bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0167_orf1
Length=150
Score E
Sequences producing significant alignments: (Bits) Value
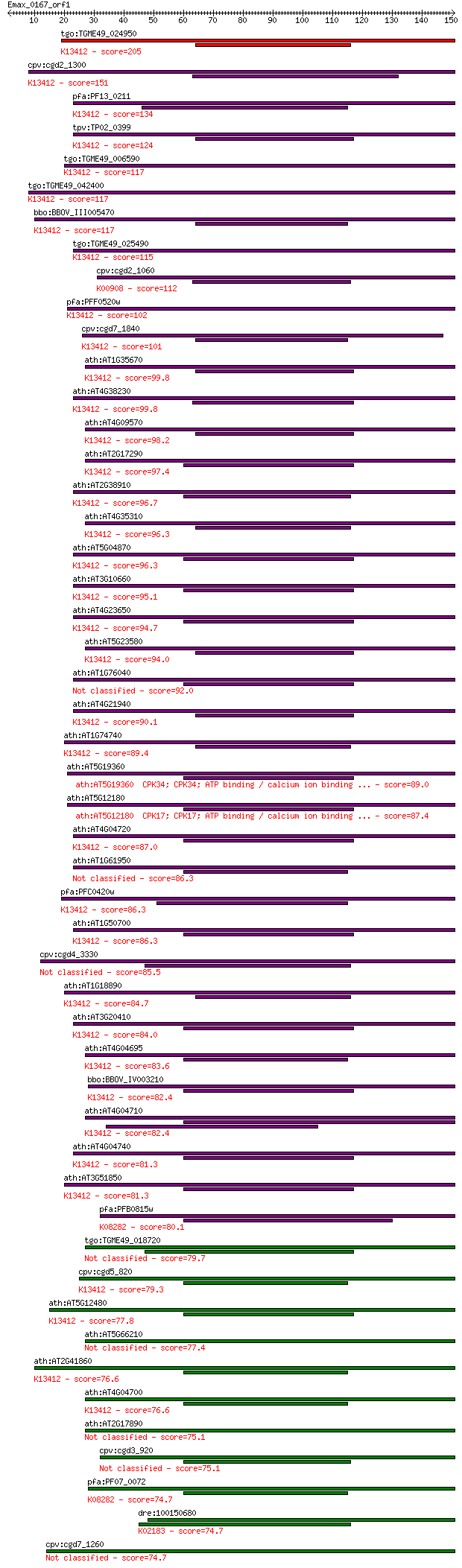
tgo:TGME49_024950 protein kinase 6, putative (EC:1.6.3.1 2.7.1... 205 4e-53
cpv:cgd2_1300 calcium/calmodulin dependent protein kinase with... 151 9e-37
pfa:PF13_0211 calcium-dependent protein kinase, putative (EC:2... 134 1e-31
tpv:TP02_0399 calcium-dependent protein kinase; K13412 calcium... 124 9e-29
tgo:TGME49_006590 calcium-dependent protein kinase, putative (... 117 1e-26
tgo:TGME49_042400 calcium-dependent protein kinase, putative (... 117 1e-26
bbo:BBOV_III005470 17.m07489; protein kinase domain containing... 117 2e-26
tgo:TGME49_025490 calcium-dependent protein kinase, putative (... 115 5e-26
cpv:cgd2_1060 calcium/calmodulin dependent protein kinase with... 112 5e-25
pfa:PFF0520w PfCDPK2; calcium-dependent protein kinase (EC:2.7... 102 3e-22
cpv:cgd7_1840 calcium/calmodulin-dependent protein kinase with... 101 1e-21
ath:AT1G35670 ATCDPK2 (CALCIUM-DEPENDENT PROTEIN KINASE 2); ca... 99.8 3e-21
ath:AT4G38230 CPK26; CPK26; ATP binding / calcium ion binding ... 99.8 3e-21
ath:AT4G09570 CPK4; CPK4; calmodulin-dependent protein kinase/... 98.2 8e-21
ath:AT2G17290 CPK6; CPK6 (CALCIUM-DEPENDENT PROTEIN KINASE 6);... 97.4 1e-20
ath:AT2G38910 CPK20; CPK20; ATP binding / calcium ion binding ... 96.7 3e-20
ath:AT4G35310 CPK5; CPK5 (calmodulin-domain protein kinase 5);... 96.3 3e-20
ath:AT5G04870 CPK1; CPK1 (CALCIUM DEPENDENT PROTEIN KINASE 1);... 96.3 4e-20
ath:AT3G10660 CPK2; CPK2 (CALMODULIN-DOMAIN PROTEIN KINASE CDP... 95.1 8e-20
ath:AT4G23650 CDPK6; CDPK6 (CALCIUM-DEPENDENT PROTEIN KINASE 6... 94.7 1e-19
ath:AT5G23580 CDPK9; CDPK9 (CALMODULIN-LIKE DOMAIN PROTEIN KIN... 94.0 2e-19
ath:AT1G76040 CPK29; CPK29; ATP binding / calcium ion binding ... 92.0 6e-19
ath:AT4G21940 CPK15; CPK15; ATP binding / calcium ion binding ... 90.1 2e-18
ath:AT1G74740 CPK30; CPK30 (CALCIUM-DEPENDENT PROTEIN KINASE 3... 89.4 4e-18
ath:AT5G19360 CPK34; CPK34; ATP binding / calcium ion binding ... 89.0 6e-18
ath:AT5G12180 CPK17; CPK17; ATP binding / calcium ion binding ... 87.4 2e-17
ath:AT4G04720 CPK21; CPK21; ATP binding / calcium ion binding ... 87.0 2e-17
ath:AT1G61950 CPK19; CPK19; ATP binding / calcium ion binding ... 86.3 3e-17
pfa:PFC0420w PfCDPK3; calcium-dependent protein kinase-3 (EC:2... 86.3 3e-17
ath:AT1G50700 CPK33; CPK33; ATP binding / calcium ion binding ... 86.3 3e-17
cpv:cgd4_3330 hypothetical protein 85.5 6e-17
ath:AT1G18890 ATCDPK1 (CALCIUM-DEPENDENT PROTEIN KINASE 1); ca... 84.7 1e-16
ath:AT3G20410 CPK9; CPK9 (calmodulin-domain protein kinase 9);... 84.0 2e-16
ath:AT4G04695 CPK31; CPK31; ATP binding / calcium ion binding ... 83.6 2e-16
bbo:BBOV_IV003210 21.m02835; calcium-dependent protein kinase ... 82.4 5e-16
ath:AT4G04710 CPK22; CPK22; ATP binding / calcium ion binding ... 82.4 5e-16
ath:AT4G04740 CPK23; CPK23; ATP binding / calcium ion binding ... 81.3 1e-15
ath:AT3G51850 CPK13; CPK13; ATP binding / calcium ion binding ... 81.3 1e-15
pfa:PFB0815w CDPK1, CPK; Calcium-dependent protein kinase 1 (E... 80.1 3e-15
tgo:TGME49_018720 calcium-dependent protein kinase, putative (... 79.7 3e-15
cpv:cgd5_820 calcium/calmodulin dependent protein kinase with ... 79.3 4e-15
ath:AT5G12480 CPK7; CPK7 (calmodulin-domain protein kinase 7);... 77.8 1e-14
ath:AT5G66210 CPK28; CPK28; ATP binding / calcium ion binding ... 77.4 2e-14
ath:AT2G41860 CPK14; CPK14; ATP binding / calcium ion binding ... 76.6 2e-14
ath:AT4G04700 CPK27; CPK27; ATP binding / calcium ion binding ... 76.6 3e-14
ath:AT2G17890 CPK16; CPK16; ATP binding / calcium ion binding ... 75.1 7e-14
cpv:cgd3_920 calmodulin-domain protein kinase 1 75.1
pfa:PF07_0072 PfCDPK4; calcium-dependent protein kinase 4 (EC:... 74.7 1e-13
dre:100150680 calmodulin 2-like; K02183 calmodulin 74.7 1e-13
cpv:cgd7_1260 calcium/calmodulin dependent protein kinase with... 74.7 1e-13
> tgo:TGME49_024950 protein kinase 6, putative (EC:1.6.3.1 2.7.11.17);
K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=681
Score = 205 bits (521), Expect = 4e-53, Method: Composition-based stats.
Identities = 96/132 (72%), Positives = 113/132 (85%), Gaps = 0/132 (0%)
Query 19 LSVRLGSDLIERFKSFQRLNKLKRLAITCVAYQLTDKEIGILHDTFSLLDKNNDGVLTLN 78
+ RLG DLIERFK+FQRL+KLK+LAITCVAYQL D +IG+LHD F+ LD N DGVLT+
Sbjct 481 IPARLGGDLIERFKAFQRLHKLKKLAITCVAYQLNDADIGMLHDAFAALDTNADGVLTVA 540
Query 79 ELLNGLQLCGVQGEEIINIIKEIDTDKNGTIDYTEFIAASLDHKIYEQESAIQNAFRVFD 138
E+ GL+ C V GEEI +I+KE+DTD NGTIDYTEFIAAS+DHKIYEQESA Q AF+VFD
Sbjct 541 EIQQGLKQCCVAGEEINDILKEMDTDGNGTIDYTEFIAASIDHKIYEQESACQAAFKVFD 600
Query 139 LDGDGKISVEEL 150
LDGDGKI+V+EL
Sbjct 601 LDGDGKITVDEL 612
Score = 36.2 bits (82), Expect = 0.042, Method: Composition-based stats.
Identities = 19/56 (33%), Positives = 33/56 (58%), Gaps = 4/56 (7%)
Query 64 FSLLDKNNDGVLTLNELLNGLQLCGVQ----GEEIINIIKEIDTDKNGTIDYTEFI 115
F + D + DG +T++EL L+ VQ E + ++KE D++ +G ID+ EF+
Sbjct 596 FKVFDLDGDGKITVDELQKVLETRCVQEAFSKEAVAEMMKEGDSNNDGCIDFDEFM 651
> cpv:cgd2_1300 calcium/calmodulin dependent protein kinase with
a kinase domain and 4 calmodulin like EF hands ; K13412 calcium-dependent
protein kinase [EC:2.7.11.1]
Length=677
Score = 151 bits (381), Expect = 9e-37, Method: Compositional matrix adjust.
Identities = 77/146 (52%), Positives = 106/146 (72%), Gaps = 3/146 (2%)
Query 8 INIYNNSSSSCLSVRLGSDLIERFKSFQRLNKLKRLAITCVAYQLTDKEIGILHDTFSLL 67
I+ N + ++ + + + L+ RF+ FQR +KLK+LA+TCVAY L D +IG L FS L
Sbjct 443 ISSPNTADATYFTNDVCNSLLARFRDFQRQSKLKKLALTCVAYHLNDADIGALQKLFSTL 502
Query 68 DKNNDGVLTLNEL---LNGLQLCGVQGEEIINIIKEIDTDKNGTIDYTEFIAASLDHKIY 124
D+N DGVLT+NE+ L+ +Q G++I N++ E+DTD NG IDYTEFIAAS+DHK+Y
Sbjct 503 DRNGDGVLTINEIRSALHKIQNVSQLGDDIDNLLMELDTDGNGRIDYTEFIAASIDHKLY 562
Query 125 EQESAIQNAFRVFDLDGDGKISVEEL 150
EQES + AF+VFDLD DG+IS +EL
Sbjct 563 EQESLCKAAFKVFDLDMDGRISPQEL 588
Score = 30.8 bits (68), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 22/75 (29%), Positives = 41/75 (54%), Gaps = 7/75 (9%)
Query 63 TFSLLDKNNDGVLTLNELLNGLQLCGVQ----GEEIINIIKEIDTDKNGTIDYTEFIAAS 118
F + D + DG ++ EL L + +Q I +++KE+D +++G ID+ EF+
Sbjct 571 AFKVFDLDMDGRISPQELSRVLNITFLQEAFEQSTIDSLLKEVDINQDGYIDFNEFMKMM 630
Query 119 LDHKIYEQ--ESAIQ 131
+ K +EQ E+ +Q
Sbjct 631 MGDK-HEQKLETKVQ 644
> pfa:PF13_0211 calcium-dependent protein kinase, putative (EC:2.7.11.17);
K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=568
Score = 134 bits (337), Expect = 1e-31, Method: Compositional matrix adjust.
