bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0169_orf1
Length=247
Score E
Sequences producing significant alignments: (Bits) Value
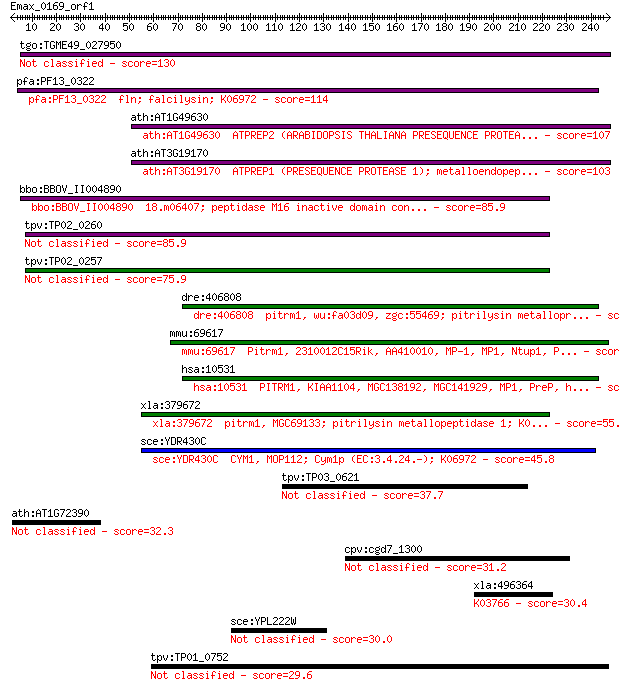
tgo:TGME49_027950 zinc metalloprotease 2, putative (EC:3.4.24.55) 130 4e-30
pfa:PF13_0322 fln; falcilysin; K06972 114 3e-25
ath:AT1G49630 ATPREP2 (ARABIDOPSIS THALIANA PRESEQUENCE PROTEA... 107 4e-23
ath:AT3G19170 ATPREP1 (PRESEQUENCE PROTEASE 1); metalloendopep... 103 7e-22
bbo:BBOV_II004890 18.m06407; peptidase M16 inactive domain con... 85.9 1e-16
tpv:TP02_0260 falcilysin 85.9 1e-16
tpv:TP02_0257 falcilysin 75.9 1e-13
dre:406808 pitrm1, wu:fa03d09, zgc:55469; pitrilysin metallopr... 66.6 8e-11
mmu:69617 Pitrm1, 2310012C15Rik, AA410010, MP-1, MP1, Ntup1, P... 64.3 3e-10
hsa:10531 PITRM1, KIAA1104, MGC138192, MGC141929, MP1, PreP, h... 61.6 2e-09
xla:379672 pitrm1, MGC69133; pitrilysin metallopeptidase 1; K0... 55.1 2e-07
sce:YDR430C CYM1, MOP112; Cym1p (EC:3.4.24.-); K06972 45.8 1e-04
tpv:TP03_0621 hypothetical protein 37.7 0.042
ath:AT1G72390 hypothetical protein 32.3 1.7
cpv:cgd7_1300 hypothetical protein 31.2 3.8
xla:496364 b3gnt5-a, b3gn-t5, b3gnt5, b3gnt5b, beta3gn-t5; UDP... 30.4 5.5
sce:YPL222W FMP40; Fmp40p 30.0 9.1
tpv:TP01_0752 hypothetical protein 29.6 9.3
> tgo:TGME49_027950 zinc metalloprotease 2, putative (EC:3.4.24.55)
Length=1728
Score = 130 bits (327), Expect = 4e-30, Method: Compositional matrix adjust.
