHVEVFRSIIGGQLLVGS 107
+ IRRI+D+LSALGN I NDY LIH DID ET EI++DVL +EVFR+ I G +L+GS
Sbjct 104 SVEIRRIDDRLSALGNVITCNDYVGLIHVDIDRETEEIVEDVLGIEVFRASIAGNVLIGS 163
Query 108 YCVLTNQGGLVHSGTSKEEMEDLSQLVQVPFTAGTVNRGSDLVGAGLMVN 157
YC N+GGLVH T+ +EME+LSQL+Q+P T+GTVNRGSD++G GL+ N
Sbjct 164 YCRFQNKGGLVHVKTTTDEMEELSQLLQIPLTSGTVNRGSDVIGGGLIAN 213
> ath:AT2G39820 eukaryotic translation initiation factor 6, putative
/ eIF-6, putative; K03264 translation initiation factor
6
Length=247
Score = 178 bits (451), Expect = 8e-45, Method: Compositional matrix adjust.
Identities = 87/155 (56%), Positives = 113/155 (72%), Gaps = 0/155 (0%)
Query 3 IPIIHTTISNTHVIGSLSVGNKHGLLLPIGTSDIEMNHIRNSLPEEIRIRRINDKLSALG 62
IPI+ T+I + IGSL VGNK+GLLL +D E+ H+R+SLP+E+ ++RI + + ALG
Sbjct 48 IPIVTTSIGGSGTIGSLCVGNKNGLLLSHTITDQELQHLRDSLPDEVVVQRIEEPICALG 107
Query 63 NCIVTNDYTALIHPDIDAETAEIIQDVLHVEVFRSIIGGQLLVGSYCVLTNQGGLVHSGT 122
N I NDY AL+HP ++ +T EII DVL VEV+R I LVGSYC L+N GG+VHS T
Sbjct 108 NAIACNDYVALVHPKLEKDTEEIISDVLGVEVYRQTIANNELVGSYCSLSNNGGMVHSNT 167
Query 123 SKEEMEDLSQLVQVPFTAGTVNRGSDLVGAGLMVN 157
+ EEM +L+ LVQVP AGTVNRGS ++ AGL VN
Sbjct 168 NVEEMVELANLVQVPLVAGTVNRGSQVISAGLTVN 202
> cel:C47B2.5 eif-6; Eukaryotic Initiation Factor family member
(eif-6); K03264 translation initiation factor 6
Length=246
Score = 177 bits (450), Expect = 9e-45, Method: Compositional matrix adjust.
Identities = 79/155 (50%), Positives = 119/155 (76%), Gaps = 0/155 (0%)
Query 3 IPIIHTTISNTHVIGSLSVGNKHGLLLPIGTSDIEMNHIRNSLPEEIRIRRINDKLSALG 62
IP++HT+I++T ++G L+VGN+HGLL+P T+D E+ H+RNSLP+E+ IRR++++LSALG
Sbjct 46 IPVVHTSIASTRIVGRLTVGNRHGLLVPNATTDQELQHLRNSLPDEVAIRRVDERLSALG 105
Query 63 NCIVTNDYTALIHPDIDAETAEIIQDVLHVEVFRSIIGGQLLVGSYCVLTNQGGLVHSGT 122
N I ND+ A++H +I AET + + +VL VEVFR + LVGSYC+L++ G LV + T
Sbjct 106 NVIACNDHVAIVHAEISAETEQALVEVLKVEVFRVSLAQNSLVGSYCILSSNGCLVAART 165
Query 123 SKEEMEDLSQLVQVPFTAGTVNRGSDLVGAGLMVN 157
E +++ L+Q+P AGT NRGS+L+GAG++VN
Sbjct 166 PPETQREIAALLQIPVVAGTCNRGSELIGAGMVVN 200
> hsa:3623 INHA; inhibin, alpha; K05500 inhibin, alpha
Length=366
Score = 30.0 bits (66), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 14/37 (37%), Positives = 18/37 (48%), Gaps = 0/37 (0%)
Query 115 GGLVHSGTSKEEMEDLSQLVQVPFTAGTVNRGSDLVG 151
GG H G+ EE ED+SQ + P T + S G
Sbjct 65 GGFTHRGSEPEEEEDVSQAILFPATDASCEDKSAARG 101
> sce:YMR208W ERG12, RAR1; Erg12p (EC:2.7.1.36); K00869 mevalonate
kinase [EC:2.7.1.36]
Length=443
Score = 28.9 bits (63), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 25/81 (30%), Positives = 38/81 (46%), Gaps = 10/81 (12%)
Query 25 HGLLLPIGTSDIEMNHIRNSLPEEIRIRRINDKLSALGNCIVTNDYTALIHPDIDAET-- 82
HGLL+ IG S + I+N L +++RI + G C +T L+ DI E
Sbjct 319 HGLLVSIGVSHPGLELIKN-LSDDLRIGSTKLTGAGGGGCSLT-----LLRRDITQEQID 372
Query 83 --AEIIQDVLHVEVFRSIIGG 101
+ +QD E F + +GG
Sbjct 373 SFKKKLQDDFSYETFETDLGG 393
> cel:E02C12.10 hypothetical protein
Length=665
Score = 28.5 bits (62), Expect = 9.9, Method: Composition-based stats.
Identities = 15/51 (29%), Positives = 27/51 (52%), Gaps = 0/51 (0%)
Query 27 LLLPIGTSDIEMNHIRNSLPEEIRIRRINDKLSALGNCIVTNDYTALIHPD 77
LLLPI +S ++ + I + RI+ + ++ + + I +DY HPD
Sbjct 363 LLLPIVSSFLDNSPISEVEKSQARIKNQKNMIAMMEDLIEVHDYNMKNHPD 413
> hsa:23299 BICD2, KIAA0699, bA526D8.1; bicaudal D homolog 2 (Drosophila)
Length=855
Score = 28.5 bits (62), Expect = 10.0, Method: Composition-based stats.
Identities = 25/94 (26%), Positives = 44/94 (46%), Gaps = 6/94 (6%)
Query 24 KHGLLLPIGTSDIEMNHIRNSLPEEI-RIRRINDKLSALGNCIVTNDYTALIHPDIDAET 82
K GLL + + ++ H R SL E+ ++ R+ + LSAL + + + + D ++
Sbjct 359 KAGLLATLQDTQKQLEHTRGSLSEQQEKVTRLTENLSALRRLQASKERQTALDNEKDRDS 418
Query 83 AEIIQDVLHVEVFRSIIGGQLLVGSYCVLTNQGG 116
E D + EV I G ++L Y V + G
Sbjct 419 HE---DGDYYEV--DINGPEILACKYHVAVAEAG 447
Lambda K H
0.318 0.137 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3516928200
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40