Identities = 67/129 (51%), Positives = 93/129 (72%), Gaps = 1/129 (0%)
Query 23 LGSDLIERFKSFQRLNKLKRLAITCVAYQLTDKEIGILHDTFSLLDKNNDGVLTLNELLN 82
L LIE+FK F +L K+K+LA+TC+AYQL +K+IG L TF D N DGVLT++E+
Sbjct 393 LNKTLIEKFKEFHKLCKIKKLAVTCIAYQLNEKDIGKLKKTFEAFDHNGDGVLTISEIFQ 452
Query 83 GLQLCGVQ-GEEIINIIKEIDTDKNGTIDYTEFIAASLDHKIYEQESAIQNAFRVFDLDG 141
L++ + E+ ++K++DTD NG IDYTEF+AA LDH I++Q+ +NAF VFDLDG
Sbjct 453 CLKVNDNEFDRELYFLLKQLDTDGNGLIDYTEFLAACLDHSIFQQDVICRNAFNVFDLDG 512
Query 142 DGKISVEEL 150
DG I+ +EL
Sbjct 513 DGVITKDEL 521
Score = 45.1 bits (105), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 27/73 (36%), Positives = 45/73 (61%), Gaps = 5/73 (6%)
Query 46 TCVAYQLTDKEIGILHDTFSLLDKNNDGVLTLNELLNGLQLCGVQ---GEEII-NIIKEI 101
C+ + + +++ I + F++ D + DGV+T +EL L VQ +EII N+IKE+
Sbjct 488 ACLDHSIFQQDV-ICRNAFNVFDLDGDGVITKDELFKILSFSAVQVSFSKEIIENLIKEV 546
Query 102 DTDKNGTIDYTEF 114
D++ +G IDY EF
Sbjct 547 DSNNDGFIDYDEF 559
> tpv:TP02_0399 calcium-dependent protein kinase; K13412 calcium-dependent
protein kinase [EC:2.7.11.1]
Length=844
Score = 124 bits (312), Expect = 9e-29, Method: Composition-based stats.
Identities = 56/128 (43%), Positives = 89/128 (69%), Gaps = 0/128 (0%)
Query 23 LGSDLIERFKSFQRLNKLKRLAITCVAYQLTDKEIGILHDTFSLLDKNNDGVLTLNELLN 82
+ L RFK F + NK+K++A+TC+AY L+++E+ L + F LDK+ DGVL+L+E+ N
Sbjct 668 ISKSLTRRFKKFLKYNKMKQMALTCLAYHLSERELAPLTNAFESLDKDGDGVLSLDEVAN 727
Query 83 GLQLCGVQGEEIINIIKEIDTDKNGTIDYTEFIAASLDHKIYEQESAIQNAFRVFDLDGD 142
GL+ I I+K IDTD++G I+YTEF+AA++D ++Y Q+ + AF +FD D D
Sbjct 728 GLKHSKQSSFHIEQIVKGIDTDQSGIIEYTEFVAAAIDARLYNQKDFFKRAFNIFDTDRD 787
Query 143 GKISVEEL 150
G+I+ E++
Sbjct 788 GRITREDM 795
Score = 32.3 bits (72), Expect = 0.55, Method: Composition-based stats.
Identities = 16/57 (28%), Positives = 31/57 (54%), Gaps = 4/57 (7%)
Query 64 FSLLDKNNDGVLTLNELLNGLQLCG----VQGEEIINIIKEIDTDKNGTIDYTEFIA 116
F++ D + DG +T ++ + E + +I++E+D D++GTI Y EF +
Sbjct 779 FNIFDTDRDGRITREDMYRVFSTESTNPRMTQEMVEDILEEVDLDRDGTISYDEFTS 835
> tgo:TGME49_006590 calcium-dependent protein kinase, putative
(EC:2.7.11.17 3.4.22.53); K13412 calcium-dependent protein
kinase [EC:2.7.11.1]
Length=761
Score = 117 bits (294), Expect = 1e-26, Method: Composition-based stats.
Identities = 61/133 (45%), Positives = 89/133 (66%), Gaps = 2/133 (1%)
Query 20 SVRLGSDLIERFKSFQRLNKLKRLAITCVAYQLTDKEIGILHDTFSLLDKNNDGVLTLNE 79
+V L + L+ K+F+ NKLK+ A+T +A +++KEI L F LD +N G L++ E
Sbjct 488 NVALPTTLMSNLKAFRAQNKLKKAALTVIAQHMSEKEIDHLRQIFMTLDVDNSGTLSVQE 547
Query 80 LLNGLQLCGVQG--EEIINIIKEIDTDKNGTIDYTEFIAASLDHKIYEQESAIQNAFRVF 137
+ GL+ G ++ II+E+D+DK+G IDYTEFIAA++D K+Y +E AFRVF
Sbjct 548 VREGLKRLGWTEIPADLQAIIEEVDSDKSGHIDYTEFIAATMDKKLYMKEDVCWAAFRVF 607
Query 138 DLDGDGKISVEEL 150
DLDG+GKIS +EL
Sbjct 608 DLDGNGKISQDEL 620
> tgo:TGME49_042400 calcium-dependent protein kinase, putative
(EC:2.7.11.17); K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=604
Score = 117 bits (293), Expect = 1e-26, Method: Composition-based stats.
Identities = 59/145 (40%), Positives = 93/145 (64%), Gaps = 2/145 (1%)
Query 8 INIYNNSSSSCLSVRLGSDLIERFKSFQRLNKLKRLAITCVAYQLTDKEIGILHDTFSLL 67
I Y ++ L S +++ F++F+ ++KLK+ A+T +A Q+ + +I L + F L
Sbjct 383 IKHYATKANPVADAPLNSKILDNFRAFRAVSKLKKAALTVIAQQMNEGQIKALKNIFLAL 442
Query 68 DKNNDGVLTLNELLNGLQLCGVQG--EEIINIIKEIDTDKNGTIDYTEFIAASLDHKIYE 125
D++ DG LT+NE+ GL G++ ++ ++ E+D+D +G IDYTEFIAASLD + Y
Sbjct 443 DEDGDGTLTINEIRVGLSKSGLKEMPSDLDALMNEVDSDGSGVIDYTEFIAASLDKRQYI 502
Query 126 QESAIQNAFRVFDLDGDGKISVEEL 150
QE AFRVFDLD +G+IS +EL
Sbjct 503 QEDVCWAAFRVFDLDNNGRISADEL 527
> bbo:BBOV_III005470 17.m07489; protein kinase domain containing
protein; K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=755
Score = 117 bits (292), Expect = 2e-26, Method: Composition-based stats.
Identities = 59/148 (39%), Positives = 101/148 (68%), Gaps = 7/148 (4%)
Query 10 IYNNSSSSCLS--VR--LGSDLIERFKSFQRLNKLKRLAITCVAYQLTDKEIGILHDTFS 65
I NN++S ++ VR L DL++RF+ F + +K+LA+TC+A+ L+D +IG L F+
Sbjct 554 ILNNATSVHVNPMVRHSLTKDLVKRFQKFDNYSTMKQLALTCIAHHLSDGDIGSLSTAFN 613
Query 66 LLDKNNDGVLTLNELLNGLQLCGVQGE---EIINIIKEIDTDKNGTIDYTEFIAASLDHK 122
+L+ DGVL +++++ GLQ V+G+ + +++++DT+ +G IDY EF+AAS+D
Sbjct 614 VLNNAGDGVLYISDIVKGLQSDKVKGQYDPAMQRLVEKLDTNGSGAIDYVEFLAASIDED 673
Query 123 IYEQESAIQNAFRVFDLDGDGKISVEEL 150
+Y+Q+ + AF+VFDLDG G I+ E +
Sbjct 674 VYKQKDFCKKAFKVFDLDGKGVITRENM 701
Score = 32.3 bits (72), Expect = 0.54, Method: Composition-based stats.
Identities = 16/55 (29%), Positives = 27/55 (49%), Gaps = 4/55 (7%)
Query 64 FSLLDKNNDGVLTLNELLNGLQL----CGVQGEEIINIIKEIDTDKNGTIDYTEF 114
F + D + GV+T + Q C + + I E+D D++G I+YT+F
Sbjct 685 FKVFDLDGKGVITRENMCKVFQCNMGGCQFTQDFVEQIFNEVDLDRDGVINYTDF 739
> tgo:TGME49_025490 calcium-dependent protein kinase, putative
(EC:1.6.3.1 2.7.11.17 3.2.1.60); K13412 calcium-dependent protein
kinase [EC:2.7.11.1]
Length=711
Score = 115 bits (288), Expect = 5e-26, Method: Composition-based stats.
Identities = 58/131 (44%), Positives = 86/131 (65%), Gaps = 3/131 (2%)
Query 23 LGSDLIERFKSFQRLNKLKRLAITCVAYQLTDKEIGILHDTFSLLDKNNDGVLTLNELLN 82
LG D++ +F+ FQ L++LK+LA+T +A L D EI L + F+ LD DGVLT+ E+
Sbjct 506 LGLDILSKFRRFQGLSRLKKLALTVIAQHLEDSEIEGLKNLFTQLDTEGDGVLTVEEIRK 565
Query 83 GLQLCGVQ---GEEIINIIKEIDTDKNGTIDYTEFIAASLDHKIYEQESAIQNAFRVFDL 139
G++ GV + ++++E+DT G+IDYTEFIAA L Y +E A + AFRV D+
Sbjct 566 GIERSGVHLPPDMVLEDVLREVDTAGTGSIDYTEFIAACLHQSHYIREEACRAAFRVLDI 625
Query 140 DGDGKISVEEL 150
+GDG +S +EL
Sbjct 626 NGDGLVSAQEL 636
> cpv:cgd2_1060 calcium/calmodulin dependent protein kinase with
a kinas domain and 4 calmodulin-like EF hands ; K00908 Ca2+/calmodulin-dependent
protein kinase [EC:2.7.11.17]
Length=718
Score = 112 bits (279), Expect = 5e-25, Method: Compositional matrix adjust.
Identities = 54/122 (44%), Positives = 83/122 (68%), Gaps = 2/122 (1%)
Query 31 FKSFQRLNKLKRLAITCVAYQLTDKEIGILHDTFSLLDKNNDGVLTLNELLNGLQLCGVQ 90
F++F + N+ ++A+T +A Q+T+ +I L + F LLD N DG LT E++ GL+ G+
Sbjct 545 FRAFHKYNRFMKVALTVIAQQMTESQISNLKEAFILLDANCDGTLTPQEIITGLKNSGIT 604
Query 91 --GEEIINIIKEIDTDKNGTIDYTEFIAASLDHKIYEQESAIQNAFRVFDLDGDGKISVE 148
+++ I+ +ID+D +G+IDYTEFIAA+LD K Y +E AF+VFD DG+GKI+
Sbjct 605 ELPSDLLAILNDIDSDGSGSIDYTEFIAATLDSKQYSKEQVCWAAFKVFDQDGNGKITAN 664
Query 149 EL 150
EL
Sbjct 665 EL 666
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 22/62 (35%), Positives = 34/62 (54%), Gaps = 9/62 (14%)
Query 63 TFSLLDKNNDGVLTLNELLNGLQLCGVQGE---------EIINIIKEIDTDKNGTIDYTE 113
F + D++ +G +T NELLN QG ++ N+IKE+D D +G ID+ E
Sbjct 649 AFKVFDQDGNGKITANELLNVFSYNSEQGSAGINDKALSDVKNMIKEVDVDGDGEIDFQE 708
Query 114 FI 115
F+
Sbjct 709 FL 710
> pfa:PFF0520w PfCDPK2; calcium-dependent protein kinase (EC:2.7.11.1);
K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=509
Score = 102 bits (255), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 54/132 (40%), Positives = 88/132 (66%), Gaps = 2/132 (1%)
Query 21 VRLGSDLIERFKSFQRLNKLKRLAITCVAYQLTDKEIGILHDTFSLLDKNNDGVLTLNEL 80
V L S L++ K+F++ N+LK++A+T +A L D EI L + F LD +N G L+ E+
Sbjct 336 VELSSTLLKNLKNFKKENELKKIALTIIAKHLCDVEINNLRNIFIALDVDNSGTLSSQEI 395
Query 81 LNGLQLCGVQ--GEEIINIIKEIDTDKNGTIDYTEFIAASLDHKIYEQESAIQNAFRVFD 138
L+GL+ G Q +I ++++ID++ +G I YT+F+AA++D + Y ++ F+ FD
Sbjct 396 LDGLKKIGYQKIPPDIHQVLRDIDSNASGQIHYTDFLAATIDKQTYLKKEVCLIPFKFFD 455
Query 139 LDGDGKISVEEL 150
+DG+GKISVEEL
Sbjct 456 IDGNGKISVEEL 467
> cpv:cgd7_1840 calcium/calmodulin-dependent protein kinase with
a kinase domain and 4 calmodulin like EF hands ; K13412 calcium-dependent
protein kinase [EC:2.7.11.1]
Length=676
Score = 101 bits (251), Expect = 1e-21, Method: Composition-based stats.