Identities = 84/248 (33%), Positives = 128/248 (51%), Gaps = 29/248 (11%)
Query 5 YPSSAEELKDWVTVSWVLNPGKKEEKEGKKEEKKDSSPPSSSSCTLLNGEERLLLQVLNF 64
+P+ E+L+D+VTVS VL+P + +R L VL
Sbjct 926 FPAPKEQLEDFVTVSMVLDP---------------------LGVAVPTPFQRQTLGVLTH 964
Query 65 LLIGDKASPLYSSLSNYTQLG-GPAYTPGLDLDLKYAMFTVGLKDVPQREG----AAKEV 119
LL+G SPLY +L T+ G G L+ LK+ +FT GLK +PQ+ +V
Sbjct 965 LLVGTSPSPLYRAL---TESGLGKQVMGELEDGLKHLIFTAGLKGIPQQSAEDSSVVDKV 1021
Query 120 EELVINTLKEIKEKGFSSLSLSAALNKIEFLLRETPRDRLPRGLIFARKMTQEIVYNKDP 179
E++V++ L++ +GFS ++ AA+N EFLLRE P+GL R+M ++DP
Sbjct 1022 EQIVLDCLEKHAREGFSEEAIEAAINSREFLLREFNTGTFPKGLAVIREMAALWTEDRDP 1081
Query 180 LNALEFEETLWKIRERLAKGEKVFEQLIQKYFSDNPHRLTLRLIGNKEIIKEREEEEKKS 239
+ L FEE ++R RL GE VF+ L+QK+F NPHR T+ L + + RE +EK
Sbjct 1082 VEGLRFEEHFEELRRRLKSGEPVFQNLLQKFFIGNPHRATIHLRADPDEEARREAQEKAE 1141
Query 240 LKKAEKEM 247
+ + +
Sbjct 1142 ISALQASL 1149
> pfa:PF13_0322 fln; falcilysin; K06972
Length=1193
Score = 114 bits (285), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 75/254 (29%), Positives = 137/254 (53%), Gaps = 17/254 (6%)
Query 4 RYPSSAEELKDWVTVSWVLNPGKKEEKEGKKEEKKD--------SSPPSSSSCTLLNGEE 55
+Y +EE ++ V+V+W+LNP + + S+ SS +L N +
Sbjct 353 KYGDHSEEKENLVSVAWLLNPKVDKTNNHNNNHSNNQSSENNGYSNGSHSSDLSLENPTD 412
Query 56 RLLLQVLNFLLIGDKASPLYSSLSNYTQLGGPAYTPGLDLDLKYAMFTVGLKDVPQREGA 115
+L ++N LLI S LY +L++ LG GL+ L +F++GLK + +
Sbjct 413 YFVLLIINNLLIHTPESVLYKALTD-CGLGNNVIDRGLNDSLVQYIFSIGLKGIKRNNEK 471
Query 116 AK-------EVEELVINTLKEIKEKGFSSLSLSAALNKIEFLLRETPRDRLPRGLIFARK 168
K EVE++++N LK++ ++GF+ ++ A++N IEF+L+E + + + F +
Sbjct 472 IKNFDKVHYEVEDVIMNALKKVVKEGFNKSAVEASINNIEFILKEANL-KTSKSIDFVFE 530
Query 169 MTQEIVYNKDPLNALEFEETLWKIRERLAKGEKVFEQLIQKYFSDNPHRLTLRLIGNKEI 228
MT ++ YN+DPL EFE+ L ++ ++ E+ ++K+F +N HR + L G++
Sbjct 531 MTSKLNYNRDPLLIFEFEKYLNIVKNKIKNEPMYLEKFVEKHFINNAHRSVILLEGDENY 590
Query 229 IKEREEEEKKSLKK 242
+E+E EK+ LKK
Sbjct 591 AQEQENLEKQELKK 604
> ath:AT1G49630 ATPREP2 (ARABIDOPSIS THALIANA PRESEQUENCE PROTEASE
2); catalytic/ metal ion binding / metalloendopeptidase/
metallopeptidase/ zinc ion binding; K06972
Length=1080
Score = 107 bits (267), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 71/199 (35%), Positives = 115/199 (57%), Gaps = 5/199 (2%)
Query 51 LNGEERLLLQVLNFLLIGDKASPLYSSLSNYTQLGGPAYTPGLDLDLKYAMFTVGLKDVP 110
L+ + +L L L+ L++G ASPL L + LG G++ +L F++GLK V
Sbjct 395 LDLQTQLALGFLDHLMLGTPASPLRKILLE-SGLGEALVNSGMEDELLQPQFSIGLKGV- 452
Query 111 QREGAAKEVEELVINTLKEIKEKGFSSLSLSAALNKIEFLLRETPRDRLPRGLIFARKMT 170
+ ++VEELV+NTL+++ ++GF + ++ A++N IEF LRE PRGL +
Sbjct 453 -SDDNVQKVEELVMNTLRKLADEGFDTDAVEASMNTIEFSLRENNTGSSPRGLSLMLQSI 511
Query 171 QEIVYNKDPLNALEFEETLWKIRERLA-KGEK-VFEQLIQKYFSDNPHRLTLRLIGNKEI 228
+ +Y+ DP L++EE L ++ R+A KG K VF LI++Y +NPH +T+ + + E
Sbjct 512 AKWIYDMDPFEPLKYEEPLKSLKARIAEKGSKSVFSPLIEEYILNNPHCVTIEMQPDPEK 571
Query 229 IKEREEEEKKSLKKAEKEM 247
E EEK L+K + M
Sbjct 572 ASLEEAEEKSILEKVKASM 590
> ath:AT3G19170 ATPREP1 (PRESEQUENCE PROTEASE 1); metalloendopeptidase;
K06972
Length=1080
Score = 103 bits (256), Expect = 7e-22, Method: Compositional matrix adjust.
Identities = 70/199 (35%), Positives = 113/199 (56%), Gaps = 5/199 (2%)
Query 51 LNGEERLLLQVLNFLLIGDKASPLYSSLSNYTQLGGPAYTPGLDLDLKYAMFTVGLKDVP 110
L+ + +L L L+ L++G ASPL L + LG + GL +L F +GLK V
Sbjct 396 LDLQTQLALGFLDHLMLGTPASPLRKILLE-SGLGEALVSSGLSDELLQPQFGIGLKGVS 454
Query 111 QREGAAKEVEELVINTLKEIKEKGFSSLSLSAALNKIEFLLRETPRDRLPRGLIFARKMT 170
+ ++VEEL+++TLK++ E+GF + ++ A++N IEF LRE PRGL +
Sbjct 455 EEN--VQKVEELIMDTLKKLAEEGFDNDAVEASMNTIEFSLRENNTGSFPRGLSLMLQSI 512
Query 171 QEIVYNKDPLNALEFEETLWKIRERLA-KGEK-VFEQLIQKYFSDNPHRLTLRLIGNKEI 228
+ +Y+ DP L++ E L ++ R+A +G K VF LI+K +N HR+T+ + + E
Sbjct 513 SKWIYDMDPFEPLKYTEPLKALKTRIAEEGSKAVFSPLIEKLILNNSHRVTIEMQPDPEK 572
Query 229 IKEREEEEKKSLKKAEKEM 247
+ E EEK L+K + M
Sbjct 573 ATQEEVEEKNILEKVKAAM 591
> bbo:BBOV_II004890 18.m06407; peptidase M16 inactive domain containing
protein (EC:3.4.24.-); K06972
Length=1166
Score = 85.9 bits (211), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 55/222 (24%), Positives = 106/222 (47%), Gaps = 15/222 (6%)
Query 5 YPSSAEELKDWVTVSWVLNPGKKEEKEGKKEEKKDSSPPSSSSCTLLNGEERLLLQVLNF 64
+ +S E +D + W+L+P E + K ++ + L ++VL
Sbjct 411 FGASGSEEEDIILTGWLLDPQTASSGETDRVTGK----------YRIDLVDALGMEVLEH 460
Query 65 LLIGDKASPLYSSLSNYTQLGGPAYTPGLDLDLKYAMFTVGLKDVPQRE----GAAKEVE 120
LL+G S LY +L + LG GL K + F +G+ + ++ A +
Sbjct 461 LLMGTSESYLYKALIK-SGLGKKVVGSGLTNYFKQSNFIIGIAGIDPKQYDKANALATFD 519
Query 121 ELVINTLKEIKEKGFSSLSLSAALNKIEFLLRETPRDRLPRGLIFARKMTQEIVYNKDPL 180
++ +TL ++ G ++ A++N IEF +RE P+GL+ M + Y KDP+
Sbjct 520 SIMNSTLLDMMNNGIKKEAIEASMNYIEFQIRELNTGTFPKGLMLVNLMQSQSQYQKDPI 579
Query 181 NALEFEETLWKIRERLAKGEKVFEQLIQKYFSDNPHRLTLRL 222
L F+ + ++++R+A K F++LI + +N H++T+ +
Sbjct 580 ECLYFDRFIAELKQRVANDSKYFQKLIDTHLVNNRHKVTVHM 621
> tpv:TP02_0260 falcilysin
Length=1181
Score = 85.9 bits (211), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 61/223 (27%), Positives = 107/223 (47%), Gaps = 24/223 (10%)