Identities = 51/122 (41%), Positives = 79/122 (64%), Gaps = 1/122 (0%)
Query 26 DLIERFKSFQRLNKLKRLAITCVAYQLTDKEIGILHDTFSLLDKNNDGVLTLNELLNGLQ 85
++I F+ FQ L++LK++A+T +A + +++I LHDTF LD + DG L+ E++ G+
Sbjct 475 NVISNFRRFQGLSRLKKIALTLIAQNIDERDILDLHDTFMELDTSRDGTLSRAEIIEGIN 534
Query 86 LCGVQGEEIIN-IIKEIDTDKNGTIDYTEFIAASLDHKIYEQESAIQNAFRVFDLDGDGK 144
G I+ ++ EID + TI YT+FIAA + + ESA + AFRVFD+DGDG+
Sbjct 535 RTGCSPTIGIDALLDEIDPEGTDTISYTDFIAACIQERQMSHESACKAAFRVFDIDGDGQ 594
Query 145 IS 146
IS
Sbjct 595 IS 596
Score = 28.9 bits (63), Expect = 5.9, Method: Composition-based stats.
Identities = 17/58 (29%), Positives = 28/58 (48%), Gaps = 7/58 (12%)
Query 64 FSLLDKNNDGVLTLNELLNGLQLCGVQ-------GEEIINIIKEIDTDKNGTIDYTEF 114
F + D + DG ++ E L + L +E+ +K D DK+GTI++ EF
Sbjct 584 FRVFDIDGDGQISNIEFLKVMSLSSKAKKSDDELAQELSEFMKSGDLDKDGTINFDEF 641
> ath:AT1G35670 ATCDPK2 (CALCIUM-DEPENDENT PROTEIN KINASE 2);
calmodulin-dependent protein kinase/ kinase; K13412 calcium-dependent
protein kinase [EC:2.7.11.1]
Length=495
Score = 99.8 bits (247), Expect = 3e-21, Method: Composition-based stats.
Identities = 49/126 (38%), Positives = 80/126 (63%), Gaps = 2/126 (1%)
Query 27 LIERFKSFQRLNKLKRLAITCVAYQLTDKEIGILHDTFSLLDKNNDGVLTLNELLNGLQL 86
++ R K F ++NK+K++A+ +A +L+++EIG L + F ++D +N G +T EL GL+
Sbjct 299 VLSRLKQFSQMNKIKKMALRVIAERLSEEEIGGLKELFKMIDTDNSGTITFEELKAGLKR 358
Query 87 CG--VQGEEIINIIKEIDTDKNGTIDYTEFIAASLDHKIYEQESAIQNAFRVFDLDGDGK 144
G + EI +++ D D +GTIDY EF+AA+L E+E + AF FD DG G
Sbjct 359 VGSELMESEIKSLMDAADIDNSGTIDYGEFLAATLHMNKMEREENLVAAFSYFDKDGSGY 418
Query 145 ISVEEL 150
I+++EL
Sbjct 419 ITIDEL 424
Score = 44.7 bits (104), Expect = 1e-04, Method: Composition-based stats.
Identities = 21/53 (39%), Positives = 33/53 (62%), Gaps = 0/53 (0%)
Query 64 FSLLDKNNDGVLTLNELLNGLQLCGVQGEEIINIIKEIDTDKNGTIDYTEFIA 116
FS DK+ G +T++EL + G+ + ++IKEID D +G ID++EF A
Sbjct 408 FSYFDKDGSGYITIDELQSACTEFGLCDTPLDDMIKEIDLDNDGKIDFSEFTA 460
> ath:AT4G38230 CPK26; CPK26; ATP binding / calcium ion binding
/ calmodulin-dependent protein kinase/ kinase/ protein kinase;
K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=340
Score = 99.8 bits (247), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 52/130 (40%), Positives = 83/130 (63%), Gaps = 2/130 (1%)
Query 23 LGSDLIERFKSFQRLNKLKRLAITCVAYQLTDKEIGILHDTFSLLDKNNDGVLTLNELLN 82
L ++ R K F +NKLK++A+ +A L+++EI L + F +D +N G +T +EL
Sbjct 149 LDPAVLSRLKQFSAMNKLKQMALRVIAESLSEEEIAGLKEMFKAMDTDNSGAITFDELKA 208
Query 83 GLQLCG--VQGEEIINIIKEIDTDKNGTIDYTEFIAASLDHKIYEQESAIQNAFRVFDLD 140
GL+ G ++ EI ++++ D DK+GTIDY EFIAA++ E+E + +AFR FD D
Sbjct 209 GLRRYGSTLKDTEIRDLMEAADIDKSGTIDYGEFIAATIHLNKLEREEHLLSAFRYFDKD 268
Query 141 GDGKISVEEL 150
G G I+++EL
Sbjct 269 GSGYITIDEL 278
Score = 42.7 bits (99), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 20/54 (37%), Positives = 32/54 (59%), Gaps = 0/54 (0%)
Query 63 TFSLLDKNNDGVLTLNELLNGLQLCGVQGEEIINIIKEIDTDKNGTIDYTEFIA 116
F DK+ G +T++EL + G+ + ++IKE+D D +G IDY EF+A
Sbjct 261 AFRYFDKDGSGYITIDELQHACAEQGMSDVFLEDVIKEVDQDNDGRIDYGEFVA 314
> ath:AT4G09570 CPK4; CPK4; calmodulin-dependent protein kinase/
kinase/ protein kinase; K13412 calcium-dependent protein
kinase [EC:2.7.11.1]
Length=501
Score = 98.2 bits (243), Expect = 8e-21, Method: Composition-based stats.
Identities = 49/126 (38%), Positives = 80/126 (63%), Gaps = 2/126 (1%)
Query 27 LIERFKSFQRLNKLKRLAITCVAYQLTDKEIGILHDTFSLLDKNNDGVLTLNELLNGLQL 86
++ R K F ++NK+K++A+ +A +L+++EIG L + F ++D +N G +T EL GL+
Sbjct 298 VLSRLKQFSQMNKIKKMALRVIAERLSEEEIGGLKELFKMIDTDNSGTITFEELKAGLKR 357
Query 87 CG--VQGEEIINIIKEIDTDKNGTIDYTEFIAASLDHKIYEQESAIQNAFRVFDLDGDGK 144
G + EI +++ D D +GTIDY EF+AA+L E+E + AF FD DG G
Sbjct 358 VGSELMESEIKSLMDAADIDNSGTIDYGEFLAATLHINKMEREENLVVAFSYFDKDGSGY 417
Query 145 ISVEEL 150
I+++EL
Sbjct 418 ITIDEL 423
Score = 44.7 bits (104), Expect = 1e-04, Method: Composition-based stats.
Identities = 21/53 (39%), Positives = 32/53 (60%), Gaps = 0/53 (0%)
Query 64 FSLLDKNNDGVLTLNELLNGLQLCGVQGEEIINIIKEIDTDKNGTIDYTEFIA 116
FS DK+ G +T++EL G+ + ++IKEID D +G ID++EF A
Sbjct 407 FSYFDKDGSGYITIDELQQACTEFGLCDTPLDDMIKEIDLDNDGKIDFSEFTA 459
> ath:AT2G17290 CPK6; CPK6 (CALCIUM-DEPENDENT PROTEIN KINASE 6);
ATP binding / calcium ion binding / calmodulin-dependent
protein kinase/ kinase/ protein kinase/ protein serine/threonine
kinase; K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=544
Score = 97.4 bits (241), Expect = 1e-20, Method: Composition-based stats.
Identities = 49/126 (38%), Positives = 81/126 (64%), Gaps = 2/126 (1%)
Query 27 LIERFKSFQRLNKLKRLAITCVAYQLTDKEIGILHDTFSLLDKNNDGVLTLNELLNGLQL 86
++ R K F +NKLK++A+ +A L+++EI L F +D +N G +T +EL GL+
Sbjct 358 VLSRLKQFSAMNKLKKMALKVIAESLSEEEIAGLRAMFEAMDTDNSGAITFDELKAGLRR 417
Query 87 CG--VQGEEIINIIKEIDTDKNGTIDYTEFIAASLDHKIYEQESAIQNAFRVFDLDGDGK 144
G ++ EI ++++ D D +GTIDY+EFIAA++ E+E + +AF+ FD DG G
Sbjct 418 YGSTLKDTEIRDLMEAADVDNSGTIDYSEFIAATIHLNKLEREEHLVSAFQYFDKDGSGY 477
Query 145 ISVEEL 150
I+++EL
Sbjct 478 ITIDEL 483
Score = 43.1 bits (100), Expect = 3e-04, Method: Composition-based stats.
Identities = 22/57 (38%), Positives = 32/57 (56%), Gaps = 0/57 (0%)
Query 60 LHDTFSLLDKNNDGVLTLNELLNGLQLCGVQGEEIINIIKEIDTDKNGTIDYTEFIA 116
L F DK+ G +T++EL G+ + +IIKE+D D +G IDY EF+A
Sbjct 463 LVSAFQYFDKDGSGYITIDELQQSCIEHGMTDVFLEDIIKEVDQDNDGRIDYEEFVA 519
> ath:AT2G38910 CPK20; CPK20; ATP binding / calcium ion binding
/ calmodulin-dependent protein kinase/ kinase/ protein kinase/
protein serine/threonine kinase; K13412 calcium-dependent
protein kinase [EC:2.7.11.1]
Length=583
Score = 96.7 bits (239), Expect = 3e-20, Method: Composition-based stats.
Identities = 52/130 (40%), Positives = 79/130 (60%), Gaps = 2/130 (1%)
Query 23 LGSDLIERFKSFQRLNKLKRLAITCVAYQLTDKEIGILHDTFSLLDKNNDGVLTLNELLN 82
L S ++ R + F +NKLK++AI +A L+++EI L + F ++D +N G +TL EL
Sbjct 403 LDSAVLSRLQQFSAMNKLKKIAIKVIAESLSEEEIAGLKEMFKMIDTDNSGHITLEELKK 462
Query 83 GLQLCG--VQGEEIINIIKEIDTDKNGTIDYTEFIAASLDHKIYEQESAIQNAFRVFDLD 140
GL G ++ EI+ +++ D D +GTIDY EFIAA + E+E + AF FD D
Sbjct 463 GLDRVGADLKDSEILGLMQAADIDNSGTIDYGEFIAAMVHLNKIEKEDHLFTAFSYFDQD 522
Query 141 GDGKISVEEL 150
G G I+ +EL
Sbjct 523 GSGYITRDEL 532
Score = 43.1 bits (100), Expect = 3e-04, Method: Composition-based stats.
Identities = 19/56 (33%), Positives = 33/56 (58%), Gaps = 0/56 (0%)
Query 60 LHDTFSLLDKNNDGVLTLNELLNGLQLCGVQGEEIINIIKEIDTDKNGTIDYTEFI 115
L FS D++ G +T +EL + G+ + +I++E+D D +G IDY+EF+
Sbjct 512 LFTAFSYFDQDGSGYITRDELQQACKQFGLADVHLDDILREVDKDNDGRIDYSEFV 567
> ath:AT4G35310 CPK5; CPK5 (calmodulin-domain protein kinase 5);
ATP binding / calcium ion binding / calmodulin-dependent
protein kinase/ kinase/ protein kinase/ protein serine/threonine
kinase; K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=556
Score = 96.3 bits (238), Expect = 3e-20, Method: Composition-based stats.
Identities = 49/126 (38%), Positives = 80/126 (63%), Gaps = 2/126 (1%)
Query 27 LIERFKSFQRLNKLKRLAITCVAYQLTDKEIGILHDTFSLLDKNNDGVLTLNELLNGLQL 86
++ R K F +NKLK++A+ +A L+++EI L + F +D +N G +T +EL GL+
Sbjct 370 VLSRLKQFSAMNKLKKMALKVIAESLSEEEIAGLREMFQAMDTDNSGAITFDELKAGLRK 429
Query 87 CG--VQGEEIINIIKEIDTDKNGTIDYTEFIAASLDHKIYEQESAIQNAFRVFDLDGDGK 144
G ++ EI +++ D D +GTIDY+EFIAA++ E+E + AF+ FD DG G
Sbjct 430 YGSTLKDTEIHDLMDAADVDNSGTIDYSEFIAATIHLNKLEREEHLVAAFQYFDKDGSGF 489
Query 145 ISVEEL 150
I+++EL
Sbjct 490 ITIDEL 495
Score = 39.7 bits (91), Expect = 0.003, Method: Composition-based stats.