Query 7 SSAEELKDWVTVSWVLNPGKKEEKEGKKEEKKDSSPPSSSSCTLLNGEERLLLQVLNFLL 66
SS + +D + +SW+L+P + K + D+ + QVL +LL
Sbjct 429 SSVDPTEDELMISWLLDPLYNGSMDKYKIDPVDN----------------VGFQVLQYLL 472
Query 67 IGDKASPLYSSLSNYTQLGGPAYTPGLDLDLKYAMFTVGLKDVPQ-----REGAAKEVEE 121
+G S LY L + + LG G K ++F+ GLK V ++ K+ EE
Sbjct 473 LGTPESVLYKGLID-SGLGKKVLVHGFLSGYKQSLFSFGLKGVDNTKFNSKDEIVKKFEE 531
Query 122 LVINTLKEIKEKGFSSLSLSAALNKIEFLLRETPRDRLPRGLIFARKMTQEIVYNKDPLN 181
+V L++IKE+GF ++ + LN +EF +RE P+GL+ ++ ++ Y +DP
Sbjct 532 VVFGILRKIKEEGFKRDAIDSGLNLVEFEMRELNSGSYPKGLMLIDQIQSQLQYGRDPFA 591
Query 182 ALEFEETLWKIRERLAKGE--KVFEQLIQKYFSDNPHRLTLRL 222
L F+ + ++R R+ F L+ K+ +N R+T+ L
Sbjct 592 LLRFDSLMKELRRRIFSDNPSNYFINLMAKHILNNATRVTVHL 634
> tpv:TP02_0257 falcilysin
Length=1119
Score = 75.9 bits (185), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 68/232 (29%), Positives = 107/232 (46%), Gaps = 34/232 (14%)
Query 7 SSAEELKDWVTVSWVLNPGKKEEKEGKKEEKKDSSPPSSSSCTLLNGEERLLLQVLNFLL 66
SS + +D + WVLNP K K K D L+ ++L L+VL++LL
Sbjct 361 SSKTKDEDMFMMGWVLNPSHKGSK------KYD-----------LDSVDKLALEVLSYLL 403
Query 67 IGDKASPLYSSLSNYTQLGGPAYTPGLDLDL---KYAMFTVGLKDVP----QREGAAKEV 119
+ S L + L + ++ PGLD +Y F G+ V R+ AK
Sbjct 404 LESSESVLLNKLVS-SKFATRRVGPGLDEYFPAYEYLSFMFGVTGVKYTEKTRDSNAKTF 462
Query 120 EELVINTLKEIKEKGFSSLSLSAALNKIEFL-------LRETPRDRLPRGLIFARKMTQE 172
E++V+ L E+ KGF+ ++ AALNK+EF ++E R PRGL R +
Sbjct 463 EKMVLEALTEVVTKGFNRKAVEAALNKVEFKHTEKKYEMKEHRRGYYPRGLALLRLVKPR 522
Query 173 IVYNKDPLNALEFEETLWKIRERLAKGEKV--FEQLIQKYFSDNPHRLTLRL 222
KDP L FE+ +++ R+ + L++K+ +N R+TL L
Sbjct 523 YQEGKDPFELLRFEQLFPELKLRVFSDDSCSYLSNLVKKHLLNNNTRVTLHL 574
> dre:406808 pitrm1, wu:fa03d09, zgc:55469; pitrilysin metalloproteinase
1 (EC:3.4.24.-); K06972
Length=1023
Score = 66.6 bits (161), Expect = 8e-11, Method: Composition-based stats.