Identities = 19/52 (36%), Positives = 30/52 (57%), Gaps = 0/52 (0%)
Query 64 FSLLDKNNDGVLTLNELLNGLQLCGVQGEEIINIIKEIDTDKNGTIDYTEFI 115
F DK+ G +T++EL G+ + +IIKE+D + +G IDY EF+
Sbjct 479 FQYFDKDGSGFITIDELQQACVEHGMADVFLEDIIKEVDQNNDGKIDYGEFV 530
> ath:AT5G04870 CPK1; CPK1 (CALCIUM DEPENDENT PROTEIN KINASE 1);
calmodulin-dependent protein kinase/ kinase; K13412 calcium-dependent
protein kinase [EC:2.7.11.1]
Length=610
Score = 96.3 bits (238), Expect = 4e-20, Method: Composition-based stats.
Identities = 50/130 (38%), Positives = 80/130 (61%), Gaps = 2/130 (1%)
Query 23 LGSDLIERFKSFQRLNKLKRLAITCVAYQLTDKEIGILHDTFSLLDKNNDGVLTLNELLN 82
L S ++ R K F +NK K++A+ +A L+++EI L + F+++D + G +T EL
Sbjct 419 LDSAVLSRMKQFSAMNKFKKMALRVIAESLSEEEIAGLKEMFNMIDADKSGQITFEELKA 478
Query 83 GLQLCG--VQGEEIINIIKEIDTDKNGTIDYTEFIAASLDHKIYEQESAIQNAFRVFDLD 140
GL+ G ++ EI+++++ D D +GTIDY EFIAA+L E+E + AF FD D
Sbjct 479 GLKRVGANLKESEILDLMQAADVDNSGTIDYKEFIAATLHLNKIEREDHLFAAFTYFDKD 538
Query 141 GDGKISVEEL 150
G G I+ +EL
Sbjct 539 GSGYITPDEL 548
Score = 44.7 bits (104), Expect = 1e-04, Method: Composition-based stats.
Identities = 20/57 (35%), Positives = 33/57 (57%), Gaps = 0/57 (0%)
Query 60 LHDTFSLLDKNNDGVLTLNELLNGLQLCGVQGEEIINIIKEIDTDKNGTIDYTEFIA 116
L F+ DK+ G +T +EL + GV+ I +++++D D +G IDY EF+A
Sbjct 528 LFAAFTYFDKDGSGYITPDELQQACEEFGVEDVRIEELMRDVDQDNDGRIDYNEFVA 584
> ath:AT3G10660 CPK2; CPK2 (CALMODULIN-DOMAIN PROTEIN KINASE CDPK
ISOFORM 2); calmodulin-dependent protein kinase/ kinase/
protein serine/threonine kinase; K13412 calcium-dependent protein
kinase [EC:2.7.11.1]
Length=646
Score = 95.1 bits (235), Expect = 8e-20, Method: Composition-based stats.
Identities = 50/130 (38%), Positives = 78/130 (60%), Gaps = 2/130 (1%)
Query 23 LGSDLIERFKSFQRLNKLKRLAITCVAYQLTDKEIGILHDTFSLLDKNNDGVLTLNELLN 82
L S ++ R K F +NK K++A+ +A L+++EI L F ++D +N G +T EL
Sbjct 455 LDSAVLSRMKQFSAMNKFKKMALRVIAESLSEEEIAGLKQMFKMIDADNSGQITFEELKA 514
Query 83 GLQLCG--VQGEEIINIIKEIDTDKNGTIDYTEFIAASLDHKIYEQESAIQNAFRVFDLD 140
GL+ G ++ EI+++++ D D +GTIDY EFIAA+L E+E + AF FD D
Sbjct 515 GLKRVGANLKESEILDLMQAADVDNSGTIDYKEFIAATLHLNKIEREDHLFAAFSYFDKD 574
Query 141 GDGKISVEEL 150
G I+ +EL
Sbjct 575 ESGFITPDEL 584
Score = 48.9 bits (115), Expect = 7e-06, Method: Composition-based stats.
Identities = 22/57 (38%), Positives = 34/57 (59%), Gaps = 0/57 (0%)
Query 60 LHDTFSLLDKNNDGVLTLNELLNGLQLCGVQGEEIINIIKEIDTDKNGTIDYTEFIA 116
L FS DK+ G +T +EL + GV+ I +++++D DK+G IDY EF+A
Sbjct 564 LFAAFSYFDKDESGFITPDELQQACEEFGVEDARIEEMMRDVDQDKDGRIDYNEFVA 620
> ath:AT4G23650 CDPK6; CDPK6 (CALCIUM-DEPENDENT PROTEIN KINASE
6); ATP binding / calcium ion binding / calmodulin-dependent
protein kinase/ kinase/ protein kinase/ protein serine/threonine
kinase; K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=529
Score = 94.7 bits (234), Expect = 1e-19, Method: Composition-based stats.
Identities = 50/130 (38%), Positives = 81/130 (62%), Gaps = 2/130 (1%)
Query 23 LGSDLIERFKSFQRLNKLKRLAITCVAYQLTDKEIGILHDTFSLLDKNNDGVLTLNELLN 82
L + ++ R K F+ +NKLK++A+ +A L+++EI L + F LD +N+G++TL EL
Sbjct 347 LDNAVLSRMKQFRAMNKLKKMALKVIAENLSEEEIIGLKEMFKSLDTDNNGIVTLEELRT 406
Query 83 GLQLCG--VQGEEIINIIKEIDTDKNGTIDYTEFIAASLDHKIYEQESAIQNAFRVFDLD 140
GL G + EI +++ D D +G+IDY EFI+A++ E+E + AF+ FD D
Sbjct 407 GLPKLGSKISEAEIRQLMEAADMDGDGSIDYLEFISATMHMNRIEREDHLYTAFQFFDND 466
Query 141 GDGKISVEEL 150
G I++EEL
Sbjct 467 NSGYITMEEL 476
Score = 43.1 bits (100), Expect = 3e-04, Method: Composition-based stats.
Identities = 21/58 (36%), Positives = 35/58 (60%), Gaps = 1/58 (1%)
Query 60 LHDTFSLLDKNNDGVLTLNELLNGLQLCGVQGEEIIN-IIKEIDTDKNGTIDYTEFIA 116
L+ F D +N G +T+ EL ++ + ++ I II E+DTD++G I+Y EF+A
Sbjct 456 LYTAFQFFDNDNSGYITMEELELAMKKYNMGDDKSIKEIIAEVDTDRDGKINYEEFVA 513
> ath:AT5G23580 CDPK9; CDPK9 (CALMODULIN-LIKE DOMAIN PROTEIN KINASE
9); ATP binding / calcium ion binding / calmodulin-dependent
protein kinase/ kinase/ protein kinase/ protein serine/threonine
kinase; K13412 calcium-dependent protein kinase
[EC:2.7.11.1]
Length=490
Score = 94.0 bits (232), Expect = 2e-19, Method: Composition-based stats.
Identities = 46/126 (36%), Positives = 78/126 (61%), Gaps = 2/126 (1%)
Query 27 LIERFKSFQRLNKLKRLAITCVAYQLTDKEIGILHDTFSLLDKNNDGVLTLNELLNGLQL 86
++ R K F +NKLK++A+ +A +L+++EIG L + F ++D + G +T EL + ++
Sbjct 295 VVSRLKKFSAMNKLKKMALRVIAERLSEEEIGGLKELFKMIDTDKSGTITFEELKDSMRR 354
Query 87 CG--VQGEEIINIIKEIDTDKNGTIDYTEFIAASLDHKIYEQESAIQNAFRVFDLDGDGK 144
G + EI +++ D D++GTIDY EF+AA++ E+E + AF FD D G
Sbjct 355 VGSELMESEIQELLRAADVDESGTIDYGEFLAATIHLNKLEREENLVAAFSFFDKDASGY 414
Query 145 ISVEEL 150
I++EEL
Sbjct 415 ITIEEL 420
Score = 46.6 bits (109), Expect = 3e-05, Method: Composition-based stats.
Identities = 21/53 (39%), Positives = 31/53 (58%), Gaps = 0/53 (0%)
Query 64 FSLLDKNNDGVLTLNELLNGLQLCGVQGEEIINIIKEIDTDKNGTIDYTEFIA 116
FS DK+ G +T+ EL + G+ + +IK+ID D +G IDY EF+A
Sbjct 404 FSFFDKDASGYITIEELQQAWKEFGINDSNLDEMIKDIDQDNDGQIDYGEFVA 456
> ath:AT1G76040 CPK29; CPK29; ATP binding / calcium ion binding
/ calmodulin-dependent protein kinase/ kinase/ protein kinase/
protein serine/threonine kinase/ protein tyrosine kinase
Length=323
Score = 92.0 bits (227), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 50/130 (38%), Positives = 78/130 (60%), Gaps = 2/130 (1%)
Query 23 LGSDLIERFKSFQRLNKLKRLAITCVAYQLTDKEIGILHDTFSLLDKNNDGVLTLNELLN 82
+ S ++ R K F+ +NKLK+LA+ +A L+++EI L TF +D + G +T +EL N
Sbjct 142 INSAVLVRMKQFRAMNKLKKLALKVIAENLSEEEIKGLKQTFKNMDTDESGTITFDELRN 201
Query 83 GLQLCG--VQGEEIINIIKEIDTDKNGTIDYTEFIAASLDHKIYEQESAIQNAFRVFDLD 140
GL G + EI +++ D DK+GTIDY EF+ A++ E+E + AF+ FD D
Sbjct 202 GLHRLGSKLTESEIKQLMEAADVDKSGTIDYIEFVTATMHRHRLEKEENLIEAFKYFDKD 261
Query 141 GDGKISVEEL 150
G I+ +EL
Sbjct 262 RSGFITRDEL 271
Score = 39.3 bits (90), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 20/58 (34%), Positives = 35/58 (60%), Gaps = 1/58 (1%)
Query 60 LHDTFSLLDKNNDGVLTLNELLNGLQLCGVQGEEIIN-IIKEIDTDKNGTIDYTEFIA 116
L + F DK+ G +T +EL + + G+ + I+ +I ++DTD +G I+Y EF+A
Sbjct 251 LIEAFKYFDKDRSGFITRDELKHSMTEYGMGDDATIDEVINDVDTDNDGRINYEEFVA 308
> ath:AT4G21940 CPK15; CPK15; ATP binding / calcium ion binding
/ calmodulin-dependent protein kinase/ kinase/ protein kinase/
protein serine/threonine kinase; K13412 calcium-dependent
protein kinase [EC:2.7.11.1]
Length=554
Score = 90.1 bits (222), Expect = 2e-18, Method: Composition-based stats.
Identities = 47/130 (36%), Positives = 79/130 (60%), Gaps = 2/130 (1%)
Query 23 LGSDLIERFKSFQRLNKLKRLAITCVAYQLTDKEIGILHDTFSLLDKNNDGVLTLNELLN 82
+ S ++ R K F+ +NKLK+LA+ +A L+++EI L F+ +D + G +T EL N
Sbjct 370 IDSAVLSRMKQFRAMNKLKKLALKVIAESLSEEEIKGLKTMFANMDTDKSGTITYEELKN 429
Query 83 GLQLCG--VQGEEIINIIKEIDTDKNGTIDYTEFIAASLDHKIYEQESAIQNAFRVFDLD 140
GL G + E+ +++ D D NGTIDY EFI+A++ ++++ + AF+ FD D
Sbjct 430 GLAKLGSKLTEAEVKQLMEAADVDGNGTIDYIEFISATMHRYRFDRDEHVFKAFQYFDKD 489
Query 141 GDGKISVEEL 150
G I+++EL
Sbjct 490 NSGFITMDEL 499
Score = 45.1 bits (105), Expect = 9e-05, Method: Composition-based stats.
Identities = 22/54 (40%), Positives = 34/54 (62%), Gaps = 1/54 (1%)
Query 64 FSLLDKNNDGVLTLNELLNGLQLCGVQGE-EIINIIKEIDTDKNGTIDYTEFIA 116
F DK+N G +T++EL + ++ G+ E I +I E+DTD +G I+Y EF A
Sbjct 483 FQYFDKDNSGFITMDELESAMKEYGMGDEASIKEVIAEVDTDNDGRINYEEFCA 536
> ath:AT1G74740 CPK30; CPK30 (CALCIUM-DEPENDENT PROTEIN KINASE
30); calmodulin-dependent protein kinase/ kinase/ protein kinase;
K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=541
Score = 89.4 bits (220), Expect = 4e-18, Method: Composition-based stats.
Identities = 49/133 (36%), Positives = 78/133 (58%), Gaps = 2/133 (1%)
Query 20 SVRLGSDLIERFKSFQRLNKLKRLAITCVAYQLTDKEIGILHDTFSLLDKNNDGVLTLNE 79
+V LG + R K F +N+LK+ A+ +A L+ +E+ ++ + F+L+D +NDG ++ E
Sbjct 325 NVPLGDIVRSRLKQFSMMNRLKKKALRVIAEHLSIQEVEVIRNMFTLMDDDNDGKISYLE 384
Query 80 LLNGLQLCGVQ-GEEIINIIKEI-DTDKNGTIDYTEFIAASLDHKIYEQESAIQNAFRVF 137
L GL+ G Q GE I ++ E+ D + NG +DY EF+A + + E + + AF F
Sbjct 385 LRAGLRKVGSQLGEPEIKLLMEVADVNGNGCLDYGEFVAVIIHLQKMENDEHFRQAFMFF 444
Query 138 DLDGDGKISVEEL 150
D DG G I EEL
Sbjct 445 DKDGSGYIESEEL 457
Score = 41.2 bits (95), Expect = 0.001, Method: Composition-based stats.
Identities = 21/54 (38%), Positives = 31/54 (57%), Gaps = 2/54 (3%)
Query 64 FSLLDKNNDGVLTLNELLNGL--QLCGVQGEEIINIIKEIDTDKNGTIDYTEFI 115
F DK+ G + EL L +L II+I++E+DTDK+G I+Y EF+
Sbjct 441 FMFFDKDGSGYIESEELREALTDELGEPDNSVIIDIMREVDTDKDGKINYDEFV 494
> ath:AT5G19360 CPK34; CPK34; ATP binding / calcium ion binding
/ calmodulin-dependent protein kinase/ kinase/ protein kinase/
protein serine/threonine kinase; K00924 [EC:2.7.1.-]
Length=523
Score = 89.0 bits (219), Expect = 6e-18, Method: Composition-based stats.
Identities = 49/132 (37%), Positives = 78/132 (59%), Gaps = 2/132 (1%)
Query 21 VRLGSDLIERFKSFQRLNKLKRLAITCVAYQLTDKEIGILHDTFSLLDKNNDGVLTLNEL 80
V L + ++ R K F+ +N K++A+ +A L+++EI L + F +D +N G +TL EL
Sbjct 335 VPLDNAVMSRLKQFKAMNNFKKVALRVIAGCLSEEEIMGLKEMFKGMDTDNSGTITLEEL 394
Query 81 LNGLQLCGVQGEE--IINIIKEIDTDKNGTIDYTEFIAASLDHKIYEQESAIQNAFRVFD 138
GL G + E + +++ D D NGTIDY EFIAA++ ++E + +AF+ FD
Sbjct 395 RQGLAKQGTRLSEYEVQQLMEAADADGNGTIDYGEFIAATMHINRLDREEHLYSAFQHFD 454
Query 139 LDGDGKISVEEL 150
D G I+ EEL
Sbjct 455 KDNSGYITTEEL 466
Score = 47.8 bits (112), Expect = 1e-05, Method: Composition-based stats.
Identities = 24/58 (41%), Positives = 34/58 (58%), Gaps = 1/58 (1%)
Query 60 LHDTFSLLDKNNDGVLTLNELLNGLQLCGVQ-GEEIINIIKEIDTDKNGTIDYTEFIA 116
L+ F DK+N G +T EL L+ G+ G +I II E+D D +G I+Y EF+A
Sbjct 446 LYSAFQHFDKDNSGYITTEELEQALREFGMNDGRDIKEIISEVDGDNDGRINYEEFVA 503
> ath:AT5G12180 CPK17; CPK17; ATP binding / calcium ion binding
/ calmodulin-dependent protein kinase/ kinase/ protein kinase/
protein serine/threonine kinase; K00924 [EC:2.7.1.-]
Length=528
Score = 87.4 bits (215), Expect = 2e-17, Method: Composition-based stats.
Identities = 48/132 (36%), Positives = 79/132 (59%), Gaps = 2/132 (1%)
Query 21 VRLGSDLIERFKSFQRLNKLKRLAITCVAYQLTDKEIGILHDTFSLLDKNNDGVLTLNEL 80
V L + ++ R K F+ +N K++A+ +A L+++EI L + F +D ++ G +TL EL
Sbjct 340 VPLDNAVMSRLKQFKAMNNFKKVALRVIAGCLSEEEIMGLKEMFKGMDTDSSGTITLEEL 399
Query 81 LNGLQLCGVQGEE--IINIIKEIDTDKNGTIDYTEFIAASLDHKIYEQESAIQNAFRVFD 138
GL G + E + +++ D D NGTIDY EFIAA++ ++E + +AF+ FD
Sbjct 400 RQGLAKQGTRLSEYEVQQLMEAADADGNGTIDYGEFIAATMHINRLDREEHLYSAFQHFD 459
Query 139 LDGDGKISVEEL 150
D G I++EEL
Sbjct 460 KDNSGYITMEEL 471
Score = 48.9 bits (115), Expect = 6e-06, Method: Composition-based stats.
Identities = 24/58 (41%), Positives = 35/58 (60%), Gaps = 1/58 (1%)
Query 60 LHDTFSLLDKNNDGVLTLNELLNGLQLCGVQ-GEEIINIIKEIDTDKNGTIDYTEFIA 116
L+ F DK+N G +T+ EL L+ G+ G +I II E+D D +G I+Y EF+A
Sbjct 451 LYSAFQHFDKDNSGYITMEELEQALREFGMNDGRDIKEIISEVDGDNDGRINYDEFVA 508
> ath:AT4G04720 CPK21; CPK21; ATP binding / calcium ion binding
/ calmodulin-dependent protein kinase/ kinase/ protein kinase/
protein serine/threonine kinase; K13412 calcium-dependent
protein kinase [EC:2.7.11.1]
Length=531
Score = 87.0 bits (214), Expect = 2e-17, Method: Composition-based stats.
Identities = 46/130 (35%), Positives = 76/130 (58%), Gaps = 2/130 (1%)
Query 23 LGSDLIERFKSFQRLNKLKRLAITCVAYQLTDKEIGILHDTFSLLDKNNDGVLTLNELLN 82
+ S ++ R K F+ +NKLK+LA+ +A L+++EI L F+ +D + G +T EL
Sbjct 348 IDSAVLSRMKQFRAMNKLKKLALKVIAESLSEEEIKGLKTMFANIDTDKSGTITYEELKT 407
Query 83 GLQLCG--VQGEEIINIIKEIDTDKNGTIDYTEFIAASLDHKIYEQESAIQNAFRVFDLD 140
GL G + E+ +++ D D NGTIDY EFI+A++ +++ + AF+ FD D
Sbjct 408 GLTRLGSRLSETEVKQLMEAADVDGNGTIDYYEFISATMHRYKLDRDEHVYKAFQHFDKD 467
Query 141 GDGKISVEEL 150
G I+ +EL
Sbjct 468 NSGHITRDEL 477
Score = 41.6 bits (96), Expect = 0.001, Method: Composition-based stats.
Identities = 21/58 (36%), Positives = 35/58 (60%), Gaps = 1/58 (1%)
Query 60 LHDTFSLLDKNNDGVLTLNELLNGLQLCGVQGE-EIINIIKEIDTDKNGTIDYTEFIA 116
++ F DK+N G +T +EL + ++ G+ E I +I E+DTD +G I++ EF A
Sbjct 457 VYKAFQHFDKDNSGHITRDELESAMKEYGMGDEASIKEVISEVDTDNDGRINFEEFCA 514
> ath:AT1G61950 CPK19; CPK19; ATP binding / calcium ion binding
/ calmodulin-dependent protein kinase/ kinase/ protein kinase/
protein serine/threonine kinase
Length=551
Score = 86.3 bits (212), Expect = 3e-17, Method: Composition-based stats.
Identities = 47/130 (36%), Positives = 78/130 (60%), Gaps = 2/130 (1%)
Query 23 LGSDLIERFKSFQRLNKLKRLAITCVAYQLTDKEIGILHDTFSLLDKNNDGVLTLNELLN 82
+ S ++ R K + +NKLK+LA +A L ++E+ L F+ +D + G +T +EL +
Sbjct 368 IDSAVLSRMKQLRAMNKLKKLAFKFIAQNLKEEELKGLKTMFANMDTDKSGTITYDELKS 427
Query 83 GLQLCG--VQGEEIINIIKEIDTDKNGTIDYTEFIAASLDHKIYEQESAIQNAFRVFDLD 140
GL+ G + E+ ++++ D D NGTIDY EFI+A+++ E+E + AF+ FD D
Sbjct 428 GLEKLGSRLTETEVKQLLEDADVDGNGTIDYIEFISATMNRFRVEREDNLFKAFQHFDKD 487
Query 141 GDGKISVEEL 150
G IS +EL
Sbjct 488 NSGFISRQEL 497
Score = 37.4 bits (85), Expect = 0.019, Method: Composition-based stats.
Identities = 20/57 (35%), Positives = 33/57 (57%), Gaps = 3/57 (5%)
Query 60 LHDTFSLLDKNNDGVLTLNELLNGLQLCGVQGEEII--NIIKEIDTDKNGTIDYTEF 114
L F DK+N G ++ EL ++ + G++I+ II E+D D +G+I+Y EF
Sbjct 477 LFKAFQHFDKDNSGFISRQELETAMKEYNM-GDDIMIKEIISEVDADNDGSINYQEF 532
> pfa:PFC0420w PfCDPK3; calcium-dependent protein kinase-3 (EC:2.7.11.17);
K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=562
Score = 86.3 bits (212), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 51/133 (38%), Positives = 85/133 (63%), Gaps = 2/133 (1%)
Query 19 LSVRLGSDLIERFKSFQRLNKLKRLAITCVAYQLTDKEIGILHDTFSLLDKNNDGVLTLN 78
+ +++ ++E FK++ L K ++LA+T +A Q D ++ L TF +LD++ G +T
Sbjct 382 MDMKMDIHVLENFKNYGLLLKFQKLAMTIIAQQSNDYDVEKLKSTFLVLDEDGKGYITKE 441
Query 79 ELLNGLQLCGVQGEEIIN-IIKEIDTDKNGTIDYTEFIAASLDHKIYEQESAIQNAFRVF 137
+L GL+ G++ + ++ +ID+D +G IDYTEFIAA+LD K ++ I AFRVF
Sbjct 442 QLKKGLEKDGLKLPYNFDLLLDQIDSDGSGKIDYTEFIAAALDRKQLSKK-LIYCAFRVF 500
Query 138 DLDGDGKISVEEL 150
D+D DG+I+ EL
Sbjct 501 DVDNDGEITTAEL 513
Score = 29.3 bits (64), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 20/71 (28%), Positives = 37/71 (52%), Gaps = 10/71 (14%)
Query 51 QLTDKEIGILHDTFSLLDKNNDGVLTLNELLNGLQLCGVQGE-------EIINIIKEIDT 103
QL+ K +++ F + D +NDG +T EL + L +G + +I+++D
Sbjct 487 QLSKK---LIYCAFRVFDVDNDGEITTAELAHILYNGNKKGNITQRDVNRVKRMIRDVDK 543
Query 104 DKNGTIDYTEF 114
+ +G ID+ EF
Sbjct 544 NNDGKIDFHEF 554
> ath:AT1G50700 CPK33; CPK33; ATP binding / calcium ion binding
/ calmodulin-dependent protein kinase/ kinase/ protein kinase/
protein serine/threonine kinase; K13412 calcium-dependent
protein kinase [EC:2.7.11.1]
Length=521
Score = 86.3 bits (212), Expect = 3e-17, Method: Composition-based stats.
Identities = 47/130 (36%), Positives = 72/130 (55%), Gaps = 2/130 (1%)
Query 23 LGSDLIERFKSFQRLNKLKRLAITCVAYQLTDKEIGILHDTFSLLDKNNDGVLTLNELLN 82
+ S ++ R K F+ +NKLK+LA+ +A + +EI L F+ +D +N G +T EL
Sbjct 342 IDSAVLSRMKQFRAMNKLKKLALKVIAENIDTEEIQGLKAMFANIDTDNSGTITYEELKE 401
Query 83 GLQLCG--VQGEEIINIIKEIDTDKNGTIDYTEFIAASLDHKIYEQESAIQNAFRVFDLD 140
GL G + E+ ++ D D NG+IDY EFI A++ E + AF+ FD D
Sbjct 402 GLAKLGSRLTEAEVKQLMDAADVDGNGSIDYIEFITATMHRHRLESNENVYKAFQHFDKD 461
Query 141 GDGKISVEEL 150
G G I+ +EL
Sbjct 462 GSGYITTDEL 471
Score = 36.6 bits (83), Expect = 0.030, Method: Composition-based stats.