Identities = 42/171 (24%), Positives = 86/171 (50%), Gaps = 5/171 (2%)
Query 72 SPLYSSLSNYTQLGGPAYTPGLDLDLKYAMFTVGLKDVPQREGAAKEVEELVINTLKEIK 131
SP Y +L + + G D + A FT+GL+ + E + V+ ++ T+ +I
Sbjct 354 SPFYKALIEPKIGSDFSSSAGFDGSTRQASFTIGLQGMA--EDDTETVKHIIAQTIDDII 411
Query 132 EKGFSSLSLSAALNKIEFLLRETPRDRLPRGLIFARKMTQEIVYNKDPLNALEFEETLWK 191
GF + A L+KIE ++ GL A + ++ DP+ L+ E++ +
Sbjct 412 ASGFEEEQIEALLHKIEIQMKHQSTSF---GLALASYIASLWNHDGDPVQLLKISESVSR 468
Query 192 IRERLAKGEKVFEQLIQKYFSDNPHRLTLRLIGNKEIIKEREEEEKKSLKK 242
R+ L + + ++ +Q YF +N H+LTL + ++ ++++ E E++ L++
Sbjct 469 FRQCLKENPRYLQEKVQHYFKNNTHQLTLSMSPDERFLEKQAEAEEQKLQQ 519
> mmu:69617 Pitrm1, 2310012C15Rik, AA410010, MP-1, MP1, Ntup1,
PreP, mKIAA1104; pitrilysin metallepetidase 1; K06972
Length=1036
Score = 64.3 bits (155), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 53/190 (27%), Positives = 95/190 (50%), Gaps = 18/190 (9%)
Query 67 IGDKASPLYSSLSNYTQLG-GPAYTP--GLDLDLKYAMFTVGLKDVPQREGAAKEVEELV 123
I SP Y +L + G G ++P G + + A F+VGL+ + +++ K V ELV
Sbjct 354 IAGPNSPFYKAL---IESGLGTDFSPDVGYNGYTREAYFSVGLQGIAEKD--VKTVRELV 408
Query 124 INTLKEIKEKGFSSLSLSAALNKIEFLLRETPRDRLPRGLIFARKMTQEIVYNKDPLNAL 183
T++E+ EKGF + A L+KIE +T GL + ++ DP+ L
Sbjct 409 DRTIEEVIEKGFEDDRIEALLHKIEI---QTKHQSASFGLTLTSYIASCWNHDGDPVELL 465
Query 184 EFEETLWKIRERLAKGEKVFEQLIQKYFSDNPHRLTLRLIGN-------KEIIKEREEEE 236
+ L + R+ L + K ++ +++YF +N H+LTL + + ++ E+ E++
Sbjct 466 QIGSQLTRFRKCLKENPKFLQEKVEQYFKNNQHKLTLSMKPDDKYYEKQTQMETEKLEQK 525
Query 237 KKSLKKAEKE 246
SL A+K+
Sbjct 526 VNSLSPADKQ 535
> hsa:10531 PITRM1, KIAA1104, MGC138192, MGC141929, MP1, PreP,
hMP1; pitrilysin metallopeptidase 1; K06972
Length=1037
Score = 61.6 bits (148), Expect = 2e-09, Method: Composition-based stats.