Identities = 19/58 (32%), Positives = 32/58 (55%), Gaps = 1/58 (1%)
Query 60 LHDTFSLLDKNNDGVLTLNELLNGLQLCGVQGEEIIN-IIKEIDTDKNGTIDYTEFIA 116
++ F DK+ G +T +EL L+ G+ + I I+ ++D D +G I+Y EF A
Sbjct 451 VYKAFQHFDKDGSGYITTDELEAALKEYGMGDDATIKEILSDVDADNDGRINYDEFCA 508
> cpv:cgd4_3330 hypothetical protein
Length=622
Score = 85.5 bits (210), Expect = 6e-17, Method: Compositional matrix adjust.
Identities = 53/141 (37%), Positives = 82/141 (58%), Gaps = 7/141 (4%)
Query 12 NNSSSSCLSVRLGSDLIERFKSFQRLNKLKRLAITCVAYQL--TDKEIGILHDTFSLLDK 69
+N+ C++ RL +L K++ + N+LK + +A+QL T +I + F LD+
Sbjct 444 SNNEPICINNRLLHNL----KNYMKQNQLKHALVNMMAHQLNVTGPQIKQITKAFKSLDQ 499
Query 70 NNDGVLTLNELLNGLQLCGVQGEEIINIIKEIDTDKNGTIDYTEFIAASLDHKIYEQESA 129
+ +GVLT EL++GLQ GV +I I++ +D D G I YTEF+AA + + E
Sbjct 500 DGNGVLTPEELISGLQSAGVPQWDINRIVQSMDVDDTGFISYTEFLAACYEWRDSEL-GV 558
Query 130 IQNAFRVFDLDGDGKISVEEL 150
I+ AF D+DGDGK+SV E
Sbjct 559 IRAAFNKMDIDGDGKLSVNEF 579
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 24/73 (32%), Positives = 42/73 (57%), Gaps = 5/73 (6%)
Query 47 CVAYQLTDKEIGILHDTFSLLDKNNDGVLTLNE----LLNGLQLCGVQGEEIINIIKEID 102
Y+ D E+G++ F+ +D + DG L++NE L +G Q V ++ +IIK D
Sbjct 546 AACYEWRDSELGVIRAAFNKMDIDGDGKLSVNEFEKVLCSGDQKLLVH-KDWDSIIKAAD 604
Query 103 TDKNGTIDYTEFI 115
T+ +G +D+ EF+
Sbjct 605 TNGDGVVDWNEFL 617
> ath:AT1G18890 ATCDPK1 (CALCIUM-DEPENDENT PROTEIN KINASE 1);
calmodulin-dependent protein kinase/ kinase/ protein kinase;
K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=545
Score = 84.7 bits (208), Expect = 1e-16, Method: Composition-based stats.
Identities = 48/133 (36%), Positives = 75/133 (56%), Gaps = 2/133 (1%)
Query 20 SVRLGSDLIERFKSFQRLNKLKRLAITCVAYQLTDKEIGILHDTFSLLDKNNDGVLTLNE 79
+V LG + R K F +N+ K+ + +A L+ +E+ ++ + FSL+D + DG +T E
Sbjct 329 NVPLGDIVRSRLKQFSMMNRFKKKVLRVIAEHLSIQEVEVIKNMFSLMDDDKDGKITYPE 388
Query 80 LLNGLQLCGVQ-GEEIINIIKEI-DTDKNGTIDYTEFIAASLDHKIYEQESAIQNAFRVF 137
L GLQ G Q GE I ++ E+ D D NG +DY EF+A + + E + + AF F
Sbjct 389 LKAGLQKVGSQLGEPEIKMLMEVADVDGNGFLDYGEFVAVIIHLQKIENDELFKLAFMFF 448
Query 138 DLDGDGKISVEEL 150
D DG I ++EL
Sbjct 449 DKDGSTYIELDEL 461
Score = 37.7 bits (86), Expect = 0.013, Method: Composition-based stats.
Identities = 19/54 (35%), Positives = 31/54 (57%), Gaps = 2/54 (3%)
Query 64 FSLLDKNNDGVLTLNELLNGL--QLCGVQGEEIINIIKEIDTDKNGTIDYTEFI 115
F DK+ + L+EL L +L + +I++E+DTDK+G I+Y EF+
Sbjct 445 FMFFDKDGSTYIELDELREALADELGEPDASVLSDIMREVDTDKDGRINYDEFV 498
> ath:AT3G20410 CPK9; CPK9 (calmodulin-domain protein kinase 9);
calmodulin-dependent protein kinase/ kinase; K13412 calcium-dependent
protein kinase [EC:2.7.11.1]
Length=541
Score = 84.0 bits (206), Expect = 2e-16, Method: Composition-based stats.
Identities = 46/130 (35%), Positives = 72/130 (55%), Gaps = 2/130 (1%)
Query 23 LGSDLIERFKSFQRLNKLKRLAITCVAYQLTDKEIGILHDTFSLLDKNNDGVLTLNELLN 82
+ S ++ R K F+ +NKLK+LA+ +A + +EI L F+ +D +N G +T EL
Sbjct 360 IDSAVLSRMKQFRAMNKLKKLALKVIAENIDTEEIQGLKAMFANIDTDNSGTITYEELKE 419
Query 83 GLQLCG--VQGEEIINIIKEIDTDKNGTIDYTEFIAASLDHKIYEQESAIQNAFRVFDLD 140
GL G + E+ ++ D D NG+IDY EFI A++ E + AF+ FD D
Sbjct 420 GLAKLGSKLTEAEVKQLMDAADVDGNGSIDYIEFITATMHRHRLESNENLYKAFQHFDKD 479
Query 141 GDGKISVEEL 150
G I+++EL
Sbjct 480 SSGYITIDEL 489
Score = 40.0 bits (92), Expect = 0.003, Method: Composition-based stats.
Identities = 19/58 (32%), Positives = 36/58 (62%), Gaps = 1/58 (1%)
Query 60 LHDTFSLLDKNNDGVLTLNELLNGLQLCGVQGEEIIN-IIKEIDTDKNGTIDYTEFIA 116
L+ F DK++ G +T++EL + L+ G+ + I ++ ++D+D +G I+Y EF A
Sbjct 469 LYKAFQHFDKDSSGYITIDELESALKEYGMGDDATIKEVLSDVDSDNDGRINYEEFCA 526
> ath:AT4G04695 CPK31; CPK31; ATP binding / calcium ion binding
/ calmodulin-dependent protein kinase/ kinase/ protein kinase/
protein serine/threonine kinase/ protein tyrosine kinase;
K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=484
Score = 83.6 bits (205), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 46/126 (36%), Positives = 75/126 (59%), Gaps = 2/126 (1%)
Query 27 LIERFKSFQRLNKLKRLAITCVAYQLTDKEIGILHDTFSLLDKNNDGVLTLNELLNGLQL 86
++ R K F+ +NKLK++A+ +A L+++EI L F+ +D + G +TL EL GL
Sbjct 304 VLSRLKQFRDMNKLKKVALKVIAANLSEEEIKGLKTLFTNIDTDKSGTITLEELKTGLTR 363
Query 87 CG--VQGEEIINIIKEIDTDKNGTIDYTEFIAASLDHKIYEQESAIQNAFRVFDLDGDGK 144
G + E+ +++ D D NGTID EFI+A++ +++ + AF+ FD D DG
Sbjct 364 LGSNLSKTEVEQLMEAADVDGNGTIDIDEFISATMHRYRLDRDDHVYQAFQHFDKDNDGH 423
Query 145 ISVEEL 150
I+ EEL
Sbjct 424 ITKEEL 429
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 23/56 (41%), Positives = 33/56 (58%), Gaps = 1/56 (1%)
Query 60 LHDTFSLLDKNNDGVLTLNELLNGLQLCGVQGE-EIINIIKEIDTDKNGTIDYTEF 114
++ F DK+NDG +T EL ++ GV E I II E+DTD +G I++ EF
Sbjct 409 VYQAFQHFDKDNDGHITKEELEMAMKEHGVGDEVSIKQIITEVDTDNDGKINFEEF 464
> bbo:BBOV_IV003210 21.m02835; calcium-dependent protein kinase
4 (EC:2.7.11.1); K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=517
Score = 82.4 bits (202), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 52/137 (37%), Positives = 77/137 (56%), Gaps = 14/137 (10%)
Query 28 IERFKSFQRLNKLKRLAITCVAYQL-TDKEIGILHDTFSLLDKNNDGVLTLNELLNG--- 83
I KSF KL + A+ + +L T +E L FS +DKN DG L +EL++G
Sbjct 339 INNMKSFYYTQKLSQAALLYIGSKLVTKEESNYLTTIFSKMDKNGDGQLDRSELIDGFSE 398
Query 84 -LQLCGVQG---------EEIINIIKEIDTDKNGTIDYTEFIAASLDHKIYEQESAIQNA 133
L+L G E++ I+++ID DKNG IDY+EF+ ++D + Q+ ++ A
Sbjct 399 YLRLKGTAADNAERMSVEEQVDQILQDIDFDKNGYIDYSEFLTVAMDRRNLMQKDRLEKA 458
Query 134 FRVFDLDGDGKISVEEL 150
F++FD DG G IS EEL
Sbjct 459 FKLFDADGSGTISSEEL 475
Score = 35.0 bits (79), Expect = 0.086, Method: Compositional matrix adjust.
Identities = 16/57 (28%), Positives = 29/57 (50%), Gaps = 0/57 (0%)
Query 60 LHDTFSLLDKNNDGVLTLNELLNGLQLCGVQGEEIINIIKEIDTDKNGTIDYTEFIA 116
L F L D + G ++ EL + V ++ ++ E+D++ +G ID+ EF A
Sbjct 455 LEKAFKLFDADGSGTISSEELGKMFGVADVSADDWQRVLHEVDSNNDGLIDFEEFTA 511
> ath:AT4G04710 CPK22; CPK22; ATP binding / calcium ion binding
/ calmodulin-dependent protein kinase/ kinase/ protein kinase/
protein serine/threonine kinase; K13412 calcium-dependent
protein kinase [EC:2.7.11.1]
Length=575
Score = 82.4 bits (202), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 47/128 (36%), Positives = 75/128 (58%), Gaps = 4/128 (3%)
Query 27 LIERFKSFQRLNKLKRLAITCVAYQLTDKEIGILHDTFSLLDKNNDGVLTLNELLNGLQL 86
++ R K F+ +NKLK+LA+ +A L+++EI L F +D + G +T EL GL
Sbjct 304 VLSRMKQFRAMNKLKKLALKVIAEGLSEEEIKGLKTMFENMDMDKSGSITYEELKMGLNR 363
Query 87 CG--VQGEEIINIIKEI--DTDKNGTIDYTEFIAASLDHKIYEQESAIQNAFRVFDLDGD 142
G + E+ +++ + D D NGTIDY EFI+A++ E++ + AF+ FD DG
Sbjct 364 HGSKLSETEVKQLMEAVSADVDGNGTIDYIEFISATMHRHRLERDEHLYKAFQYFDKDGS 423
Query 143 GKISVEEL 150
G I+ EE+
Sbjct 424 GHITKEEV 431
Score = 33.9 bits (76), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 31/118 (26%), Positives = 48/118 (40%), Gaps = 27/118 (22%)
Query 60 LHDTFSLLDKNNDGVLTLNELLNGLQLCGVQGEE-IINIIKEIDTDKNGTIDYTEFIA-- 116
L+ F DK+ G +T E+ ++ G+ E ++I E D + +G IDY EF
Sbjct 411 LYKAFQYFDKDGSGHITKEEVEIAMKEHGMGDEANAKDLISEFDKNNDGKIDYEEFCTMM 470
Query 117 --------ASLDHKIYE----------------QESAIQNAFRVFDLDGDGKISVEEL 150
L ++Y E+ I AF+ FD D G I+ +EL
Sbjct 471 RNGILQPQGKLLKRLYMNLEELKTGLTRLGSRLSETEIDKAFQHFDKDNSGHITRDEL 528
Score = 30.8 bits (68), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 23/72 (31%), Positives = 39/72 (54%), Gaps = 5/72 (6%)
Query 34 FQRLNKLKRLAITCVAYQLTDKEIGILHDTFSLLDKNNDGVLTLNELLNGLQLCGVQGE- 92
+ L +LK +T + +L++ EI F DK+N G +T +EL + ++ G+ E
Sbjct 486 YMNLEELK-TGLTRLGSRLSETEID---KAFQHFDKDNSGHITRDELESAMKEYGMGDEA 541
Query 93 EIINIIKEIDTD 104
I +I E+DTD
Sbjct 542 SIKEVISEVDTD 553
> ath:AT4G04740 CPK23; CPK23; ATP binding / calcium ion binding
/ calmodulin-dependent protein kinase/ kinase/ protein kinase/
protein serine/threonine kinase; K13412 calcium-dependent
protein kinase [EC:2.7.11.1]
Length=520
Score = 81.3 bits (199), Expect = 1e-15, Method: Composition-based stats.
Identities = 44/130 (33%), Positives = 72/130 (55%), Gaps = 2/130 (1%)
Query 23 LGSDLIERFKSFQRLNKLKRLAITCVAYQLTDKEIGILHDTFSLLDKNNDGVLTLNELLN 82
+ S ++ R K F+ +NKLK+LA+ A L+++EI L F+ +D N G +T +L
Sbjct 337 IDSTVLSRMKQFRAMNKLKKLALKVSAVSLSEEEIKGLKTLFANMDTNRSGTITYEQLQT 396
Query 83 GLQ--LCGVQGEEIINIIKEIDTDKNGTIDYTEFIAASLDHKIYEQESAIQNAFRVFDLD 140
GL + E+ +++ D D NGTIDY EFI+A++ + + AF+ D D
Sbjct 397 GLSRLRSRLSETEVQQLVEASDVDGNGTIDYYEFISATMHRYKLHHDEHVHKAFQHLDKD 456
Query 141 GDGKISVEEL 150
+G I+ +EL
Sbjct 457 KNGHITRDEL 466
Score = 43.1 bits (100), Expect = 3e-04, Method: Composition-based stats.
Identities = 22/58 (37%), Positives = 36/58 (62%), Gaps = 1/58 (1%)
Query 60 LHDTFSLLDKNNDGVLTLNELLNGLQLCGVQGE-EIINIIKEIDTDKNGTIDYTEFIA 116
+H F LDK+ +G +T +EL + ++ G+ E I +I E+DTD +G I++ EF A
Sbjct 446 VHKAFQHLDKDKNGHITRDELESAMKEYGMGDEASIKEVISEVDTDNDGKINFEEFRA 503
> ath:AT3G51850 CPK13; CPK13; ATP binding / calcium ion binding
/ calmodulin-dependent protein kinase/ kinase/ protein kinase/
protein serine/threonine kinase/ protein tyrosine kinase;
K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=528
Score = 81.3 bits (199), Expect = 1e-15, Method: Composition-based stats.
Identities = 46/133 (34%), Positives = 76/133 (57%), Gaps = 2/133 (1%)
Query 20 SVRLGSDLIERFKSFQRLNKLKRLAITCVAYQLTDKEIGILHDTFSLLDKNNDGVLTLNE 79
+V LG + R K F +N+ KR A+ +A L+ +E+ + F+ +D +NDG++++ E
Sbjct 320 NVPLGDVVKSRLKQFSVMNRFKRKALRVIAEFLSTEEVEDIKVMFNKMDTDNDGIVSIEE 379
Query 80 LLNGLQLCGVQ--GEEIINIIKEIDTDKNGTIDYTEFIAASLDHKIYEQESAIQNAFRVF 137
L GL+ Q E+ +I+ +DT GT+DY EF+A SL + + ++ AF F
Sbjct 380 LKAGLRDFSTQLAESEVQMLIEAVDTKGKGTLDYGEFVAVSLHLQKVANDEHLRKAFSYF 439
Query 138 DLDGDGKISVEEL 150
D DG+G I +EL
Sbjct 440 DKDGNGYILPQEL 452
Score = 40.4 bits (93), Expect = 0.002, Method: Composition-based stats.
Identities = 23/61 (37%), Positives = 35/61 (57%), Gaps = 6/61 (9%)
Query 60 LHDTFSLLDKNNDGVLTLNELLNGLQLCGVQGEEIIN----IIKEIDTDKNGTIDYTEFI 115
L FS DK+ +G + EL + L+ G G++ ++ I +E+DTDK+G I Y EF
Sbjct 432 LRKAFSYFDKDGNGYILPQELCDALKEDG--GDDCVDVANDIFQEVDTDKDGRISYEEFA 489
Query 116 A 116
A
Sbjct 490 A 490
> pfa:PFB0815w CDPK1, CPK; Calcium-dependent protein kinase 1
(EC:2.7.11.1); K08282 non-specific serine/threonine protein
kinase [EC:2.7.11.1]
Length=524
Score = 80.1 bits (196), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 53/131 (40%), Positives = 75/131 (57%), Gaps = 13/131 (9%)
Query 32 KSFQRLNKLKRLAITCVAYQLTD-KEIGILHDTFSLLDKNNDGVLTLNELLNGL------ 84
+ F+ KL + AI + +LT +E L D F LDKN DG L EL+ G
Sbjct 348 RKFEGSQKLAQAAILFIGSKLTTLEERKELTDIFKKLDKNGDGQLDKKELIEGYNILRSF 407
Query 85 -----QLCGVQGEEIINIIKEIDTDKNGTIDYTEFIAASLDHKIYEQESAIQNAFRVFDL 139
+L V+ EE+ NI+KE+D DKNG I+Y+EFI+ +D +I E +++AF +FD
Sbjct 408 KNELGELKNVE-EEVDNILKEVDFDKNGYIEYSEFISVCMDKQILFSEERLRDAFNLFDT 466
Query 140 DGDGKISVEEL 150
D GKI+ EEL
Sbjct 467 DKSGKITKEEL 477
Score = 38.1 bits (87), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 22/70 (31%), Positives = 38/70 (54%), Gaps = 2/70 (2%)
Query 60 LHDTFSLLDKNNDGVLTLNELLNGLQLCGVQGEEIINIIKEIDTDKNGTIDYTEFIAASL 119
L D F+L D + G +T EL N L + + ++ E D +K+ ID+ EF+ ++
Sbjct 457 LRDAFNLFDTDKSGKITKEELANLFGLTSISEQMWNEVLGEADKNKDNMIDFDEFV--NM 514
Query 120 DHKIYEQESA 129
HKI + +S+
Sbjct 515 MHKICDNKSS 524
> tgo:TGME49_018720 calcium-dependent protein kinase, putative
(EC:2.7.11.17)
Length=557
Score = 79.7 bits (195), Expect = 3e-15, Method: Composition-based stats.
Identities = 48/126 (38%), Positives = 71/126 (56%), Gaps = 3/126 (2%)
Query 27 LIERFKSFQRLNKLKRLAITCVAYQL--TDKEIGILHDTFSLLDKNNDGVLTLNELLNGL 84
+ E K + R + LK + +A+QL T ++I ++ F LDKN DG+L+ EL GL
Sbjct 389 ICENMKRYMRQSHLKNALVNLMAHQLNVTGQQIRHINQIFRQLDKNGDGLLSHQELTEGL 448
Query 85 QLCGVQGEEIINIIKEIDTDKNGTIDYTEFIAASLDHKIYEQESAIQNAFRVFDLDGDGK 144
GV +I I++ ID D +G + YTEF+AA + E + + AF+ D DGDG+
Sbjct 449 AEAGVPQWDINRILQSIDVDDSGNVSYTEFLAACYCWQETEL-NVVWTAFQKIDKDGDGR 507
Query 145 ISVEEL 150
ISV E
Sbjct 508 ISVREF 513
Score = 37.7 bits (86), Expect = 0.015, Method: Composition-based stats.
Identities = 22/73 (30%), Positives = 41/73 (56%), Gaps = 5/73 (6%)
Query 47 CVAYQLTDKEIGILHDTFSLLDKNNDGVLTLNE---LLNGLQLCGVQGEEIINIIKEIDT 103
C +Q T E+ ++ F +DK+ DG +++ E L+ G + EE+ ++ ++D
Sbjct 482 CYCWQET--ELNVVWTAFQKIDKDGDGRISVREFCDLVLGRDNKLIPEEELRAMVAQMDR 539
Query 104 DKNGTIDYTEFIA 116
D +G ID+ EF+A
Sbjct 540 DGDGQIDWDEFVA 552
> cpv:cgd5_820 calcium/calmodulin dependent protein kinase with
a kinase domain and 4 calmodulin like EF hands ; K13412 calcium-dependent
protein kinase [EC:2.7.11.1]
Length=523
Score = 79.3 bits (194), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 52/139 (37%), Positives = 78/139 (56%), Gaps = 15/139 (10%)
Query 25 SDLIERFKSFQRLNKLKRLAITCVAYQLTDKE-IGILHDTFSLLDKNNDGVLTLNELLNG 83
++ IE + FQ KL + A+ +A +LT +E L D F +DKN DG L EL++G
Sbjct 335 ANAIENMRKFQNSQKLAQAALLYMASKLTSQEETKELTDIFRHIDKNGDGQLDRQELIDG 394
Query 84 LQLCGVQGEEII-----NIIKEIDT-------DKNGTIDYTEFIAASLDHKIYEQESAIQ 131
+ GEE+ I E+D D+NG IDY+EF+ ++D K + ++
Sbjct 395 Y--SKLSGEEVAVFDLPQIESEVDAILGAADFDRNGYIDYSEFVTVAMDRKSLLSKDKLE 452
Query 132 NAFRVFDLDGDGKISVEEL 150
+AF+ FD DG+GKISV+EL
Sbjct 453 SAFQKFDQDGNGKISVDEL 471
Score = 29.6 bits (65), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 14/55 (25%), Positives = 31/55 (56%), Gaps = 0/55 (0%)
Query 60 LHDTFSLLDKNNDGVLTLNELLNGLQLCGVQGEEIINIIKEIDTDKNGTIDYTEF 114
L F D++ +G ++++EL + L ++ + +I ID++ +G +D+ EF
Sbjct 451 LESAFQKFDQDGNGKISVDELASVFGLDHLESKTWKEMISGIDSNNDGDVDFEEF 505
> ath:AT5G12480 CPK7; CPK7 (calmodulin-domain protein kinase 7);
ATP binding / calcium ion binding / calmodulin-dependent
protein kinase/ kinase/ protein kinase/ protein serine/threonine
kinase/ protein tyrosine kinase; K13412 calcium-dependent
protein kinase [EC:2.7.11.1]
Length=441
Score = 77.8 bits (190), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 46/138 (33%), Positives = 74/138 (53%), Gaps = 2/138 (1%)
Query 15 SSSCLSVRLGSDLIERFKSFQRLNKLKRLAITCVAYQLTDKEIGILHDTFSLLDKNNDGV 74
+ +V LG + R K F +NKLK+ A+ +A L+ +E + + F ++D N G
Sbjct 226 AKKAPNVSLGETVKARLKQFSVMNKLKKRALRVIAEHLSVEEAAGIKEAFEMMDVNKRGK 285
Query 75 LTLNELLNGLQLCGVQ-GEEIINIIKE-IDTDKNGTIDYTEFIAASLDHKIYEQESAIQN 132
+ L EL GLQ G Q + + I+ E D D +GT++Y+EF+A S+ K + +
Sbjct 286 INLEELKYGLQKAGQQIADTDLQILMEATDVDGDGTLNYSEFVAVSVHLKKMANDEHLHK 345
Query 133 AFRVFDLDGDGKISVEEL 150
AF FD + G I ++EL
Sbjct 346 AFNFFDQNQSGYIEIDEL 363
Score = 48.1 bits (113), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 24/60 (40%), Positives = 37/60 (61%), Gaps = 3/60 (5%)
Query 60 LHDTFSLLDKNNDGVLTLNELLNGL--QLCGVQGEEIIN-IIKEIDTDKNGTIDYTEFIA 116
LH F+ D+N G + ++EL L +L EE+I I++++DTDK+G I Y EF+A
Sbjct 343 LHKAFNFFDQNQSGYIEIDELREALNDELDNTSSEEVIAAIMQDVDTDKDGRISYEEFVA 402
> ath:AT5G66210 CPK28; CPK28; ATP binding / calcium ion binding
/ calmodulin-dependent protein kinase/ protein kinase/ protein
serine/threonine kinase/ protein tyrosine kinase
Length=488
Score = 77.4 bits (189), Expect = 2e-14, Method: Composition-based stats.