Identities = 45/173 (26%), Positives = 87/173 (50%), Gaps = 9/173 (5%)
Query 72 SPLYSSLSNYTQLGGPAYTP--GLDLDLKYAMFTVGLKDVPQREGAAKEVEELVINTLKE 129
SP Y +L G ++P G + + A F+VGL+ + +++ + V L+ T+ E
Sbjct 359 SPFYKALIESGL--GTDFSPDVGYNGYTREAYFSVGLQGIAEKD--IETVRSLIDRTIDE 414
Query 130 IKEKGFSSLSLSAALNKIEFLLRETPRDRLPRGLIFARKMTQEIVYNKDPLNALEFEETL 189
+ EKGF + A L+KIE ++ GL+ + ++ DP+ L+ L
Sbjct 415 VVEKGFEDDRIEALLHKIEIQMKHQSTSF---GLMLTSYIASCWNHDGDPVELLKLGNQL 471
Query 190 WKIRERLAKGEKVFEQLIQKYFSDNPHRLTLRLIGNKEIIKEREEEEKKSLKK 242
K R+ L + K ++ +++YF +N H+LTL + + + +++ + E LK+
Sbjct 472 AKFRQCLQENPKFLQEKVKQYFKNNQHKLTLSMRPDDKYHEKQAQVEATKLKQ 524
> xla:379672 pitrm1, MGC69133; pitrilysin metallopeptidase 1;
K06972
Length=1027
Score = 55.1 bits (131), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 39/170 (22%), Positives = 83/170 (48%), Gaps = 9/170 (5%)
Query 55 ERLLLQVLNFLLIGDKASPLYSSLSNYTQLGGPAYTP--GLDLDLKYAMFTVGLKDVPQR 112
E L +L+ L++ SP Y +L G ++P G + + F++GL+ + +
Sbjct 336 EAFTLSLLSSLMVDGPNSPFYKALIEANL--GTDFSPDTGFNNYTRETYFSIGLQGINKE 393
Query 113 EGAAKEVEELVINTLKEIKEKGFSSLSLSAALNKIEFLLRETPRDRLPRGLIFARKMTQE 172
+ +++V+ ++ T+ EI E+G + A L+K+E ++ GL A +
Sbjct 394 D--SEKVKHIINRTINEIAEQGIEPERIEALLHKLEIQMKH---QSTSFGLTLASYIASC 448
Query 173 IVYNKDPLNALEFEETLWKIRERLAKGEKVFEQLIQKYFSDNPHRLTLRL 222
+ DP++ L+ + + + R+ L + K + +++YF N HR+ L +
Sbjct 449 WNHEGDPVDLLKIGDKISRFRQCLKENPKFLQDKVKQYFQVNQHRMMLSM 498
> sce:YDR430C CYM1, MOP112; Cym1p (EC:3.4.24.-); K06972
Length=989
Score = 45.8 bits (107), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 48/188 (25%), Positives = 82/188 (43%), Gaps = 7/188 (3%)
Query 55 ERLLLQVLNFLLIGDKASPLYSSLSNYTQLGGPAYTPGLDLDLKYAMFTVGLKDVPQREG 114
+ LL+VL LL+ +S +Y L + G++ + TVG++ V E
Sbjct 317 DTFLLKVLGNLLMDGHSSVMYQKLIESGIGLEFSVNSGVEPTTAVNLLTVGIQGVSDIEI 376
Query 115 AAKEVEELVINTLKEIKEKGFSSLSLSAALNKIEFLLRETPRDRLPRGLIFARKMTQEIV 174
V + N L+ E F + A + ++E ++ D GL +
Sbjct 377 FKDTVNNIFQNLLE--TEHPFDRKRIDAIIEQLELSKKDQKADF---GLQLLYSILPGWT 431
Query 175 YNKDPLNALEFEETLWKIRERL-AKGEKVFEQLIQKYFSDNPHRLTLRLIGNKEIIKERE 233
DP +L FE+ L + R L KG+ +F+ LI+KY P T + G++E K +
Sbjct 432 NKIDPFESLLFEDVLQRFRGDLETKGDTLFQDLIRKYIVHKP-CFTFSIQGSEEFSKSLD 490
Query 234 EEEKKSLK 241
+EE+ L+
Sbjct 491 DEEQTRLR 498
> tpv:TP03_0621 hypothetical protein
Length=551
Score = 37.7 bits (86), Expect = 0.042, Method: Compositional matrix adjust.
Identities = 26/106 (24%), Positives = 58/106 (54%), Gaps = 14/106 (13%)
Query 113 EGAAKEVEELVINTLKEIKEKGFSSLSLSAALNKIEFLLRETPRDRLPRGLIFARKMTQE 172
E E +++N +K E+ F +L++++AL + +LL+ T +++ ++ +K+
Sbjct 310 ENLYAECGNMLLNMVKRCFEEKFVNLNMNSALPTLTYLLQLTNSNKIK--MLMLKKL--- 364
Query 173 IVYNKDPLNALEFEE-----TLWKIRERLAKGEKVFEQLIQKYFSD 213
+K+ +N L+F + ++K R L K ++ QL++ YF+D
Sbjct 365 ---DKNSVNQLDFPQFHHLLIVFK-RLNLVKNREIVAQLVENYFND 406
> ath:AT1G72390 hypothetical protein
Length=1325
Score = 32.3 bits (72), Expect = 1.7, Method: Composition-based stats.
Identities = 15/36 (41%), Positives = 23/36 (63%), Gaps = 0/36 (0%)
Query 2 EYRYPSSAEELKDWVTVSWVLNPGKKEEKEGKKEEK 37
+ R P +A ++ SW +NPG++ EKE KKEE+
Sbjct 492 QSRMPHNAFIRSNFPQTSWNVNPGQQIEKEPKKEEQ 527
> cpv:cgd7_1300 hypothetical protein
Length=1483
Score = 31.2 bits (69), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 26/93 (27%), Positives = 46/93 (49%), Gaps = 16/93 (17%)
Query 139 SLSAALNKIE-FLLRETPRDRLPRGLIFARKMTQEIVYNKDPLNALEFEETLWKIRERLA 197
+LS LN ++ F L+ET + LIF + + + + +P EF + +K+++ +
Sbjct 507 TLSQTLNTLQDFFLKET------KNLIFGNNIVSDWITSSNPDRISEFSDYSFKLKDSIL 560
Query 198 KGEKVFEQLIQKYFSDNPHRLTLRLIGNKEIIK 230
K K+ SDN R + LI N E++K
Sbjct 561 KKLKI--------HSDNWDRY-VDLITNDEVLK 584
> xla:496364 b3gnt5-a, b3gn-t5, b3gnt5, b3gnt5b, beta3gn-t5; UDP-GlcNAc:betaGal
beta-1,3-N-acetylglucosaminyltransferase 5
(EC:2.4.1.-); K03766 beta-1,3-N-acetylglucosaminyltransferase
5 [EC:2.4.1.206]
Length=377
Score = 30.4 bits (67), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 15/32 (46%), Positives = 21/32 (65%), Gaps = 0/32 (0%)
Query 192 IRERLAKGEKVFEQLIQKYFSDNPHRLTLRLI 223
I++ L K F+ LIQ+ FSD H LTL+L+
Sbjct 142 IQQDLVNENKRFKDLIQQDFSDTFHNLTLKLL 173
> sce:YPL222W FMP40; Fmp40p
Length=688
Score = 30.0 bits (66), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 14/39 (35%), Positives = 23/39 (58%), Gaps = 0/39 (0%)
Query 92 GLDLDLKYAMFTVGLKDVPQREGAAKEVEELVINTLKEI 130
G+DLDL+ M + LKD+ AKE ++++ L +I
Sbjct 455 GVDLDLEKCMSSTNLKDIEHAAEKAKEFCDVIVEPLLDI 493
> tpv:TP01_0752 hypothetical protein
Length=771
Score = 29.6 bits (65), Expect = 9.3, Method: Compositional matrix adjust.
Identities = 51/208 (24%), Positives = 86/208 (41%), Gaps = 33/208 (15%)
Query 59 LQVLNFLLIGDKASPLYSSLSNYTQLGGPAYTPGLDLDLKYAMFTVGLKDVPQREGAAKE 118
+ V+ L+ ASPL +L + G ++L K F + +KDVP + +
Sbjct 330 ISVMGSYLVDSTASPLEKALIHTDNPYGSCVDFSMEL-FKETYFQLTVKDVPLSKNSKNP 388
Query 119 VE---ELVINTLKEIKEKGFS------------SLSLSAALNKIEFLLRETPRDRLPRGL 163
E E++ N +K + E + L +IE + ET + + +
Sbjct 389 TEDKIEVLGNKVKSLLENIYKEEFDMERMRMLIKRCYHNYLRQIETIAHETLIENVIGYI 448
Query 164 IFARKMTQ--EIVYNKDPLNALEFEETLWKIRERLAKGEKVFEQLIQKYFSDNPH---RL 218
I+ K Q +I+ + D + L L K EK +++L+ KYF + P +
Sbjct 449 IYGSKREQLEKIIVDHDLVKNL------------LEKDEKYWKELLLKYFIEPPSVSVKC 496
Query 219 TLRLIGNKEIIKEREEEEKKSLKKAEKE 246
L +K I KE +E + LKK KE
Sbjct 497 MPSLERSKVIEKEEKELMRTQLKKYGKE 524
Lambda K H
0.311 0.132 0.365
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8764825436
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40