Identities = 44/133 (33%), Positives = 76/133 (57%), Gaps = 9/133 (6%)
Query 27 LIERFKSFQRLNKLKRLAITCVAYQLTDKEIGILHDTFSLLDKNNDGVLTLNELLNGLQL 86
++ + F R ++LK+ A+ +A L + EI L D F +D + +GV++L E+ L
Sbjct 337 VLNNLRQFVRYSRLKQFALRALASTLDEAEISDLRDQFDAIDVDKNGVISLEEMRQALAK 396
Query 87 ---CGVQGEEIINIIKEIDTDKNGTIDYTEFIAASLD-HKIYEQESA-----IQNAFRVF 137
++ + I++ ID++ +G +D+TEF+AA+L H++ E +S + AF F
Sbjct 397 DLPWKLKDSRVAEILEAIDSNTDGLVDFTEFVAAALHVHQLEEHDSEKWQLRSRAAFEKF 456
Query 138 DLDGDGKISVEEL 150
DLD DG I+ EEL
Sbjct 457 DLDKDGYITPEEL 469
> ath:AT2G41860 CPK14; CPK14; ATP binding / calcium ion binding
/ calmodulin-dependent protein kinase/ kinase/ protein kinase/
protein serine/threonine kinase; K13412 calcium-dependent
protein kinase [EC:2.7.11.1]
Length=530
Score = 76.6 bits (187), Expect = 2e-14, Method: Composition-based stats.
Identities = 48/143 (33%), Positives = 79/143 (55%), Gaps = 4/143 (2%)
Query 10 IYNNSSSSCLSVRLGSDLIERFKSFQRLNKLKRLAITCVAYQLTDKEIGILHDTFSLLDK 69
I N ++S +V LG + R K F +NKLK+ A+ +A L+ +E + + F ++D
Sbjct 312 IQNGKNAS--NVSLGETVRARLKQFSVMNKLKKRALRVIAEHLSVEETSCIKERFQVMDT 369
Query 70 NNDGVLTLNELLNGLQLCG--VQGEEIINIIKEIDTDKNGTIDYTEFIAASLDHKIYEQE 127
+N G +T+ EL GLQ G V ++I ++ D DK+G +D EF+A S+ + +
Sbjct 370 SNRGKITITELGIGLQKLGIVVPQDDIQILMDAGDVDKDGYLDVNEFVAISVHIRKLGND 429
Query 128 SAIQNAFRVFDLDGDGKISVEEL 150
++ AF FD + G I +EEL
Sbjct 430 EHLKKAFTFFDKNKSGYIEIEEL 452
Score = 37.7 bits (86), Expect = 0.013, Method: Composition-based stats.
Identities = 21/57 (36%), Positives = 31/57 (54%), Gaps = 2/57 (3%)
Query 60 LHDTFSLLDKNNDGVLTLNELLNGL-QLCGVQGEEIIN-IIKEIDTDKNGTIDYTEF 114
L F+ DKN G + + EL + L EE++ II ++DT+K+G I Y EF
Sbjct 432 LKKAFTFFDKNKSGYIEIEELRDALADDVDTTSEEVVEAIILDVDTNKDGKISYDEF 488
> ath:AT4G04700 CPK27; CPK27; ATP binding / calcium ion binding
/ kinase/ protein kinase/ protein serine/threonine kinase/
protein tyrosine kinase; K13412 calcium-dependent protein kinase
[EC:2.7.11.1]
Length=485
Score = 76.6 bits (187), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 44/126 (34%), Positives = 72/126 (57%), Gaps = 2/126 (1%)
Query 27 LIERFKSFQRLNKLKRLAITCVAYQLTDKEIGILHDTFSLLDKNNDGVLTLNELLNGLQL 86
++ R K F+ NK K++ + +A L+++EI L F+ +D + G +TL EL GL
Sbjct 304 VLSRLKRFRDANKFKKVVLKFIAANLSEEEIKGLKTLFTNIDTDKSGNITLEELKTGLTR 363
Query 87 CG--VQGEEIINIIKEIDTDKNGTIDYTEFIAASLDHKIYEQESAIQNAFRVFDLDGDGK 144
G + E+ +++ D D NGTID EFI+A++ +++ + AF+ FD D DG
Sbjct 364 LGSNLSKTEVEQLMEAADMDGNGTIDIDEFISATMHRYKLDRDEHVYKAFQHFDKDNDGH 423
Query 145 ISVEEL 150
I+ EEL
Sbjct 424 ITKEEL 429
Score = 36.2 bits (82), Expect = 0.044, Method: Compositional matrix adjust.
Identities = 21/56 (37%), Positives = 31/56 (55%), Gaps = 1/56 (1%)
Query 60 LHDTFSLLDKNNDGVLTLNELLNGLQLCGVQGE-EIINIIKEIDTDKNGTIDYTEF 114
++ F DK+NDG +T EL ++ G E I II + DTD +G I++ EF
Sbjct 409 VYKAFQHFDKDNDGHITKEELEMAMKEDGAGDEGSIKQIIADADTDNDGKINFEEF 464
> ath:AT2G17890 CPK16; CPK16; ATP binding / calcium ion binding
/ calmodulin-dependent protein kinase/ protein kinase/ protein
serine/threonine kinase
Length=571
Score = 75.1 bits (183), Expect = 7e-14, Method: Composition-based stats.
Identities = 41/135 (30%), Positives = 77/135 (57%), Gaps = 13/135 (9%)
Query 27 LIERFKSFQRLNKLKRLAITCVAYQLTDKEIGILHDTFSLLDKNNDGVLTLNELLNGL-- 84
++ + F + ++LK+ A+ +A L ++E+ L D F +D + +GV++L E+ L
Sbjct 383 VLNNMRQFVKFSRLKQFALRALATTLDEEELADLRDQFDAIDVDKNGVISLEEMRQALAK 442
Query 85 -QLCGVQGEEIINIIKEIDTDKNGTIDYTEFIAASL--------DHKIYEQESAIQNAFR 135
++ + I++ ID++ +G +D+ EF+AA+L D + ++Q S + AF
Sbjct 443 DHPWKLKDARVAEILQAIDSNTDGFVDFGEFVAAALHVNQLEEHDSEKWQQRS--RAAFE 500
Query 136 VFDLDGDGKISVEEL 150
FD+DGDG I+ EEL
Sbjct 501 KFDIDGDGFITAEEL 515
> cpv:cgd3_920 calmodulin-domain protein kinase 1
Length=538
Score = 75.1 bits (183), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 48/136 (35%), Positives = 76/136 (55%), Gaps = 17/136 (12%)
Query 32 KSFQRLNKLKRLAITCVAYQLTD-KEIGILHDTFSLLDKNNDGVLTLNELLNG----LQL 86
+ FQ KL + A+ +A +LT E L + F LD NNDG+L +EL+ G ++L
Sbjct 356 RQFQAEKKLAQAALLYMASKLTTLDETKQLTEIFRKLDTNNDGMLDRDELVRGYHEFMRL 415
Query 87 CGVQGEEII------------NIIKEIDTDKNGTIDYTEFIAASLDHKIYEQESAIQNAF 134
GV +I +++ +D D +G+I+Y+EFIA+++D I ++ AF
Sbjct 416 KGVDSNSLIQNEGSTIEDQIDSLMPLLDMDGSGSIEYSEFIASAIDRTILLSRERMERAF 475
Query 135 RVFDLDGDGKISVEEL 150
++FD DG GKIS +EL
Sbjct 476 KMFDKDGSGKISTKEL 491
Score = 38.1 bits (87), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 17/58 (29%), Positives = 33/58 (56%), Gaps = 2/58 (3%)
Query 60 LHDTFSLLDKNNDGVLTLNELLNGLQLC--GVQGEEIINIIKEIDTDKNGTIDYTEFI 115
+ F + DK+ G ++ EL +Q EE+ +II+++D +K+G +D+ EF+
Sbjct 471 MERAFKMFDKDGSGKISTKELFKLFSQADSSIQMEELESIIEQVDNNKDGEVDFNEFV 528
> pfa:PF07_0072 PfCDPK4; calcium-dependent protein kinase 4 (EC:2.7.1.11);
K08282 non-specific serine/threonine protein kinase
[EC:2.7.11.1]
Length=528
Score = 74.7 bits (182), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 50/137 (36%), Positives = 72/137 (52%), Gaps = 14/137 (10%)
Query 28 IERFKSFQRLNKLKRLAITCVAYQLTD-KEIGILHDTFSLLDKNNDGVLTLNELLNGL-Q 85
I + FQ KL + A+ + +LT E L F +DKN DG L NEL+ G +
Sbjct 348 IANIRQFQSTQKLAQAALLYMGSKLTTIDETKELTKIFKKMDKNGDGQLDRNELIIGYKE 407
Query 86 LCGVQGE------------EIINIIKEIDTDKNGTIDYTEFIAASLDHKIYEQESAIQNA 133
L ++GE E+ I+ ID D+NG I+Y+EF+ S+D K+ ++ A
Sbjct 408 LLKLKGEDTSDLDNAAIEYEVDQILNSIDLDQNGYIEYSEFLTVSIDRKLLLSTERLEKA 467
Query 134 FRVFDLDGDGKISVEEL 150
F++FD DG GKIS EL
Sbjct 468 FKLFDKDGSGKISANEL 484
Score = 38.9 bits (89), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 20/55 (36%), Positives = 29/55 (52%), Gaps = 0/55 (0%)
Query 60 LHDTFSLLDKNNDGVLTLNELLNGLQLCGVQGEEIINIIKEIDTDKNGTIDYTEF 114
L F L DK+ G ++ NEL L V E ++KE+D + +G ID+ EF
Sbjct 464 LEKAFKLFDKDGSGKISANELAQLFGLSDVSSECWKTVLKEVDQNNDGEIDFKEF 518
> dre:100150680 calmodulin 2-like; K02183 calmodulin
Length=229
Score = 74.7 bits (182), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 44/106 (41%), Positives = 63/106 (59%), Gaps = 3/106 (2%)
Query 48 VAYQLTDKEIGILHDTFSLLDKNNDGVLTLNELLNGLQLCGVQG--EEIINIIKEIDTDK 105
+A QLT+++I + FSL DK+ DG +T EL ++ G E+ ++I E+D D
Sbjct 81 LADQLTEEQIAEFKEAFSLFDKDGDGTITTKELGTVMRSLGQNPTEAELQDMINEVDADG 140
Query 106 NGTIDYTEFIA-ASLDHKIYEQESAIQNAFRVFDLDGDGKISVEEL 150
NGTID+ EF+ + K + E I+ AFRVFD DG+G IS EL
Sbjct 141 NGTIDFPEFLTMMARKMKDTDSEEEIREAFRVFDKDGNGYISAAEL 186
Score = 35.0 bits (79), Expect = 0.090, Method: Compositional matrix adjust.
Identities = 23/75 (30%), Positives = 43/75 (57%), Gaps = 5/75 (6%)
Query 45 ITCVAYQL--TDKEIGILHDTFSLLDKNNDGVLTLNELLNGLQLCG--VQGEEIINIIKE 100
+T +A ++ TD E I + F + DK+ +G ++ EL + + G + EE+ +I+E
Sbjct 150 LTMMARKMKDTDSEEEI-REAFRVFDKDGNGYISAAELRHVMTNLGEKLTDEEVDEMIRE 208
Query 101 IDTDKNGTIDYTEFI 115
D D +G ++Y EF+
Sbjct 209 ADIDGDGQVNYEEFV 223
> cpv:cgd7_1260 calcium/calmodulin dependent protein kinase with
an EF hand N-terminal to the kinase domain and 4 calmodulin
like EF hands at the C-terminus
Length=1042
Score = 74.7 bits (182), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 50/140 (35%), Positives = 74/140 (52%), Gaps = 5/140 (3%)
Query 14 SSSSCLSVRLGSDLIERFKSFQRLNKLKRLAITCVAYQLT--DKEIGILHDTFSLLDKNN 71
S SS V +G L+ K + R + L+ + I +++QL +I + F LD +N
Sbjct 776 SHSSYCEVDIGMPLLAHLKVYTRQSDLRHILIHMLSHQLALDTTQINMATSVFKSLDTDN 835
Query 72 DGVLTLNELLNGLQLCGVQGEEIINIIKEIDTDKNGTIDYTEFIAASLDHKIYEQE-SAI 130
DGVL+++EL +GLQ G+ E IIK +D D NG I Y+EFI A H E +
Sbjct 836 DGVLSISELGSGLQKLGISSRESSMIIKAMDIDGNGIISYSEFITAC--HIWRRNEIRQL 893
Query 131 QNAFRVFDLDGDGKISVEEL 150
++ F D + GKI+ EE
Sbjct 894 KSFFMKIDRNNQGKITREEF 913
Lambda K H
0.317 0.139 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3134054208
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40